Abstract
Voltage-gated ion channels underlie electrical activity of neurons and are dynamically regulated by diverse cell signaling pathways that alter their phosphorylation state. Recent global mass spectrometric–based analyses of the mouse brain phosphoproteome have yielded a treasure trove of new data as to the extent and nature of phosphorylation of numerous ion channel principal or α subunits in mammalian brain. Here we compile and review data on 347 phosphorylation sites (261 unique) on 42 different voltage-gated ion channel α subunits that were identified in these recent studies. Researchers in the ion channel field can now begin to explore the role of these novel in vivo phosphorylation sites in the dynamic regulation of the localization, activity, and expression of brain ion channels through multisite phosphorylation of their principal subunits.
INTRODUCTION
Voltage-dependent ion channels underlie the electrical signaling that allows for neurotransmission within neurons, and allow for various modes of Ca2+ influx critical to coupling this electrical activity to diverse physiological events (Hille, 2001). As such, voltage-gated ion channel activity is highly regulated by diverse neuronal signaling pathways. Neuronal ion channels exist as supramolecular protein complexes composed of pore-forming transmembrane principal or α subunits (Yu et al., 2005), auxiliary or regulatory subunits (Hanlon and Wallace, 2002), and a diverse array of interacting proteins (Dai et al., 2009). Diverse posttranslational events acting on each of these components dynamically regulate the expression, localization, and function of neuronal ion channels (Levitan, 2006). While numerous noncovalent mechanisms such as ligand binding, sensing of transmembrane voltage, and interaction with other proteins are known to play prominent roles in regulating neuronal ion channels, direct covalent modification of the component subunits of these multiprotein ion channel complexes by phosphorylation has long been recognized as a widely used and potent mechanism for neurons to achieve dynamic and reversible changes in ion channel function, and to impact their contribution to neuronal signaling (Levitan, 1985).
Phosphorylation constitutes a common covalent post-translational modification in eukaryotes (Cohen, 2001), with (as of early 2009) up to 25,000 described phosphorylation sites (or phosphosites) on 7,000 human proteins, out of an estimated 500,000 potential phosphosites that exist in a cellular proteome (Lemeer and Heck, 2009). In neurons, reversible activity-dependent phosphorylation represents a major mechanism of dynamic regulation of synaptic development (Saneyoshi et al., 2010), as well as synaptic potentiation, depression, and homeostatic plasticity (Turrigiano, 2008), through phosphorylation of a large number of synaptic proteins including ligand-gated ion channels (Collins and Grant, 2007). Neurons also exhibit cellular plasticity at the level of intrinsic excitability, achieved through phosphorylation of components of ion channel subunits, for example of voltage-gated sodium or Nav (Cantrell and Catterall, 2001) and potassium or Kv (Schulz et al., 2008) channels, which localize in distinct neuronal compartments (Vacher et al., 2008). As opposed to the classical approaches of in vivo or in vitro radiolabeling with 32P, peptide mapping and/or sequencing, and site-directed mutagenesis (e.g., Costa et al., 1982; Costa and Catterall, 1984), mass spectrometry (MS)–based phosphoproteomic techniques have recently emerged as the primary tool for the identification of phosphorylation on ion channel subunits (Cerda and Trimmer, 2010). While many of these studies continue to rely on effective purification of the target ion channel before analysis, a set of recent studies from the proteomics field, aimed at defining the global phosphoproteome of mouse brain samples with high complexity, and without a focus on ion channels per se, have yielded a dataset that is extremely valuable to the ion channel community. Here we provide an overview of these studies, as well as the subset of these databases that pertain to voltage-gated ion channel α subunits. These studies provide important insights to the ion channel community on the extent and nature of phosphorylation of mammalian brain ion channels, and a wealth of phosphosites that can be tested for their specific role in regulating these ion channels through dynamic and reversible phosphorylation of their principal pore-forming and voltage-sensing α subunits.
Recent innovations in proteomics and bioinformatics have expanded our knowledge to include nearly 10,000 mammalian brain proteins (Wang et al., 2006). Data from such high-throughput proteomic approaches represents information on ion channel expression patterns that could be of great use to the ion channel community, but may not be as readily accessible to the average “channelologist.” However, the number of publications describing such “global” analyses is large and ever increasing, and sifting through huge databases to gain information on the spatial and temporal expression patterns of Your Favorite Channel can be tedious and time consuming, although the resultant information can reveal important insights. More recently, analogous high-throughput studies have provided an even rarer gem, the determination of brain peptides chemically modified with phosphate, and the site of phosphorylation within these peptides (Lemeer and Heck, 2009). Such in vivo studies have yielded an enormous dataset of phosphosites, including those on mammalian brain ion channels. (Hereafter, we will refer to such sites as “in vivo phosphosites.”) What in the past would take a tremendous amount of effort in purifying the ion channel proteins from brain preparations, and then identifying the phosphosites (using techniques that often used multiple millicuries of 32P) is now available to the average channelologist at the click of the mouse. That said, one first needs to be aware of the existence of these studies, then search through the accompanying datasets for the presence of Your Favorite Channel, and then perform analyses to determine the site of phosphorylation within specific channel isoforms. This can present a challenge for those more accustomed to a patch clamp rig than a large and complex proteomics dataset. In this brief review, we attempt to first bring these recent studies to the attention of members of the ion channel community, to highlight the wealth of the data being generated by these approaches, and second to detail the subset of the data related to voltage-gated ion channels. The goal is to ensure that the existing data, and others that will emerge from future studies, can be effectively used to gain mechanistic insights into how neuronal ion channels are regulated by reversible phosphorylation at these in vivo phosphosites. Note that this synopsis is assuredly not comprehensive, as there are likely other high-throughput studies that have yielded data leading to identification of ion channel phosphosites that we have not included in our analyses, which focuses only on data from four recent global analyses from mouse brain (Munton et al., 2007; Trinidad et al., 2008; Tweedie-Cullen et al., 2009; Wisniewski et al., 2010). Moreover, we should mention that the recent establishment of open source curated databases of posttranslational modifications, such as PhosphositePlus (http://www.phosphosite.org), Phosida (http://141.61.102.18/phosida/index.aspx, which currently contains 23,387 proteins and 69,790 identified phosphorylation sites across eukaryotes; Gnad et al., 2007), Phospho.ELM (http://phospho.elm.eu.org; current version 9.0 contains 8,718 substrate proteins from different species covering 42,573 phosphorylation sites; Diella et al., 2004; Diella et al., 2008), and Human Protein Reference Database (http://www.hprd.org, containing 30,047 proteins and 93,710 posttranslational modifications; Amanchy et al., 2007; Prasad et al., 2009), provides investigators additional opportunities for mining large phospho-sites databases for ion channel phosphorylation sites.
Recent high-throughput analyses of the mouse brain phosphoproteome
Wisniewski et al. (2010) analyzed phosphosites on whole brain tissue with parallel whole protein and peptide level fractionation methods. A crude brain homogenate solubilized in detergent buffer was digested using a recently developed Filter-Assisted Sample Preparation method, rather than conventional in-solution or in-gel digestion methods (Wisniewski et al., 2009). The resultant peptides were directly subjected to protein size exclusion chromatography, or applied to anti-phosphotyrosine antibody beads. Phosphopeptides were then enriched by binding to TiO2 beads in the presence of dihydroxybenzoic acid to avoid nonspecific binding, and/or further fractionated by strong anion exchange (SAX) chromatography. Using an LTQ-Orbitrap MS instrument, the 10 most intense precursors at a resolution of 60,000 were selected, and the precursors were fragmented through collision-induced dissociation (CID), one of the most widely used peptide fragmentation methods. Peptide identification is accomplished by matching the acquired CID mass information with a segment of the sequence from a protein database. For all of the CID tandem mass spectra, the MASCOT search engine was used to search a mouse protein sequence database (i.e., mouse IPI database, version 3.46). Note that such large-scale peptide and protein identification approaches often contain nonspecific matching with a false segment of protein sequence, resulting in incorrect peptide identification. To assess the identification in large-scale data, a modified protein sequence database was applied, which contains forward (real) and reverse (virtual) protein sequences (total ∼110,572 entries). A false-positive rate was calculated, based on the proportion of incorrect identifications assigned to reverse sequences relative to the total identifications (forward and reverse databases). In this study, false-positive rates for both peptides and proteins were 1%. The search yielded 12,035 unique phosphosites on 4,579 proteins, including 231 different phosphosites on 42 different voltage-gated ion channel α subunits, as detailed in Table I.
