The Parkin ubiquitin ligase marks the mitofusins Mfn1 and Mfn2 for proteasome-dependent degradation, promoting disposal of damaged mitochondria by preventing their fusion with healthy organelles.
Abstract
Damage to mitochondria can lead to the depolarization of the inner mitochondrial membrane, thereby sensitizing impaired mitochondria for selective elimination by autophagy. However, fusion of uncoupled mitochondria with polarized mitochondria can compensate for damage, reverse membrane depolarization, and obviate mitophagy. Parkin, an E3 ubiquitin ligase that is mutated in monogenic forms of Parkinson’s disease, was recently found to induce selective autophagy of damaged mitochondria. Here we show that ubiquitination of mitofusins Mfn1 and Mfn2, large GTPases that mediate mitochondrial fusion, is induced by Parkin upon membrane depolarization and leads to their degradation in a proteasome- and p97-dependent manner. p97, a AAA+ ATPase, accumulates on mitochondria upon uncoupling of Parkin-expressing cells, and both p97 and proteasome activity are required for Parkin-mediated mitophagy. After mitochondrial fission upon depolarization, Parkin prevents or delays refusion of mitochondria, likely by the elimination of mitofusins. Inhibition of Drp1-mediated mitochondrial fission, the proteasome, or p97 prevents Parkin-induced mitophagy.
Introduction
Parkin (PARK2) and PINK1 (PARK6), gene products mutated in autosomal recessive forms of Parkinson’s disease (Kitada et al., 1998; Valente et al., 2004), appear to be involved in mitochondrial maintenance in fly (Greene et al., 2003; Clark et al., 2006; Park et al., 2006; Yang et al., 2006) and mammalian models (Büeler, 2009; Li and Guo, 2009). Genetic studies in Drosophila show that PINK1, a kinase located in mitochondria, functions upstream of Parkin, an E3 ubiquitin (Ub) ligase located in the cytosol, in the same pathway that preserves mitochondrial integrity (Clark et al., 2006; Park et al., 2006; Yang et al., 2006). Interestingly, perturbing mitochondrial dynamics by either promoting fission or suppressing fusion can compensate for pink1 and parkin mutations (Deng et al., 2008; Poole et al., 2008; Yang et al., 2008; Park et al., 2009). Although these studies suggest that PINK1- and Parkin-mediated mitochondrial integrity is tightly linked to the regulation of mitochondrial fission, how such fission protects mitochondria remains unknown.
Dysfunctional mitochondria may be selectively eliminated by autophagy, termed mitophagy (Kim et al., 2007), through pathways distinct from bulk autophagy that provide starved cells with nutrients. One pathway of mitophagy appears to be activated by Parkin after its translocation from the cytosol specifically to dysfunctional mitochondria (Narendra et al., 2008). Consistent with genetic studies in flies that indicated that they work in the same pathway, Parkin translocation and mitophagy induction require PINK1 activity (Geisler et al., 2010; Matsuda et al., 2010; Narendra et al., 2010; Vives-Bauza et al., 2010). Recent studies further show that upon Parkin translocation to damaged mitochondria, Parkin E3 Ub ligase activity increases (Matsuda et al., 2010), and mitochondrial substrates such as VDAC1 become ubiquitinated (Geisler et al., 2010), followed by recruitment of p62 and aggregation of mitochondria by the HDAC6 deacetylase (Lee et al., 2010) .
Mitochondria function in a dynamic network constantly fusing and dividing through the activity of large GTPases and auxiliary proteins. When damaged mitochondria lose membrane potential, fission, or lack of fusion, can segregate them from the mitochondrial network, where they can be engulfed by autophagosomes (Twig et al., 2008). Here we show that Parkin induces the ubiquitination of mitofusins Mfn1 and Mfn2, large GTPases that mediate mitochondrial fusion, leading to their degradation in both a proteasome- and a AAA+ ATPase p97-dependent manner upstream of mitophagy. Upon depolarization, Parkin prevents or delays refusion of mitochondria, likely by the elimination of mitofusins. These findings illuminate how Parkin may stimulate mitophagy by the manipulation of mitochondrial dynamics and suggest how decreasing mitofusin expression in the fly compensates for loss of Parkin or PINK1. Consistent with our results in mammalian cells, it was recently shown that the expression level of endogenous Marf, a fly mitofusin orthologue, was altered by Parkin and PINK1 expression (Poole et al., 2010), and Marf (Ziviani et al., 2010) was found to be ubiquitinated dependent on Parkin and PINK1 expression.
Results
Parkin and PINK1 mediate Mitofusin ubiquitination and proteasomal degradation
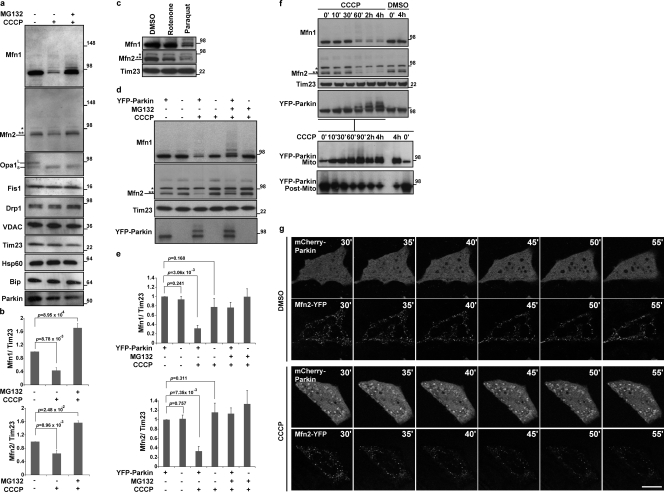
Most known E3 Ub ligase substrates of Parkin have been identified in the cytosol, where Parkin normally localizes (Matsuda and Tanaka, 2010). To identify potential Parkin substrates on mitochondria after depolarization and Parkin translocation, we examined the level of various mitochondrial proteins in the human neuroblastoma cell line SH-SY5Y, which expresses endogenous Parkin (Lutz et al., 2009). 2 h after adding the mitochondrial uncoupler carbonyl cyanide m-chlorophenylhydrazone (CCCP) to depolarize the mitochondria, we observed the selective reduction in expression of endogenous Mfn1 and Mfn2, human homologues of yeast Fzo1 that is known to be degraded by the proteasome (Fig. 1 a and b; Neutzner and Youle, 2005). None of the other mitochondrial proteins examined displayed a reduction in protein levels, whereas Opa1 was cleaved as previously described (Ishihara et al., 2006; Griparic et al., 2007). Incubation of cells with a proteasome inhibitor, MG132, prevented the CCCP-induced decrease in Mfn1 and Mfn2 levels, which suggests that they are degraded by the proteasome. As the mitochondrial respiratory chain inhibitor rotenone has been used to generate an animal model of Parkinsonism (Betarbet et al., 2000) and the oxidizing agent and herbicide paraquat has been linked to human Parkinsonism (Cochemé and Murphy, 2008; Brooks et al., 1999), we examined their effects on Mfn degradation. After a 24-h exposure to paraquat, SH-SY5Y cells display a clear decrease in levels of Mfn1 and 2 (Fig. 1 c). Rotenone treatment causes a more minor effect consistent with its weaker effect on Parkin translocation to the mitochondria (unpublished data). To specifically test the role of Parkin in mitofusin elimination, we compared Mfn levels in HeLa cells, which express little or no endogenous Parkin (Denison et al., 2003; Pawlyk et al., 2003), and in HeLa cells stably expressing YFP-Parkin (HeLa: YFP-Parkin). Endogenous Mfn1 and Mfn2 expression was not affected by YFP-Parkin in untreated cells, whereas their expression levels were greatly decreased after the addition of CCCP to HeLa: YFP-Parkin cells (Fig. 1, d and e). Furthermore, MG132 inhibited the elimination of Mfn1 and Mfn2, and also generated higher molecular weight bands of Mfn1, which is suggestive of ubiquitination (Fig. 1 a and d, MG132 plus lanes). Mfn levels were not reduced upon CCCP addition to HeLa cells not expressing YFP-Parkin. These data indicate that Parkin expression, when combined with mitochondrial uncoupling, induces mitofusin degradation via the Ub–proteasome system (UPS).
Figure 1.