Table I.
Phosphorylation sites identified on voltage-gated ion channel α subunits
| Subunit/location | Study/accession no./site | Study/accession no./site | Study/accession no./site | Study/accession no./site |
| Cav1.2/CACNA1C | W:Q0PCR6-1 | Tr: Q01815 | M: Q0PCR6-1 | |
| ID II-III | S845 | S845 | ||
| C-term | S1721 | S1691 | ||
| Cav2.1/CACNA1A | W: P97445 | Tr: P97445 | M: P97445 | T:P97445 |
| ID I-II | T411 | |||
| ID I-II | S468 | |||
| ID II-III | S752 | S752 | S752 | |
| ID II-III | S755 | S755 | S755 | |
| ID II-III | S792 | |||
| ID II-III | S867 | |||
| ID II-III | S1038 | S1038 | ||
| C-term | T1935 | |||
| C-term | S1946 | |||
| C-term | T1948 | |||
| C-term | S1981 | S1981 | ||
| C-term | Y2027 | |||
| C-term | S2028 | |||
| C-term | S2030 | |||
| C-term | S2068 | |||
| C-term | T2070 | |||
| C-term | S2071 | |||
| C-term | S2078 | |||
| C-term | S2091 | S2091 | ||
| C-term | S2200 | S2220 | S2220 | |
| C-term | S2252 | T2241/T2244/S2250/S2252 | ||
| C-term | S2273 | S2273 | S2273 | |
| C-term | S2303 | S2303 | ||
| C-term | S2329 | |||
| Cav2.2/CACNA1B | W: O55017 | Tr: A2AIR7 | M: A2AIR9 | T: A2AIR9 |
| ID I-II | S424 | |||
| ID I-II | S446 | |||
| ID II-III | S745 | S746 | S745 | S745 |
| ID II-III | S748 | S749 | ||
| ID II-III | S753 | |||
| ID II-III | S783 | S784 | S783 | S783 |
| ID II-III | S892 | |||
| ID II-III | S915 | |||
| ID II-III | T920 | |||
| C-term | S1927 | |||
| C-term | S1951 | |||
| C-term | S2014 | |||
| C-term | S2221 | |||
| C-term | S2244 | S2245 | S2242 | S2242 |
| Cav2.3/CACNA1E | W: IPI00331064 | Tr: O88405 | M: Q61290 | T: Q61290 |
| N-term | S15 | S15 | ||
| N-term | S20 | |||
| N-term | T29 | T29 | ||
| ID I-II | S428 | |||
| ID I-II | T441 | T441 | ||
| ID II-III | S737 | S737 | S737 | S737 |
| ID II-III | S740 | S740 | ||
| ID II-III | S746 | S746 | ||
| ID II-III | S793 | |||
| ID II-III | S794 | |||
| ID II-III | S856 | S856 | S856 | |
| ID II-III | T865 | |||
| ID II-III | S866 | |||
| ID II-III | S873 | |||
| ID II-III | S876 | |||
| ID II-III | S1051 | S1051 | ||
| ID II-III | S1056 | |||
| ID II-III | T1094 | |||
| C-term | S2054 | S2054 | ||
| C-term | T2067 | |||
| C-term | S2073 | S2073 | ||
| C-term | S2097 | |||
| Cav3.1/CACNA1G | W: Q5SUF7 | Tr: Q5SUF5 | T: Q5SUF5 | |
| ID I-II | S467 | S466/S467 | S467 | |
| ID II-III | S1148 | |||
| ID II-III | S1149 | |||
| ID II-III | S1171 | |||
| ID II-III | S1172 | |||
| C-term | S2089 | |||
| C-term | S2145 | |||
| C-term | S2273 | |||
| C-term | S2281 | |||
| C-term | S2283 | |||
| C-term | S2361 | |||
| Cav3.3/CACNA1I | W: NP_001037773.1 | |||
| ID II-III | S948 | |||
| ID II-III | S951 | |||
| Nav1.1/SCN1A | W: A2APX8 | Tr: A2APX6 | ||
| ID I-II | T465 | |||
| ID I-II | S467 | |||
| ID I-II | S551 | |||
| ID I-II | S565 | |||
| ID I-II | S607 | |||
| ID I-II | S620 | |||
| ID I-II | T721 | |||
| ID I-II | T723 | |||
| ID I-II | S730 | |||
| C-term | S1928 | |||
| Nav1.2/SCN2A | W: B1AWN6 | Tr: A2AJZ2 | T: B1AWN6 | |
| ID I-II | S468 | |||
| ID I-II | S471 | |||
| ID I-II | S475 | |||
| ID I-II | S484 | |||
| ID I-II | S486 | |||
| ID I-II | S488 | |||
| ID I-II | S526 | |||
| ID I-II | S528 | |||
| ID I-II | S540 | S540 | ||
| ID I-II | S554 | |||
| ID I-II | S558 | |||
| ID I-II | S561 | |||
| ID I-II | S568 | |||
| ID I-II | S573 | |||
| ID I-II | S576 | |||
| ID I-II | S579 | S579 | ||
| ID I-II | S610 | |||
| ID I-II | S623 | |||
| ID I-II | S626 | |||
| ID I-II | T713 | |||
| ID I-II | T715 | |||
| ID I-II | S722 | |||
| C-term | T1944 | |||
| Nav1.6/SCN8A | W:Q9WTU3 | |||
| ID I-II | S504 | |||
| ID I-II | S518 | |||
| ID I-II | S520 | |||
| ID I-II | S522 | |||
| ID I-II | S600 | |||
| Nav1.9/SCN9A | W: Q62205 | |||
| ID I-II | S502 | |||
| ID I-II | S504 | |||
| C-term | S1062 | |||
| C-term | S1064 | |||
| HCN1 | W: O88704 | Tr: O88704 | M:O88704 | |
| N-term | T39 | T39 | T39 | |
| N-term | S69 | S69 | ||
| C-term | S588 | |||
| HCN2 | W:O88703 | Tr: O88703 | M: O88703 | T: O88703 |
| N-term | S90 | S90 | ||
| N-term | S119 | S119 | S119 | |
| N-term | S131 | |||
| N-term | S134 | |||
| C-term | S726 | |||
| C-term | S743 | |||
| C-term | S750 | |||
| C-term | S756 | |||
| C-term | S757 | |||
| C-term | S771 | S771 | ||
| C-term | S795 | |||
| C-term | S834 | |||
| C-term | S840 | S840 | ||
| C-term | S842 | S842 | S842 | |
| HCN3 | W: O88705 | Tr:O88705 | M: O88705 | |
| C-term | S633 | S633 | S633 | |
| C-term | S720 | |||
| C-term | S936 | |||
| HCN4 | W: O70507 | M: O70507 | ||
| N-term | S139 | |||
| C-term | S921 | |||
| Kv1.1/KCNA1 | W: IPI00133719 | |||
| N-term | S23 | |||
| Kv1.2/KCNA2 | W: P63141 | M: P63141 | T: P63141 | |
| C-term | T421 | |||
| C-term | T433 | |||
| C-term | S434 | S434 | ||
| C-term | S440 | |||
| C-term | S441 | S441 | ||
| C-term | S447 | |||
| Kv1.4/KCNA4 | W: Q61423 | Tr:Q8CBF8 | M: Q61423 | |
| N-term | S101 | |||
| N-term | S113 | |||
| N-term | S122 | S122 | S122 | |
| Kv1.5/KCNA5 | W: Q61762 | |||
| C-term | S535 | |||
| Kv1.6/KCNA6 | W: Q61923 | M: Q61923 | ||
| N-term | S6 | |||
| N-term | T8 | T8 | ||
| Kv2.1/KCNB1 | W: Q8K0D1 | TR: Q03717 | M: Q8K0D1 | T: Q8K0D1 |
| N-term | S12 | |||
| N-term | S15 | |||
| C-term | S444 | |||
| C-term | S484 | |||
| C-term | S517 | |||
| C-term | S518 | |||
| C-term | S519 | |||
| C-term | S520 | |||
| C-term | S567 | |||
| C-term | S655 | S655 | S655 | S655 |
| C-term | S782 | |||
| C-term | T803 | T803 | ||
| C-term | S804 | S804 | ||
| Kv2.2/KCNB2 | W: A6H8H5 | |||
| C-term | T478 | |||
| C-term | S481 | |||
| C-term | S488 | |||