Mitochondrial depolarization induces selective degradation of mitofusins by Parkin. (a) Whole cell lysates of SH-SY5Y cells (30 µg of proteins) were subjected to SDS-PAGE after treatment with DMSO (control), CCCP (depolarized), or CCCP with a proteasomal inhibitor, MG132. Immunoblotting results show the stability of mitofusin 1 (Mfn1), mitofusin 2 (Mfn2), Opa1, Fis1, Drp1, VDAC, Tim23, Hsp60, Bip, and Parkin. CCCP, 20 µM for 2 h; MG132, 30 µM, 30 min prior and with CCCP. A single asterisk on the Mfn2 panel indicates a nonspecific band and the double asterisks indicates a cross-reactive band to Mfn1 (Fig. S1, a and b). Opa1 shows conversion from a long form (L) to a shorter form (S) upon depolarization. (b) The average of the Mfn1 or Mfn2 levels normalized to the level of Tim23 is presented with standard deviation indicated by the error bars. Each graph was generated from three independent experiments as described for panel a. The protein levels in control (nontreated) cells were set as 1. (c) Whole cell lysates (30 µg of proteins) of SH-SY5Y cells subjected to SDS-PAGE after treatments with DMSO, Rotenone, or Paraquat are shown. An asterisk on the Mfn2 panel indicates a nonspecific band and the double asterisks indicate a cross-reactive band to Mfn1. Rotenone, 100 nM for 24 h; Paraquat, 2 mM for 24 h. (d) Whole cell lysates (10 µg of proteins) of HeLa cells or HeLa cells stably transfected with YFP-Parkin were subjected to SDS-PAGE after treatment with DMSO, CCCP, or CCCP with MG132. An asterisk on the Mfn2 panel indicates a nonspecific band and the double asterisks indicate a cross-reactive band to Mfn1. CCCP, 10 µM for 90 min; MG132, 30 µM, 30 min prior and with CCCP. (e) The average of Mfn1 or Mfn2 levels normalized to the level of Tim23 is presented with standard deviation indicated by the error bars. Each graph was generated from three independent experiments from d. The protein levels in control (nontreated, HeLa cells stably transfected with YFP-Parkin) cells were set as 1. Molecular mass is indicated in kilodaltons next to the gel blots. (f) Whole cell lysates of HeLa cells stably transfected with YFP-Parkin (top, 10 µg of proteins) were fractionated into mitochondrial and the postmitochondrial fractions (bottom). The asterisk on the Mfn2 panel indicates a nonspecific band, and the double asterisks indicate a cross-reactive band to Mfn1. CCCP, 10 µM. (g) HeLa cells transiently expressing Mfn2-YFP and mCherry-Parkin were treated with DMSO or CCCP. After 30 min of CCCP or DMSO treatment (time 30’), live cell images were captured every 5 min by confocal microscopy. CCCP, 10 µM. Bar, 10 µm.
We compared the time course of YFP-Parkin translocation to mitochondria with that of mitofusin degradation after mitochondrial uncoupling. As shown in Fig. 1 f, 60 min after triggering mitochondrial depolarization with CCCP, the proportion of mitochondrial YFP-Parkin increased substantially, whereas mitofusin protein levels were drastically reduced. Both Mfn1 and Mfn2 are degraded in a Parkin-dependent manner, although Mfn1 degradation appears to be generally more robust than Mfn2 degradation. Interestingly, higher molecular weight bands of YFP-Parkin were also detected coincident with the time of mitochondrial translocation of Parkin. These high molecular weight Parkin bands were detected with YFP-Parkin and not with untagged Parkin, which is consistent with recent studies suggesting that the GFP portion of GFP-Parkin is a pseudo-substrate for Parkin (Matsuda et al., 2010). This also suggests that the E3 ligase activity of Parkin is activated upon mitochondrial translocation. Simultaneous imaging of mCherry-Parkin and Mfn2-YFP expressed in HeLa cells after CCCP treatment also revealed a decrease in Mfn2-YFP signal, whereas untreated cells exhibited stable Mfn2-YFP intensity (Fig. 1 g and Fig. S2 a), which further indicated that CCCP-induced mitochondrial translocation of Parkin initiated degradation of mitofusin protein.
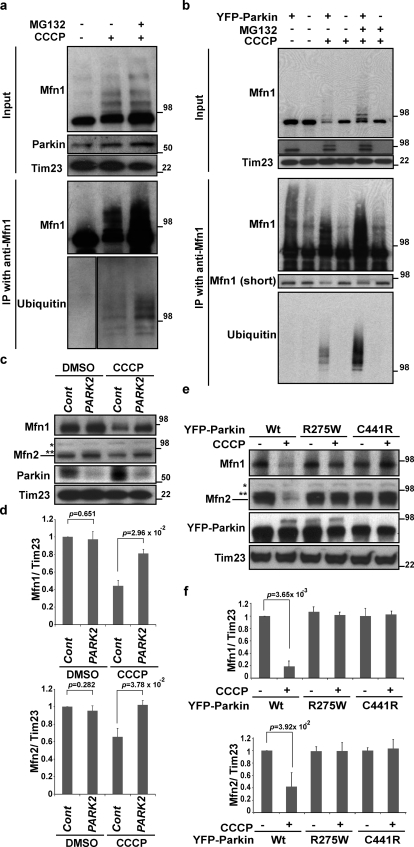
We also determined whether endogenous Mfn1 becomes ubiquitinated upon Parkin translocation to mitochondria. To this end, we applied denatured Mfn1 immunoprecipitation (IP) from control and CCCP-treated SH-SY5Y cells. The data show that CCCP treatment induces high molecular weight species of Mfn1 (Fig. 2 a) that accumulated to even greater extents upon MG132 and CCCP cotreatment. Importantly, these high molecular weight species of Mfn1 cross-reacted with anti-Ub antibodies (Fig. 2 a, bottom). We also examined Mfn1 ubiquitination in HeLa cells with and without stable Parkin expression. As with SH-SY5Y cells, ubiquitinated Mfn1 was detected specifically after CCCP treatment in HeLa: YFP-Parkin cells, whereas no Mfn1 ubiquitination was detected after CCCP treatment of control HeLa cells (Fig. 2 b).
Figure 2.
Mitofusin ubiquitination requires Parkin ligase activity. (a and b) SH-SY5Y (a), HeLa cells, or HeLa cells stably transfected with YFP-Parkin (b) were collected as cell lysates with denaturing buffer (see Materials and methods), then subjected to IP with anti-Mfn1 antibody. IP products were detected with anti-Mfn1 and anti-Ub (P4D1). CCCP, 10 µM for 2 h for SH-SY5Y cells, 90 min for HeLa cells; MG132, 30 µM, 30 min prior and with CCCP. (c) Whole cell lysates of SH-SY5Y cells transfected with control siRNA or PARK2 siRNA were treated with DMSO or CCCP. An asterisk on the Mfn2 panel indicates a nonspecific band and the double asterisks indicate a cross-reactive band to Mfn1. CCCP, 20 µM for 2 h. (d) The average of the Mfn1 or Mfn2 level normalized to the level of Tim23 is presented with standard deviation indicated by the error bars. Each graph was generated from three independent experiments from c. The protein levels in control (nontreated, control siRNA) cells were set as 1. (e) HeLa cells transiently transfected with wild-type or two patient mutations of YFP-Parkin were treated with or without CCCP. Whole cell lysates were subjected to immunoblotting with anti-Mfn1, Mfn2, Parkin, and Tim23. CCCP, 10 µM for 90 min. An asterisk on the Mfn2 panel indicates a nonspecific band and the double asterisks indicates a cross-reactive band to Mfn1. Molecular mass is indicated in kilodaltons next to the gel blots. (f) The average of the Mfn1 or Mfn2 levels normalized to the level of Tim23 is presented with standard deviation indicated by the error bars. Each graph was generated from three independent experiments from e. The protein levels in control (nontreated, with each YFP-Parkin expression) cells were set as 1.
Overall, these data indicate that Mfn1 is ubiquitinated and degraded in Parkin-dependent manner. To test this possibility further by knocking down PARK2 (Parkin) in SH-SY5Y cells, we examined whether Mfn degradation requires endogenous Parkin expression. We found that PARK2 knockdown inhibits Mfn degradation upon CCCP treatment (Fig. 2, c and d, PARK2 siRNA), which confirmed the essential role of Parkin in CCCP-induced Mfn degradation. We also determined whether pathogenic variants of Parkin affect the Mfn1/2 degradation process. To achieve this, we applied two Parkin mutants: ParkinR275W, which translocates to depolarized mitochondria but fails to induce mitophagy; and ParkinC441R, which fails to translocate to mitochondria (Narendra et al., 2010). In contrast to transient expression of wild-type YFP-Parkin, expression of neither YFP-ParkinR275W nor YFP-ParkinC441R induced Mfn1/2 elimination upon CCCP treatment (Fig. 2, e and f).
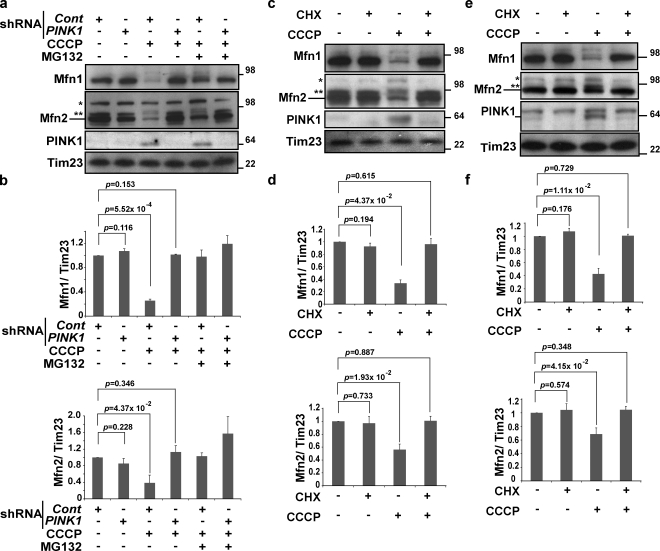
PINK1, a mitochondrial kinase, is essential for the Parkin translocation to depolarized mitochondria and for Parkin-mediated mitophagy (Geisler et al., 2010; Matsuda et al., 2010; Narendra et al., 2010; Vives-Bauza et al., 2010). Therefore, we determined using PINK1 RNAi whether PINK1 is also required for CCCP-induced Mfn proteasomal degradation in the context of endogenous Parkin expression. The data show that RNAi-mediated depletion of PINK1 expression in Parkin-expressing M17 human neuroblastoma cells inhibited degradation of Mfn1 and Mfn2 (Fig. 3, a and b).