| C-term | S490 | |||
| C-term | S503 | |||
| C-term | S507 | |||
| C-term | S530 | |||
| Kv3.1/KCNC1 | W: P15388 | Tr: Q3TR92 | T: P15388 | |
| N-term | S158 | |||
| N-term | S160 | S160 | ||
| C-term | T421 | |||
| C-term | S468 | |||
| C-term | T483 | |||
| Kv3.2/KCNC2 | W: P70311 | |||
| C-term | S509 | |||
| C-term | S557 | |||
| C-term | S604 | |||
| C-term | S619 | |||
| Kv3.3/KCNC3 | W: Q63959 | Tr: Q63959 | ||
| C-term | S717 | S717 | ||
| C-term | S732 | |||
| C-term | S740 | |||
| C-term | T751 | |||
| C-term | S755 | |||
| Kv3.4/KCNC4 | W: Q8R1C0 | |||
| C-term | S555 | |||
| Kv4.1/KCND1 | W: Q03719 | |||
| C-term | S460 | |||
| C-term | S555 | |||
| Kv4.2/KCND2 | W: Q9Z0V2 | M: Q9Z0V2 | T: Q9Z0V2 | |
| N-term | T154 | |||
| C-term | S548 | S548 | ||
| C-term | S552 | S552 | ||
| Kv4.3/KCND3 | W: Q9Z0V1 | |||
| N-term | S153 | |||
| Kv5.1/KCNF1 | W: Q7TSH7 | |||
| C-term | S444 | |||
| C-term | S470 | |||
| C-term | S472 | |||
| Kv7.2/KCNQ2 | W: NP_034741 | Tr: NP_034741 | M: NP_034741 | T: NP_034741 |
| N-term | S52 | |||
| C-term | S352 | S352 | ||
| C-term | S457 | |||
| C-term | T462 | |||
| C-term | S466 | S466 | ||
| C-term | S468 | S468 | ||
| C-term | S476 | |||
| C-term | S485 | |||
| C-term | S507 | S507 | ||
| C-term | Y671 | |||
| C-term | S673 | S673 | ||
| C-term | T728 | |||
| C-term | S729 | |||
| C-term | S799 | |||
| C-term | S801 | |||
| Kv7.3/KCNQ3 | W: Q8K3F6 | Tr: Q14B66 | M: Q8K3F6 | |
| N-term | T82 | |||
| C-term | S579 | |||
| C-term | T580 | T577 | ||
| C-term | S596 | |||
| C-term | S599 | S599 | ||
| Kv7.5/KCNQ5 | W: Q9JK45 | |||
| C-term | S469 | |||
| C-term | S477 | |||
| Kv10.1/KCNH1 | W: Q32MR7 | |||
| C-term | S899 | |||
| C-term | S904 | |||
| Kv10.2/KCNH2 | W: XP_001477398.1 | Tr: 035219 | ||
| N-term | S281 | |||
| N-term | S320/T321/S322 | |||
| N-term | S362 | |||
| N-term | S364 | |||
| C-term | S913 | |||
| C-term | S916 | |||
| C-term | S1180 | |||
| Kv10.5/KCNH5 | W: Q920E3 | |||
| C-term | S883 | |||
| C-term | S888 | |||
| Kv10.7/KCNH7 | W: A2AUY8 | |||
| N-term | S174 | |||
| N-term | T318 | |||
| N-term | S319 | |||
| C-term | S891 | |||
| C-term | S1007 | |||
| C-term | S1135 | |||
| C-term | S1169 | |||
| C-term | S1174 | |||
| C-term | S1188 | |||
| Slo1/KCNMA1 | W: Q08460-1 | Tr: Q08460-1 | M: Q08460-1 | T: Q08460-1 |
| N-term | S136 | |||
| C-term | T709 | T709 | ||
| C-term | S711 | S711 | S711 | S711 |
| C-term | S923 | S923 | ||
| C-term | S924 | S924 | ||
| C-term | S928 | |||
| C-term | S938 | |||
| C-term | S1060 | |||
| C-term | T1061 | T1061 | T1061 | T1061 |
| C-term | T1064 | |||
| TrpC3/TRPC3 | W:B1ATV3-1 | |||
| C-term | S807 | |||
| TrpC6/TRPC6 | W: Q61143 | |||
| C-term | S814 | |||
| TrpM2/TRPM2 | W: Q91YD4 | |||
| N-term | S601 | |||
| TrpM3/TRPM3 | W: Q5F4S9 | Tr: Q5F4S9 | ||
| C-term | T1299 | |||
| C-term | S1301 | |||
| C-term | S1314 | |||
| C-term | S1317 | |||
| C-term | T1336/S1338/T1340/S1341 | |||
| C-term | S1605 | |||
| C-term | S1608 | |||
| C-term | S1609 | |||
| TrpV2/TRPV2 | W: Q9WTR1 | |||
| N-term | T14 | |||
| N-term | S15 |
Note additional sites identified on Slo1/KCNMA1 Q08460-2: T670 (W) and S672 (W, T), corresponding to T709 and S711 in Q08460-1. W, Wisniewski et al., 2010; Tr, Trinidad et al., 2008; M, Munton et al., 2007; T, Tweedie-Cullen et al., 2009.
Trinidad et al. (2008) analyzed phosphosites on postsynaptic density fractions generated from four different mouse brain regions by differential centrifugation and sucrose gradient density centrifugation. After in-solution digestions, the digested peptide samples from cortex, midbrain, cerebellum, and hippocampus were individually labeled with four different iTRAQ reagents (114, 115, 116, and 117 D, respectively), to allow for the subsequent identification of peptides originating from the different brain regions. A mixture with equal amounts of each sample was subjected to strong cation exchange (SCX) chromatography, which yielded over 90 fractions. Binding to TiO2 beads further enriched the phosphopeptides. Eluted phosphopeptides were analyzed on a hybrid quadrupole time-of-flight MS (QSTAR Pulsar) instrument. All CID tandem mass spectra were searched by Analyst QS software against a Uniprot Mus musculus database containing original and randomized protein sequences (64,717 entries). Phosphopeptide identification and site assignment was manually performed with diagnostic peaks (e.g., high abundant neutral loss peak) when appropriate, and quantification of iTRAQ-labeled phosphopeptides was performed using “Proteinprospector” software. The false-positive rate was 0.12%. This study yielded 1,564 unique phosphosites on 831 proteins, including 52 different phosphosites on 19 different voltage-gated ion channel α subunits (Table I).
Munton et al. (2007) analyzed phosphosites on naive and stimulated mouse brain synaptosomes. Tryptic peptides generated by in-solution digestion were subjected to SCX chromatography, and five of the SCX fractions were further enriched by immobilized metal ion chromatography (IMAC). Eluted material was analyzed on MALDI TOF-TOF MS, ion trap LCQ, and LTQ-FT MS instruments with a reject mass list for common air contaminants. All CID tandem mass spectra from these synaptosomal phosphoproteomes were searched using Mascot against an EBI mouse protein database (32,849 entries). The false-positive rate was not reported. This study yielded 974 unique phosphosites on 499 proteins, including 32 different phosphosites on 16 different voltage-gated ion channel α subunits (Table I).