Figure 3.
Mitofusin degradation requires PINK1 expression. (a) M17 cells expressing control or PINK1 shRNA were treated with CCCP or CCCP plus MG132. Lysates were immunoblotted with anti-Mfn1, Mfn2, PINK1, and Tim23. CCCP, 20 µM for 4 h; MG132, 30 µM, 30 min prior and with CCCP. (b) The average of Mfn1 or Mfn2 levels normalized to the level of Tim23 is presented with the standard deviation indicated by the error bars. Each graph was generated from three independent experiments from panel a. The protein levels in control (nontreated, control shRNA) cells were set as 1. (c) HeLa cells stably transfected with YFP-Parkin were pretreated with CHX before treatment with CCCP, as indicated on top. Lysates were immunoblotted with anti-Mfn1, Mfn2, PINK1, and Tim23. CCCP, 10 µM for 90 min; CHX, 100 µM, 30 min prior as a pretreatment and during incubation with or without CCCP. (d) The average of the Mfn1 or Mfn2 level normalized to the level of Tim23 is presented with the standard deviation indicated by the error bars. Each graph was generated from three independent experiments from c. The protein levels in control (nontreated) cells were set as 1. (e) SH-SY5Y cells were pretreated with CHX before the treatments of CCCP, as indicated on top. Lysates were immunoblotted with anti-Mfn1, Mfn2, PINK1, and Tim23. CCCP, 20 µM for 2 h; CHX, 100 µM, 30 min prior as a pretreatment and during incubation with or without CCCP. Molecular mass is indicated in kilodaltons next to the gel blots. (f) The average of the Mfn1 or Mfn2 level normalized to the level of Tim23 is presented with the standard deviation indicated by the error bars. Each graph was generated from three independent experiments from e. The protein levels in control (nontreated) cells were set as 1.
To further test the significance of Parkin activity for CCCP-induced Mfn degradation, we used an alternative strategy to inhibit endogenous Parkin activation. In contrast to inhibition of transcription, inhibition of translation by treatment with cycloheximide (CHX) prevents both the CCCP-dependent induction of PINK1 expression and Parkin translocation to CCCP-uncoupled mitochondria (Narendra et al., 2010). Thus, we tested whether CHX would block Mfn1/2 degradation in CCCP-treated HeLa: YFP-Parkin cells and SHSY5Y cells. The data show that cotreatment of cells with CHX and CCCP completely prevents Mfn degradation (Fig. 3, c–f), which further supports the scenario in which Mfn ubiquitination and degradation not only requires endogenous PINK1 but also PINK1 accumulation and PINK1-dependent mitochondrial accumulation of Parkin during CCCP treatment.
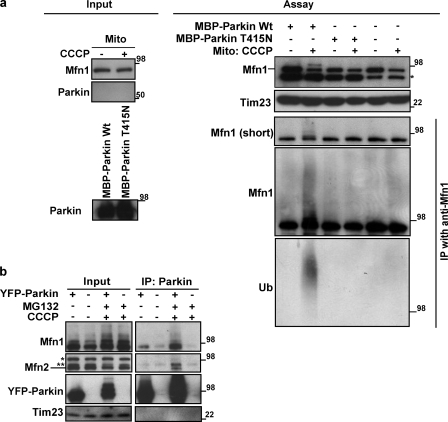
To establish more directly that the ubiquitination of Mfn1 is mediated by Parkin, we reconstituted the ubiquitination process using a cell free system. As Mfn1 has two hydrophobic domains spanning the outer mitochondrial membrane (OMM), we used isolated mitochondria as a source of Mfn1 substrate to combine with purified recombinant maltose-binding protein (MBP)-tagged Parkin (MBP-Parkin) and purified UbcH7, an E2 Ub-conjugating enzyme found to function with Parkin (Matsuda et al., 2006). Consistent with the in vivo data, we found that a high molecular weight ladder of Mfn1 appeared more prominent when the mitochondria were isolated from CCCP-treated HeLa cells, as compared with control HeLa cells (Fig. 4 a). Furthermore, high molecular species of Mfn1 were detectable in mitochondrial fractions from CCCP-treated HeLa cells incubated with wild-type MBP-Parkin, but not recombinant MBP-ParkinT415N (Fig. 4 a), which is a Parkinson’s disease patient mutation lacking E3 ligase activity (Matsuda et al., 2006). Using IP with the anti-Mfn1 antibody under denaturing conditions, we confirmed that these high molecular weight bands of Mfn1 were also detected with an anti-Ub antibody. Collectively, these results indicate that Parkin promotes ubiquitination of Mfn1 and that this ubiquitination can be recapitulated under in vitro conditions, which suggests a direct role of Parkin in ubiquitination of Mfn1. However, as mitochondria express at least one membrane-spanning E3 Ub ligase, MARCH5 (Karbowski et al., 2007), Parkin could be activating another E3 ligase to ubiquitinate mitofusins. To determine whether MARCH5 is required for Parkin-dependent Mfn ubiquitination, we used wild-type HCT116 or MARCH5−/− HCT116 cell lines that express abundant endogenous Parkin (Fig. S3 b). After CCCP treatment for 3 h, Mfn1 and Mfn2 levels decreased comparably in HCT116 or HCT116 MARCH5−/− cells (Fig. S3 b), which suggests that Parkin-induced Mfn degradation occurs in a MARCH5-independent manner.
Figure 4.
Mitofusin ubiquitination requires Parkin and depolarization of mitochondria. (a) Mitochondrial fractions (30 µg) from HeLa cells treated with or without CCCP and recombinant MBP-Parkin wild-type or MBP-Parkin T415N proteins (left) were subjected to the in vitro ubiquitination assay (right). Isolated mitochondria were incubated with MBP-Parkin wild type or T415N. Reaction mixtures were subjected to the denatured IP with anti-Mfn1 antibody as in Fig. 2. IP products were detected with anti-Mfn1 and anti-Ub (P4D1). UbcH7 was used as an E2 enzyme. The asterisk indicates a nonspecific band from MBP proteins or other reaction components. (b) HeLa cells or HeLa cells stably transfected with YFP-Parkin were subjected to nondenaturing IP with anti-Parkin antibody (PRK8). IP products were detected by anti-Parkin, Mfn1, and Mfn2. CCCP, 10 µM for 90 min; MG132, 30 µM, 30 min prior and with CCCP. An asterisk on the Mfn2 panel indicates a nonspecific band and the double asterisks indicate a cross-reactive band to Mfn1. Molecular mass is indicated in kilodaltons next to the gel blots.
We also asked whether Parkin might bind to Mfn1/2. The data show that in HeLa: YFP-Parkin cells, YFP-Parkin coimmunoprecipitated with endogenous Mfn1 and Mfn2, and that these interactions increased upon CCCP-induced depolarization of mitochondria (Fig. 4 b). In sum, molecular interactions of Parkin with Mfn1 and Mfn2 as well as cell free Parkin-dependent ubiquitination of Mfn proteins indicate that Parkin-mediated ubiquitination of Mfn1 and Mfn2 might be direct.
Role of p97 in Mfn proteasomal degradation
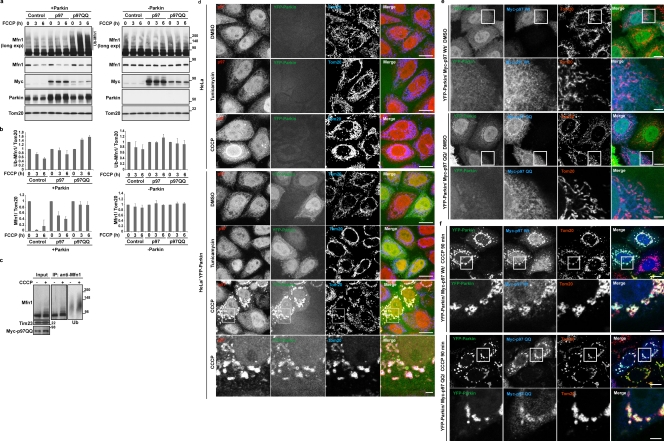
How membrane-spanning proteins such as the mitofusins are extracted from the OMM before their proteasomal degradation is currently unknown. We therefore examined if Mfn degradation requires the activity of p97, a AAA+ ATPase involved in the retrotranslocation of ER membrane-spanning proteins after ubiquitination and en route to proteasomal degradation (Ye et al., 2001; Rabinovich et al., 2002). To test this notion, we overexpressed wild-type p97 and the dominant-negative mutant p97 (E305Q/E578Q; p97QQ; Ye et al., 2003) in HeLa cells with or without transient Parkin expression. We found that Parkin-mediated Mfn1 ubiquitination was more apparent, and Mfn1 degradation was largely prevented by expression of the p97QQ (Fig. 5, a–c, p97QQ), which suggests that p97 mediates degradation of ubiquitinated Mfn1. Additionally, we found that overexpression of p97QQ slowed down the turnover rate of Mfn1 (Fig. S4, c and d) in control HeLa cells, which suggests that the UPS, through the p97 activity, might also mediate steady-state Mfn1 ubiquitination and degradation in the absence of mitochondrial uncoupling and Parkin expression.
Figure 5.