Tweedie-Cullen et al., (2009) analyzed phosphosites on a synaptic fraction prepared by differential centrifugation and sucrose gradient density centrifugation. Isolated nuclear, synaptic, and histone proteins were digested in solution, and the resultant tryptic peptides were subjected to SCX chromatography, and the early eluting phosphopeptide-rich fractions applied to TiO2 or IMAC beads. Eluted phosphopeptide samples were analyzed by three different types of MS instruments: MALDI TOF/TOF, LTQ-FT, or LTQ-Orbitrap. In the case of the mass analysis with the LTQ-Orbitrap instrument, lock mass was used to increase mass accuracy. Database analysis of the total CID tandem mass spectra was performed against an EBI mouse protein database (42,656 entries). The false-positive rate was 0.53%. All assigned tandem spectra with phosphorylation modifications were manually inspected and filtered out with strict criteria (MASCOT expected value below 0.05). Normalized delta ion scores were calculated with scores between two top ranking peptides and used for precise site assignment (<0.4). This study yielded 1,341 unique phosphosites on 638 proteins, including 32 different phosphosites on 14 different voltage-gated ion channel α subunits (Table I).
While the phosphosites identified in the above studies were confirmed using the most strict proteomic criteria and manual inspections, the bulk of the phosphosites obtained from these high-throughput approaches were not experimentally validated by other means. Recent proteomic technological innovations using MS have lead to a wealth of newly identified phosphosites on cellular proteins but, to date, there exist few alternative approaches to effectively cross-validate phosphosite identification. Therefore, the current method of determining the false-positive rate of these high-throughput data are based entirely on the dataset from that particular mass spectrometric analysis, and not any independent evaluation. This may not be sufficient for data validation of all phosphosites. For example, a recent study of the yeast phosphoproteome showed that applying a single false discovery rate for all peptide identifications significantly overestimates the occurrence of rare modifications such as tyrosine phosphorylation (there are no dedicated tyrosine kinases in yeast) (Gnad et al., 2009). An appropriate false discovery rate threshold confined to phosphopeptides, among all peptides identified, is adjustable for more sensitive or more reliable data analysis. Whether the identified phosphosites are located in structurally accessible protein regions may be a further consideration when determining false positives; this is rarely an option for ion channel α subunits, for which few structures are available, and those that are (e.g., Kv1.2; Long et al., 2005) lack the cytoplasmic domains targeted by phosphorylation. Effective validation of the biological role of any identified phosphosites still involves using biochemical, cell biological, and electrophysiological analyses of individual proteins expressing site-specific mutations in the identified phosphosites.
Analysis of the obtained datasets for ion channel phosphosites
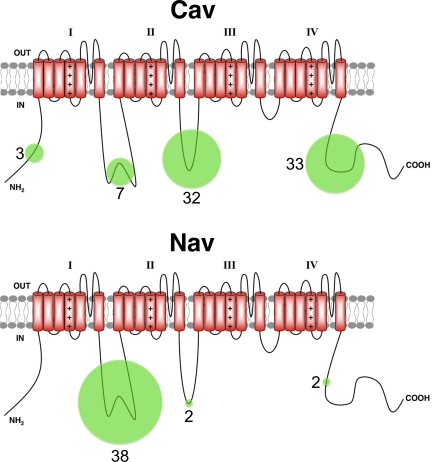
An overview of the dataset (Table I) focusing on the location of the identified phosphosites on the 24 transmembrane segment ion channel α subunits reveals that for Cav channel α subunits, phosphorylation is primarily split between the ID II-III linker and the C terminus (Fig. 1), whereas for Nav channel α subunits, phosphorylation is primarily (90%) on the ID I-II linker (Fig. 1), as suggested by previous studies (Cantrell and Catterall, 2001). All of the six transmembrane ion channel α subunits (Fig. 2) have the majority of their phosphosites on the C terminus, although this varies from ≈90% for Kv7 and Slo1-BK channel family members, to ≈65% for Kv10 and HCN family members.
Figure 1.
Schematic of phosphosites identified in high-throughput studies on 24 transmembrane segment ion channel α subunits. Areas of the green circles represent the percentage of phosphosites found within a specific cytoplasmic domain (N-terminal, ID I-II, ID-II-III, or C-terminal). Numbers adjacent to circles represent the total number of unique phosphosites identified within that domain for all members of that family (Cav: six members; Nav: four members).
Figure 2.
Schematic of phosphosites identified in high-throughput studies on six transmembrane segment ion channel α subunits. Areas of the green circles represent the percentage of phosphosites found within a specific cytoplasmic domain (N or C terminus). Numbers adjacent to circles represent the total number of unique phosphosites identified within that domain for all members of that family (Kv1-4: 14 members; Kv7: 3 members; Kv10: 4 members; Slo1-BK: 1 member; HCN: 4 members; TRP: 5 members).
An overall number of unique phosphosites identified shows that the largest ion channel α subunits, namely those within the 24 transmembrane segment family of Cav and Nav channel α subunits, have the most extensive phosphorylation, with Cav2.1 (24 phosphosites), Nav1.2 (23 phosphosites), and Cav2.3 (22 phosphosites) topping the list (Table I). This may be expected given the large size of these polypeptides (2,000–2,400 amino acids) and that only one α subunit is present per channel complex. Among the six transmembrane segment family members, which are smaller polypeptides and assemble as tetramers in the final channel complex, Kv7.2 (15 phosphosites), HCN2 (14 phosphosites), Kv2.1 (13 phosphosites), and Slo1-BK (10 phosphosites) are notable in their extensive representation, suggesting a high degree of phosphorylation-dependent modulation of these channels. Note that while these data suggest extensive multisite phosphorylation exists on individual molecules of ion channel α subunits, these studies have not provided conclusive data as these phosphosites come from relatively small tryptic fragments of the larger polypeptides. As such, except for the small subset of neighboring phosphosites found within the same multiply phosphorylated tryptic peptide, it is formally possible that the multiple phospho-sites identified in these ion channel α subunits are stochastically distributed in different molecules.
Note that representation on the list of “most phosphorylated” ion channel α subunits is likely a combination of not only the actual extent of phosphorylation, but also nature of the phosphorylation, as the techniques employed (MS-based analyses of tryptic peptides) bias the analysis toward phosphosites on tryptic peptides with a chemical nature (e.g., size, charge, hydropathy, etc.) that makes them amenable to detection by mass spectrometers. Of course, the overall abundance of a specific ion channel α subunit in a given sample, whether it is whole brain or a subcellular fraction thereof, will also affect its representation in samples with high proteomic and phosphoproteomic complexity, such as those used for global analyses. As such, enriching for ion channel α subunits is an attractive approach to increase coverage of their phosphoproteome.
These high-throughput approaches focused exclusively on adult mouse brain. This raises questions as to the implications for the rat and human brain orthologues of these mouse brain ion channel α subunits. If conservation of the phosphoacceptor site itself is any indication, then these mouse brain data are likely to have strong implications for studies in other mammals. Focusing on the ion channel α subunits within a subfamily that exhibit the highest number of identified phosphosites (Cav2.1, Nav1.2, Kv2.1, Kv7.2, and TRPM3), we found that, with the exception of mouse and human Cav2.1 (87% overall identity), overall sequence identity between mouse, rat, and human ion channel α subunit orthologues was 93% or higher. As to the phosphosites themselves (85 in total), there was 100% conservation of every site in mouse, rat, and human Nav1.2 (23 phosphosites), Kv2.1 (13 phosphosites), Kv7.2 (15 phosphosites), and TrpM3 (8 phosphosites), and for human Cav2.1, 92% conservation (22/24). It should be noted that conservation of the phospho-acceptor site itself may not in itself suggest that the site is phosphorylated, as changes in neighboring amino acids in the primary, secondary, or tertiary structure may alter protein kinase consensus motifs (Ubersax and Ferrell, 2007). Also note that with the exception of the study by Munton et al., these studies focused on samples prepared from animals under basal conditions, and may not contain examples of sites whose phosphorylation requires specific stimuli.