AAA+ ATPase p97 mediates mitofusin degradation and accumulates on mitochondria after depolarization. (a) HeLa cells or HeLa cells transiently expressing Parkin were transfected with wild-type Myc-p97 or dominant-negative Myc-p97QQ (E305Q/E578Q). After treatment with 10 µM FCCP for the indicated times, cells and mitochondrial fractions were collected as in Fig. 1 f. (b) The average of ubiquitinated (top) or nonubiquitinated (bottom) Mfn1 or Mfn2 normalized to the level of Tom20 is presented with the standard deviation indicated by the error bars. Each graph was generated from three independent experiments from panel a. The protein levels at time 0 were set as 1. (c) HeLa cells stably transfected with YFP-Parkin were transiently transfected with Myc-p97QQ and collected as cell lysates with denaturing buffer, then subjected to IP with anti-Mfn1 antibody. IP products were detected with anti-Mfn1 and anti-Ub (P4D1). CCCP, 10 µM for 90 min. Molecular mass is indicated in kilodaltons next to the gel blots. (d) HeLa cells or HeLa cells transiently transfected with YFP-Parkin were treated with DMSO, Tunicamycin, or CCCP. Cells were immunostained with anti-p97 and anti-Tom20. The boxed regions are magnified in the bottom panels. Tunicamycin, 5 µg/ml for 6 h; CCCP, 10 µM for 90 min. (e and f) HeLa cells transiently transfected with YFP-Parkin and wild-type Myc-p97 or Myc-p97QQ were treated with DMSO (e) or CCCP (f). Cells were fixed and immunostained with anti-Myc and anti-Tom20 antibodies. The boxed regions are magnified in the bottom panels. CCCP, 10 µM for 90 min. Bars: (top panels) 10 µm; (bottom enlarged panels) 1 µm.
To further test the role and mechanism of p97 in Parkin-dependent degradation of Mfn proteins, we examined the subcellular localization of p97 under conditions of inducing Parkin translocation to mitochondria. After 90 min of CCCP treatment, endogenous p97 accumulated on mitochondria in HeLa cells transiently expressing YFP-Parkin (Fig. 5 d) but not in control HeLa cells or cells treated with tunicamycin, a stressor of the ER. The accumulation p97 on mitochondria upon Parkin translocation was confirmed by the data showing that Myc-p97 also translocated to the mitochondria in CCCP-treated Parkin-expressing cells. Interestingly, both wild-type Myc-p97 and Myc-p97QQ accumulated on mitochondria upon CCCP treatment in HeLa cells transiently expressing YFP-Parkin, but not on mitochondria in control HeLa cells lacking Parkin expression (Fig. 5, e and f). These data indicate that p97 accumulation on mitochondria requires Parkin expression and mitochondrial depolarization, and occurs independently of p97 ATPase activity.
Next, we examined whether PINK1 is required for mitochondrial accumulation of p97. In PINK1 knockout mouse embryonic fibroblasts (PINK1−/−MEFs), mitochondrial accumulation of p97 was not detectable upon CCCP treatment (Fig. 6 a), which indicates that p97 translocation is triggered by the activation of PINK1–Parkin activity upon depolarization of mitochondria. This was further supported by data showing that p97 accumulated specifically on uncoupled, Parkin-positive, and Ub-positive mitochondria (Fig. 6 b).
Figure 6.
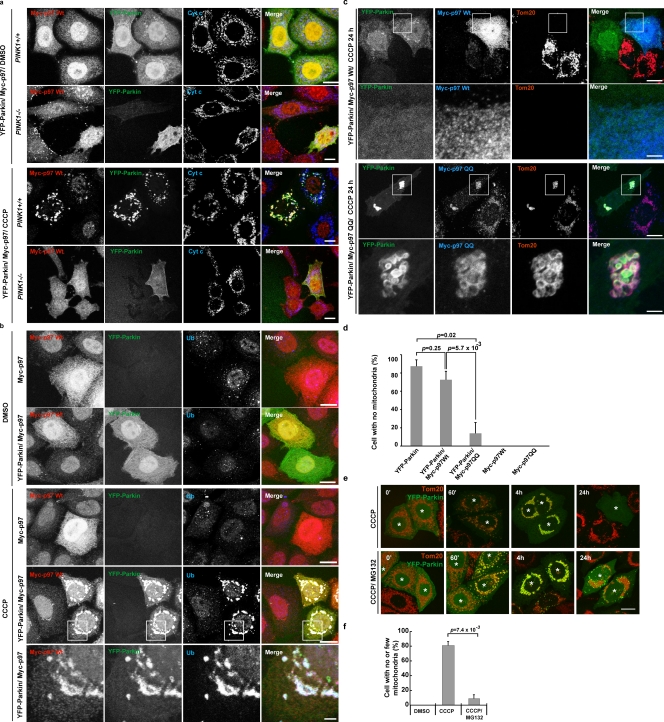