Functional implication of phosphosites identified in high-throughput approaches
There are numerous examples of phosphorylation at defined sites on specific ion channels dynamically regulating how these ion channels impact cell function. One way that phosphorylation can act is through altering the abundance of specific ion channels in specific membrane domains. This can occur via changes in the proportion of ion channels in intracellular membranes versus the plasma membrane, by impacting the rates of channel exocytosis and endocytosis, or by affecting the density of channels in a specific plasma membrane subdomain via effects on channel clustering. Phosphorylation can also affect the gating of ion channels, altering the probability that an ion channel will be open under a given set of conditions.
A large literature exists providing a large number of examples of stimuli impacting ion channel abundance, distribution, and gating. As many of these stimuli are known to act through signaling pathways leading to activation of protein kinases or protein phosphatases, the assumption has been that the effects are meditated through changes in the phosphorylation state of the ion channel subunits themselves. In some cases, regulation can be obtained for recombinant ion channels expressed in heterologous cells, in which case candidate phosphosites can be identified using phosphosite prediction software (e.g., http://www.cbs.dtu.dk/services/NetPhosK/; http://scansite.mit.edu/motifscan_seq.phtml; http://www.hprd.org/PhosphoMotif_finder), and mutations generated that lock specific candidate phosphosites in either a dephospho- or phospho-mimetic state (e. g., Ser to Ala or Asp mutations, respectively). Data obtained from these approaches have allowed for the identification of specific sites critical to phosphorylation-dependent modulation of channel abundance, distribution, or function. However, rarely have these studies directly shown that the impact of the mutations is via altering phosphorylation state, and that the mutation only altered phosphorylation at the mutated site. Moreover, there are few cases where the candidate phosphosite being studied is known to be modified by phosphorylation in native cells. As such, we are left with a large number of candidate phosphosites whose mutation impacts the channel, but whose physiological relevance is unknown.
The recent high-throughput approaches described here have yielded, for the most part, a dataset completely complementary to these types of studies, in providing a large amount of information as to which sites are found phosphorylated in native cells, but without any functional insights. In a few cases, which we detail below, phosphosites identified in these recent phosphoproteomics studies have provided in vivo relevance to studies already performed in heterologous cells. Future studies aimed at determining the functional impact of the phosphosites we highlight here will serve to increase the number of examples of phosphosites that appear in vivo, and functional relevance is known.
Phosphorylation of Cav2.2 α1 subunits, which regulate neurotransmitter release from nerve terminals, in their DI-DII linker region has been proposed to play a crucial role in diverse aspects of Cav channel function. For example, mutation of rat Cav2.2 at the site identified in mouse brain (using accession no. O55017) as S424 mediates PKC-dependent Cav2.2 inhibition (Fang et al., 2006; Rajagopal et al., 2009). ERK-dependent up-regulation of Cav2.2 channels is mediated through phosphorylation of the site identified in mouse brain as S446 (Martin et al., 2006). In the DII-DIII linker, PKC phosphorylates the sites identified in mouse brain as S773 and S892, and CaMKII at S783, and impacts syntaxin1A–Cav2.2 interaction (Yokoyama et al., 2005). PKC-dependent phosphorylation on S892 was also found to disrupt interaction with syntaxin 1A and abolish the hyperpolarizing shift in the steady-state inactivation caused by this protein–protein interaction (Jarvis and Zamponi, 2001).
The predominant sodium channel in brain, Nav1.2, is a substrate for PKA phosphorylation in its DI-DII linker domain (Cantrell et al., 1997; Smith and Goldin, 1997). PKA phosphorylates Nav1.2 at the in vivo mouse brain sites S573, S610, and S623, calcineurin dephosphorylates these three phosphosites, and protein phosphatase 2A (PP2A) dephosphorylates S610 (Smith and Goldin, 1997). PKA causes a decrease of the Nav peak current triggered by dopaminergic stimulation of rat hippocampal neurons, but does not alter the voltage dependence or kinetic parameters. The S573A mutation abolishes the dopamine-induced PKA effects (Cantrell et al., 1997).
Kv1.2 channel subunits are expressed in axons and nerve terminals, and modulate action potential propagation and nerve terminal excitation. Site-directed mutagenesis studies showed that the S440A and S441A mutations diminish Kv1.2 surface expression and whole cell current (Yang et al., 2007). The S449A substitution causes a decrease in phosphorylation at S440 and S441, again leading to diminished surface expression (Yang et al., 2007). PKA phosphorylates Kv1.2 at S449 (Johnson et al., 2009).
Kv2.1 is expressed in large clusters in the somatodendritic membrane of most neurons (Trimmer, 1991). Kv2.1 is extensively phosphorylated in vivo, and increased neuronal excitability leads to calcineurin-mediated Kv2.1 dephosphorylation, a loss of clustering, and hyperpolarizing shifts in voltage-dependent gating (Misonou et al., 2004, 2005, 2006). A mass spectrometry–based analysis of Kv2.1 purified from rat brain identified 16 phosphosites (Park et al., 2006; Mohapatra et al., 2007), of which five (S15, S484, S520, S567, S655, and S804) were also identified in high-throughput approaches detailed here (Table I). Of the phosphosites identified in the high throughput studies, mutations at S15, S567, and S655 partially mimic and occlude the effects of dephosphorylation in causing hyperpolarizing shifts in the voltage-dependent gating. In addition, phosphorylation on S804 via p38MAPK increases the surface expression of Kv2.1, leading to neuronal apoptosis (Redman et al., 2007).
Kv channels containing Kv4.2 α subunits mediate transient A-type K+ currents in neurons, which are critically involved in the regulation of dendritic excitability and plasticity (Shah et al., 2010). In hippocampal slices, PKA activation by forskolin and other pharmacological agents results in Kv4.2 phosphorylation at S552 (Anderson et al., 2000). PKA-dependent phosphorylation of this residue results in a depolarizing shift of Kv4.2 activation, but only in presence of the auxiliary subunit KChIP3 (Schrader et al., 2002). PKA phosphorylation at S552 also mediates AMPA-stimulated Kv4.2 internalization from dendritic spines (Hammond et al., 2008). Note that a previous mass spectrometry–based analysis of Kv4.2 purified from rat brain (Seikel and Trimmer, 2009) identified four phosphosites (S548, S552, S572, and S575) of which two (S548 and S552) were found in the high-throughput analyses of mouse brain (Table I).
Kv7 (KCNQ) subunits form neuronal Kv channels that underlie the M-current (so-called due to its modulation by muscarinic signaling). These channels localize in axon initial segments, nodes of Ranvier, and somata throughout the nervous system. Of the residues found phosphorylated in mouse brain (Table I), PKA phosphorylation of KCNQ2 at S52 has been shown to enhance KCNQ2 currents in heterologous cells (Schroeder et al., 1998). A previous mass spectrometric–based analysis identified phosphorylation at either S578 or T579 on recombinant human KCNQ3 expressed in heterologous cells (Surti et al., 2005). Mutations at these positions in KCNQ3 eliminate functional expression of KCNQ2/3 channels (Surti et al., 2005). These sites are analogous to in vivo phosphosites identified in mouse brain proteomic studies on Kv7.3 at S579 and T580.
Large conductance Ca2+-activated K+ (Slo1-BK) channels are involved in neuronal repolarization and rapid afterhyperpolarization. PKC-dependent phosphorylation on S695 of the bovine Slo1-BK α subunit (corresponding to S711 of the mouse isoform shown in Table I) decreases the open probability of the channel, an effect countered by protein phosphatase 1 (Zhou et al., 2010). PKG-dependent phosphorylation on S855 together with S869 (S869 corresponding to S924 in the mouse isoform used here) enhances the current amplitude of the human Slo1-BK channel in Xenopus oocytes (Nara et al., 2000).
The overall nature of ion channel phosphorylation revealed by high-throughput analyses
To gain insights into whether these data provide insights into the predominance of ion channel phosphorylation by a particular protein kinase, we have analyzed the phosphosites on the ion channel α subunits with the most extensive phosphorylation (Cav2.1, Nav1.2, Kv2.1, Kv7.2, and TRPM3) for consensus motifs using NetPhosK (http://www.cbs.dtu.dk/services/NetPhosK/). Of the 83 phosphosites analyzed, the most predominant motifs recognized were for the proline-directed protein kinases p38MAP kinase, GSK3 and Cdk5, with 30 phosphosites being scored for phosphorylation by one or more of these protein kinases. There were 13 phosphosites that scored as consensus motifs for PKA, and 10 for PKC. There were 19 phosphosites that did not score as sites for any consensus protein kinase motif. These results are somewhat consistent with our previous analyses of phosphosites found on Nav1.2 (Berendt et al., 2010) and Kv2.1 (Park et al., 2006) purified from rat brain, which revealed a predominance of phosphosites for proline-directed protein kinases, as well as several sites not recognized by any prediction program.
In this regard it is interesting to compare the data obtained from these high throughput global analyses versus those from studies on purified ion channel α subunits (Table II). Recent studies from our laboratory have used this approach for Nav1.2 (Berendt et al., 2010), Kv2.1 (Park et al., 2006), and Slo1-BK (in collaboration with Fakler et al.) (Yan et al., 2008), among others. Note that in each case, the analyses of purified ion channel α subunits were from rat brain, while the global analyses were performed on mouse brain. For Nav1.2, of the 23 phosphosites identified in these global analyses, 10 were found in the single-target analysis of purified Nav1.2 (Berendt et al., 2010). However, the MS-based analyses of purified Nav1.2 identified six phosphosites not seen in the global analyses. In the case of Kv2.1, of the 13 phosphosites identified in the global analyses of mouse brain, six were observed in the analysis of Kv2.1 purified from rat brain, which also revealed the presence of 10 additional phosphosites not seen in the global analyses (Park et al., 2006). For Slo1-BK, of the 10 unique phosphosites identified in the global analyses, all but two were also observed in the analyses of the purified Slo1-BK, which also provided an additional 20 phosphosites (Yan et al., 2008).
Table II.
Results from high versus low-throughput analyses
| Subunit/location | High accession no./site | Low accession no./site |
| Nav1.2/SCN2A | B1AWN6/Mus | P04775-1/rat |
| N-term | S4 | |
| ID I-II | S468 | S468 |
| ID I-II | S471 | S471 |
| ID I-II | S475 | |
| ID I-II | S484 | S484 |
| ID I-II | S486 | S486 |
| ID I-II | S488 | |
| ID I-II | S526 | |
| ID I-II | S528 | S528 |
| ID I-II | S540 | |
| ID I-II | S554 | S554 |
| ID I-II | S558 | |
| ID I-II | S561 | |
| ID I-II | S568 | |
| ID I-II | S573 | |
| ID I-II | S576 | |
| ID I-II | S579 | S579 |
| ID I-II | S610 | S610 |
| ID I-II | S623 | S623 |
| ID I-II | S626 | |
| ID I-II | S687 | |
| ID I-II | S688 | |
| ID I-II | T713 | |
| ID I-II | T715 | |
| ID I-II | S722 | S722 |
| C-term | S1966 | |
| C-term | T1944 | T1944 |
| Kv2.1/KCNB1 | Q8K0D1/mus | P15387/rat |
| N-term | S12 | |
| N-term | S15 | S15 |
| C-term | S444 | |
| C-term | S457 | |
| C-term | S484 | |
| C-term | S496 | |
| C-term | S503 | |
| C-term | S517 | |
| C-term | S518 | |
| C-term | S519 | |
| C-term | S520 | S520 |
| C-term | S541 | |
| C-term | S567 | S567 |
| C-term | S590 | |
| C-term | S607 | |
| C-term | S655 | S655 |
| C-term | S719 | |
| C-term | S771 | |
| C-term | S782 | |
| C-term | S799 | |
| C-term | T803 | |
| C-term | S804 | S804 |
| C-term | T836 | |
| Slo1/KCNMA1 | Q08460-1/Mus | Q62976-2/Rat |
| N-term | S136 | S106 |
| N-term | S107 | |
| C-term | S551 | |
| C-term | T709 | T676 |
| C-term | S711 | S678 |
| C-term | S691 | |
| C-term | T694 | |
| C-term | S695 | |
| C-term | S813 | |
| C-term | S818 | |
| C-term | S879 | |
| C-term | T883 | |
| C-term | S923 | S890 |
| C-term | S924 | S891 |
| C-term | S928 | S895 |
| C-term | S938 | S905 |
| C-term | S1060 | |
| C-term | T1061 | T1001 |
| C-term | T1064 | |
| C-term | S1059 | |
| C-term | S1101 | |
| C-term | S1106 | |
| C-term | S1112 | |
| C-term | S1113 | |
| C-term | S1116 | |
| C-term | S1117 | |
| C-term | S1118 | |
| C-term | S1121 | |
| C-term | S1124 | |
| C-term | T1125 |
These comparisons suggest that while the dataset from these global analyses represents a wealth of information for ion channel researchers, parallel approaches using enrichment of the target ion channel α subunits provide datasets that are oftentimes more extensive, revealing additional phosphosites that may play important roles in the modulation of these channels. What these datasets do suggest is that neither approach used in isolation has yet to provide a comprehensive view of phosphosites on any given ion channel α subunit. Of course, phosphorylation is by nature dynamic, such that variations in the exact state of the animal subject, details of sample preparation, and other factors that are difficult to precisely control can influence the dataset obtained. Moreover, these analyses were primarily performed on samples derived from whole adult rat or mouse brains, such that phosphosites highly represented in other temporal and spatial contexts may be underrepresented. It is also important to stress that any site identified in phosphoproteomic screens needs to be directly confirmed by at least one independent method, for example multiple reaction monitoring (which can also provide quantitative information; Rinehart et al., 2009), immunoblotting with phosphospecific antibodies (Park et al., 2006), or other approaches.
The wealth of data obtained from these studies raises the question as to whether we in the ion channel field should simply sit back and wait as these types of global analyses, being performed by our colleagues in the proteomics field, provide us with intriguing and extensive sets of in vivo phosphosites. We would argue that we should accept such bountiful gifts with gratitude and appreciation, but that we in the ion channel field need to continue our complementary efforts to identify in vivo phosphosites through focused analyses of individual ion channel subunits purified from brain and other tissues. Together, these approaches are likely to yield a comprehensive dataset that will allow for a more complete picture of the nature and extent of in vivo phosphosites crucial to dynamic regulation of native ion channel expression, localization, and function.
Acknowledgments
We thank Dr. Ry Tweedie-Cullen for providing data sets. We are grateful to Dr. Jon Sack and Ms. Ashleigh Evans for critical comments on the manuscript.
The work cited from our laboratory was supported by National Institutes of Health grants NS38343, NS42225, NS64428, and NS64957.
Angus C. Nairn served as editor.
Footnotes
Abbreviations used in this paper:
- CID
- collision-induced dissociation
- IMAC
- immobilized metal affinity chromatography
- MS
- mass spectrometry
- SAX
- strong anion exchange
- SCX
- strong cation exchange
References
- Amanchy R., Periaswamy B., Mathivanan S., Reddy R., Tattikota S.G., Pandey A. 2007. A curated compendium of phosphorylation motifs. Nat. Biotechnol. 25:285–286 10.1038/nbt0307-285 [DOI] [PubMed] [Google Scholar]
- Anderson A.E., Adams J.P., Qian Y., Cook R.G., Pfaffinger P.J., Sweatt J.D. 2000. Kv4.2 phosphorylation by cyclic AMP-dependent protein kinase. J. Biol. Chem. 275:5337–5346 10.1074/jbc.275.8.5337 [DOI] [PubMed] [Google Scholar]
- Berendt F.J., Park K.S., Trimmer J.S. 2010. Multisite phosphorylation of voltage-gated sodium channel alpha subunits from rat brain. J. Proteome Res. 9:1976–1984 10.1021/pr901171q [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantrell A.R., Catterall W.A. 2001. Neuromodulation of Na+ channels: an unexpected form of cellular plasticity. Nat. Rev. Neurosci. 2:397–407 10.1038/35077553 [DOI] [PubMed] [Google Scholar]
- Cantrell A.R., Smith R.D., Goldin A.L., Scheuer T., Catterall W.A. 1997. Dopaminergic modulation of sodium current in hippocampal neurons via cAMP-dependent phosphorylation of specific sites in the sodium channel alpha subunit. J. Neurosci. 17:7330–7338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerda O., Trimmer J.S. 2010. Analysis and functional implications of phosphorylation of neuronal voltage-gated potassium channels. Neurosci. Lett. 486:60–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen P. 2001. The role of protein phosphorylation in human health and disease. The Sir Hans Krebs Medal Lecture. Eur. J. Biochem. 268:5001–5010 10.1046/j.0014-2956.2001.02473.x [DOI] [PubMed] [Google Scholar]
- Collins M.O., Grant S.G. 2007. Supramolecular signalling complexes in the nervous system. Subcell. Biochem. 43:185–207 10.1007/978-1-4020-5943-8_9 [DOI] [PubMed] [Google Scholar]
- Costa M.R., Catterall W.A. 1984. Phosphorylation of the alpha subunit of the sodium channel by protein kinase C. Cell. Mol. Neurobiol. 4:291–297 10.1007/BF00733592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M.R., Casnellie J.E., Catterall W.A. 1982. Selective phosphorylation of the alpha subunit of the sodium channel by cAMP-dependent protein kinase. J. Biol. Chem. 257:7918–7921 [PubMed] [Google Scholar]
- Dai S., Hall D.D., Hell J.W. 2009. Supramolecular assemblies and localized regulation of voltage-gated ion channels. Physiol. Rev. 89:411–452 10.1152/physrev.00029.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diella F., Cameron S., Gemund C., Linding R., Via A., Kuster B., Sicheritz-Ponten T., Blom N., Gibson T.J. 2004. Phospho.ELM: a database of experimentally verified phosphorylation sites in eukaryotic proteins. BMC Bioinformatics. 5:79 10.1186/1471-2105-5-79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diella F., Gould C.M., Chica C., Via A., Gibson T.J. 2008. Phospho.ELM: a database of phosphorylation sites–update 2008. Nucleic Acids Res. 36:D240–D244 10.1093/nar/gkm772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang H., Patanavanich S., Rajagopal S., Yi X., Gill M.S., Sando J.J., Kamatchi G.L. 2006. Inhibitory role of Ser-425 of the alpha1 2.2 subunit in the enhancement of Cav 2.2 currents by phorbol-12-myristate, 13-acetate. J. Biol. Chem. 281:20011–20017 10.1074/jbc.M601776200 [DOI] [PubMed] [Google Scholar]
- Gnad F., Ren S.B., Cox J., Olsen J.V., Macek B., Oroshi M., Mann M. 2007. PHOSIDA (phosphorylation site database): management, structural and evolutionary investigation, and prediction of phosphosites. Genome Biol. 8:R250 10.1186/gb-2007-8-11-r250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnad F., de Godoy L.M., Cox J., Neuhauser N., Ren S., Olsen J.V., Mann M. 2009. High-accuracy identification and bioinformatic analysis of in vivo protein phosphorylation sites in yeast. Proteomics. 9:4642–4652 10.1002/pmic.200900144 [DOI] [PubMed] [Google Scholar]
- Hammond R.S., Lin L., Sidorov M.S., Wikenheiser A.M., Hoffman D.A. 2008. Protein kinase a mediates activity-dependent Kv4.2 channel trafficking. J. Neurosci. 28:7513–7519 10.1523/JNEUROSCI.1951-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanlon M.R., Wallace B.A. 2002. Structure and function of voltage-dependent ion channel regulatory beta subunits. Biochemistry. 41:2886–2894 10.1021/bi0119565 [DOI] [PubMed] [Google Scholar]
- Hille B. 2001. Ionic channels of excitable membranes. Third ed Sinauer, Sunderland, MA: 814 pp [Google Scholar]
- Jarvis S.E., Zamponi G.W. 2001. Distinct molecular determinants govern syntaxin 1A-mediated inactivation and G-protein inhibition of N-type calcium channels. J. Neurosci. 21:2939–2948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R.P., El-Yazbi A.F., Hughes M.F., Schriemer D.C., Walsh E.J., Walsh M.P., Cole W.C. 2009. Identification and functional characterization of protein kinase A-catalyzed phosphorylation of potassium channel Kv1.2 at serine 449. J. Biol. Chem. 284:16562–16574 10.1074/jbc.M109.010918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemeer S., Heck A.J. 2009. The phosphoproteomics data explosion. Curr. Opin. Chem. Biol. 13:414–420 10.1016/j.cbpa.2009.06.022 [DOI] [PubMed] [Google Scholar]
- Levitan I.B. 1985. Phosphorylation of ion channels. J. Membr. Biol. 87:177–190 10.1007/BF01871217 [DOI] [PubMed] [Google Scholar]
- Levitan I.B. 2006. Signaling protein complexes associated with neuronal ion channels. Nat. Neurosci. 9:305–310 10.1038/nn1647 [DOI] [PubMed] [Google Scholar]
- Long S.B., Campbell E.B., Mackinnon R. 2005. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 309:897–903 10.1126/science.1116269 [DOI] [PubMed] [Google Scholar]
- Martin S.W., Butcher A.J., Berrow N.S., Richards M.W., Paddon R.E., Turner D.J., Dolphin A.C., Sihra T.S., Fitzgerald E.M. 2006. Phosphorylation sites on calcium channel alpha1 and beta subunits regulate ERK-dependent modulation of neuronal N-type calcium channels. Cell Calcium. 39:275–292 10.1016/j.ceca.2005.11.002 [DOI] [PubMed] [Google Scholar]
- Misonou H., Mohapatra D.P., Park E.W., Leung V., Zhen D., Misonou K., Anderson A.E., Trimmer J.S. 2004. Regulation of ion channel localization and phosphorylation by neuronal activity. Nat. Neurosci. 7:711–718 10.1038/nn1260 [DOI] [PubMed] [Google Scholar]
- Misonou H., Mohapatra D.P., Menegola M., Trimmer J.S. 2005. Calcium- and metabolic state-dependent modulation of the voltage-dependent Kv2.1 channel regulates neuronal excitability in response to ischemia. J. Neurosci. 25:11184–11193 10.1523/JNEUROSCI.3370-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misonou H., Menegola M., Mohapatra D.P., Guy L.K., Park K.S., Trimmer J.S. 2006. Bidirectional activity-dependent regulation of neuronal ion channel phosphorylation. J. Neurosci. 26:13505–13514 10.1523/JNEUROSCI.3970-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohapatra D.P., Park K.S., Trimmer J.S. 2007. Dynamic regulation of the voltage-gated Kv2.1 potassium channel by multisite phosphorylation. Biochem. Soc. Trans. 35:1064–1068 10.1042/BST0351064 [DOI] [PubMed] [Google Scholar]
- Munton R.P., Tweedie-Cullen R., Livingstone-Zatchej M., Weinandy F., Waidelich M., Longo D., Gehrig P., Potthast F., Rutishauser D., Gerrits B., et al. 2007. Qualitative and quantitative analyses of protein phosphorylation in naive and stimulated mouse synaptosomal preparations. Mol. Cell. Proteomics. 6:283–293 [DOI] [PubMed] [Google Scholar]
- Nara M., Dhulipala P.D.K., Ji G.J., Kamasani U.R., Wang Y.X., Matalon S., Kotlikoff M.I. 2000. Guanylyl cyclase stimulatory coupling to K-Ca channels. Am. J. Physiol. Cell Physiol. 279:C1938–C1945 [DOI] [PubMed] [Google Scholar]
- Park K.S., Mohapatra D.P., Misonou H., Trimmer J.S. 2006. Graded regulation of the Kv2.1 potassium channel by variable phosphorylation. Science. 313:976–979 10.1126/science.1124254 [DOI] [PubMed] [Google Scholar]
- Prasad T.S.K., Goel R., Kandasamy K., Keerthikumar S., Kumar S., Mathivanan S., Telikicherla D., Raju R., Shafreen B., Venugopal A., et al. 2009. Human Protein Reference Database-2009 update. Nucleic Acids Res. 37:D767–D772 10.1093/nar/gkn892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopal S., Fang H., Oronce C.I., Jhaveri S., Taneja S., Dehlin E.M., Snyder S.L., Sando J.J., Kamatchi G.L. 2009. Site-specific regulation of CA(V)2.2 channels by protein kinase C isozymes betaII and epsilon. Neuroscience. 159:618–628 10.1016/j.neuroscience.2008.12.047 [DOI] [PubMed] [Google Scholar]
- Redman P.T., He K., Hartnett K.A., Jefferson B.S., Hu L., Rosenberg P.A., Levitan E.S., Aizenman E. 2007. Apoptotic surge of potassium currents is mediated by p38 phosphorylation of Kv2.1. Proc. Natl. Acad. Sci. USA. 104:3568–3573 10.1073/pnas.0610159104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinehart J., Maksimova Y.D., Tanis J.E., Stone K.L., Hodson C.A., Zhang J., Risinger M., Pan W., Wu D., Colangelo C.M., et al. 2009. Sites of regulated phosphorylation that control K-Cl cotransporter activity. Cell. 138:525–536 10.1016/j.cell.2009.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saneyoshi T., Fortin D.A., Soderling T.R. 2010. Regulation of spine and synapse formation by activity-dependent intracellular signaling pathways. Curr. Opin. Neurobiol. 20:108–115 10.1016/j.conb.2009.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader L.A., Anderson A.E., Mayne A., Pfaffinger P.J., Sweatt J.D. 2002. PKA modulation of Kv4.2-encoded A-type potassium channels requires formation of a supramolecular complex. J. Neurosci. 22:10123–10133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder B.C., Kubisch C., Stein V., Jentsch T.J. 1998. Moderate loss of function of cyclic-AMP-modulated KCNQ2/KCNQ3 K+ channels causes epilepsy. Nature. 396:687–690 10.1038/25367 [DOI] [PubMed] [Google Scholar]
- Schulz D.J., Temporal S., Barry D.M., Garcia M.L. 2008. Mechanisms of voltage-gated ion channel regulation: from gene expression to localization. Cell. Mol. Life Sci. 65:2215–2231 10.1007/s00018-008-8060-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seikel E., Trimmer J.S. 2009. Convergent modulation of Kv4.2 channel alpha subunits by structurally distinct DPPX and KChIP auxiliary subunits. Biochemistry. 48:5721–5730 10.1021/bi802316m [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah M.M., Hammond R.S., Hoffman D.A. 2010. Dendritic ion channel trafficking and plasticity. Trends Neurosci. 33:307–316 10.1016/j.tins.2010.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R.D., Goldin A.L. 1997. Phosphorylation at a single site in the rat brain sodium channel is necessary and sufficient for current reduction by protein kinase A. J. Neurosci. 17:6086–6093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surti T.S., Huang L., Jan Y.N., Jan L.Y., Cooper E.C. 2005. Identification by mass spectrometry and functional characterization of two phosphorylation sites of KCNQ2/KCNQ3 channels. Proc. Natl. Acad. Sci. USA. 102:17828–17833 10.1073/pnas.0509122102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimmer J.S. 1991. Immunological identification and characterization of a delayed rectifier K+ channel polypeptide in rat brain. Proc. Natl. Acad. Sci. USA. 88:10764–10768 10.1073/pnas.88.23.10764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinidad J.C., Thalhammer A., Specht C.G., Lynn A.J., Baker P.R., Schoepfer R., Burlingame A.L. 2008. Quantitative analysis of synaptic phosphorylation and protein expression. Mol. Cell. Proteomics. 7:684–696 [DOI] [PubMed] [Google Scholar]
- Turrigiano G.G. 2008. The self-tuning neuron: synaptic scaling of excitatory synapses. Cell. 135:422–435 10.1016/j.cell.2008.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tweedie-Cullen R.Y., Reck J.M., Mansuy I.M. 2009. Comprehensive mapping of post-translational modifications on synaptic, nuclear, and histone proteins in the adult mouse brain. J. Proteome Res. 8:4966–4982 10.1021/pr9003739 [DOI] [PubMed] [Google Scholar]
- Ubersax J.A., Ferrell J.E., Jr 2007. Mechanisms of specificity in protein phosphorylation. Nat. Rev. Mol. Cell Biol. 8:530–541 10.1038/nrm2203 [DOI] [PubMed] [Google Scholar]
- Vacher H., Mohapatra D.P., Trimmer J.S. 2008. Localization and targeting of voltage-dependent ion channels in mammalian central neurons. Physiol. Rev. 88:1407–1447 10.1152/physrev.00002.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Qian W.J., Chin M.H., Petyuk V.A., Barry R.C., Liu T., Gritsenko M.A., Mottaz H.M., Moore R.J., Camp Ii D.G., et al. 2006. Characterization of the mouse brain proteome using global proteomic analysis complemented with cysteinyl-peptide enrichment. J. Proteome Res. 5:361–369 10.1021/pr0503681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski J.R., Zougman A., Nagaraj N., Mann M. 2009. Universal sample preparation method for proteome analysis. Nat. Methods. 6:359–362 10.1038/nmeth.1322 [DOI] [PubMed] [Google Scholar]
- Wisniewski J.R., Nagaraj N., Zougman A., Gnad F., Mann M. 2010. Brain phosphoproteome obtained by a FASP-based method reveals plasma membrane protein topology. J. Proteome Res. 9:3280–3289 10.1021/pr1002214 [DOI] [PubMed] [Google Scholar]
- Yan J., Olsen J.V., Park K.S., Li W., Bildl W., Schulte U., Aldrich R.W., Fakler B., Trimmer J.S. 2008. Profiling the phospho-status of the BKCa channel alpha subunit in rat brain reveals unexpected patterns and complexity. Mol. Cell. Proteomics. 7:2188–2198 10.1074/mcp.M800063-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J.W., Vacher H., Park K.S., Clark E., Trimmer J.S. 2007. Trafficking-dependent phosphorylation of Kv1.2 regulates voltage-gated potassium channel cell surface expression. Proc. Natl. Acad. Sci. USA. 104:20055–20060 10.1073/pnas.0708574104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama C.T., Myers S.J., Fu J., Mockus S.M., Scheuer T., Catterall W.A. 2005. Mechanism of SNARE protein binding and regulation of Cav2 channels by phosphorylation of the synaptic protein interaction site. Mol. Cell. Neurosci. 28:1–17 10.1016/j.mcn.2004.08.019 [DOI] [PubMed] [Google Scholar]
- Yu F.H., Yarov-Yarovoy V., Gutman G.A., Catterall W.A. 2005. Overview of molecular relationships in the voltage-gated ion channel superfamily. Pharmacol. Rev. 57:387–395 10.1124/pr.57.4.13 [DOI] [PubMed] [Google Scholar]
- Zhou X.B., Wulfsen I., Utku E., Sausbier U., Sausbier M., Wieland T., Ruth P., Korth M. 2010. Dual role of protein kinase C on BK channel regulation. Proc. Natl. Acad. Sci. USA. 107:8005–8010 10.1073/pnas.0912029107 [DOI] [PMC free article] [PubMed] [Google Scholar]