p97 recruitment to ubiquitinated mitochondria mediates PINK1–Parkin-induced mitochondrial elimination. (a) MEFs from PINK1+/+ or PINK1−/− mice were transiently transfected with YFP-Parkin and Myc-p97. Cells were treated with DMSO or CCCP, then immunostained with anti-Myc and anti–cytochrome c. CCCP, 20 µM for 2 h. Bar, 10 µm. (b) HeLa cells transiently expressing Myc-p97 or YFP-Parkin/Myc-p97 were treated with DMSO or CCCP. Cells were immunostained with anti-Myc and anti-Ub (fk1). CCCP, 10 µM for 90 min. The boxed regions are magnified in the bottom panels. (c) HeLa cells were transiently transfected with YFP-Parkin and wild-type Myc-p97 or Myc-p97QQ. After treatment with CCCP for 24 h, cells were fixed and immunostained with anti-Myc and anti-Tom20 antibodies. CCCP, 10 µM. The boxed regions are magnified in the bottom panels. Bars: (top panels) 10 µm; (bottom enlarged panels) 1 µm. (d) Parkin-induced mitophagy in cells shown as in c was quantified (n > 50). CCCP, 10 µM for 24 h. Data represent the mean ± SD with P values of at least three replicates. (e) HeLa cells or HeLa cells expressing YFP-Parkin were treated with CCCP or CCCP plus MG132. Mitochondria were immunostained with anti-Tom20. Cells expressing YFP-Parkin are indicated with asterisks. CCCP, 10 µM for 24 h; MG132, 30 µM 30 min prior and with CCCP. Bar, 10 µm. (f) MG132 blocks Parkin-induced mitophagy in HeLa cells as in d. A quantification is shown. Data represent the mean ± SD with P values of at least three replicates.
p97 is required for PINK1–Parkin-mediated mitophagy
We examined if p97 activity is required for PINK1–Parkin -mediated mitophagy after mitochondrial depolarization. After 24 h of treatment with CCCP, overexpression of p97QQ strongly inhibited mitochondrial elimination, which suggests that degradation of Mfn (or other ubiquitinated mitochondrial proteins) is required for Parkin-induced mitophagy (Fig. 6, c and d). We also investigated whether proteasome activity is required for Parkin-mediated mitophagy. To achieve this, HeLa cells transiently expressing YFP-Parkin were either treated with CCCP alone or cotreated with CCCP and MG132. Indeed, after 24 h of CCCP treatment, YFP-Parkin eliminated all detectable mitochondria from ∼80% of cells, whereas <10% of CCCP- and MG132-cotreated cells displayed complete mitochondrial loss (Fig. 6, e and f). Because YFP-Parkin showed a similar efficiency of translocation to mitochondria after CCCP with and without MG132 treatment (Fig. 6 e, 60’ and 4 h), the inhibition of mitochondrial elimination by MG132 suggests that Parkin-dependent degradation of dysfunctional mitochondria requires not only autophagy, but also the UPS.
Mitochondrial fusion inhibition and Parkin-mediated mitophagy
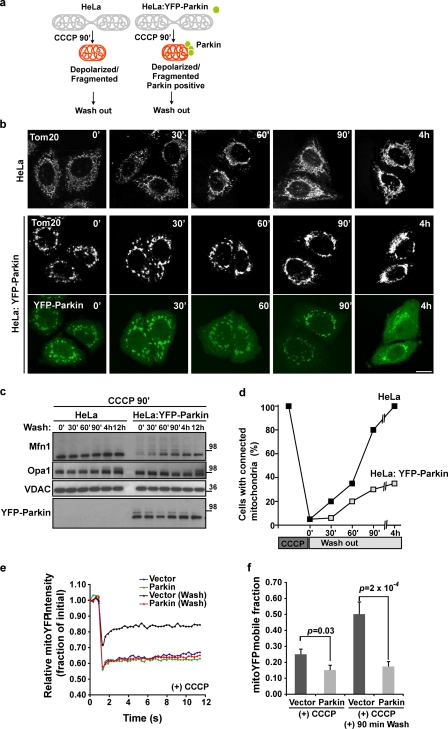
In Drosophila, either promoting fission of mitochondria or suppressing fusion partially rescues the swollen mitochondrial phenotype of the pink1 and parkin mutant flies (Deng et al., 2008; Poole et al., 2008; Yang et al., 2008; Park et al., 2009). This suggests that the PINK1–Parkin pathway may normally promote fission of mitochondria. In addition, recent work indicates that mitochondrial fission is required for the progression of mitophagy (Nowikovsky et al., 2007; Twig et al., 2008; Kanki et al., 2009), which suggests that promoting mitochondrial fission could compensate for a mitophagy deficit in Parkin-defective flies. Loss of mitofusins would be expected to inhibit mitochondrial fusion, perhaps fostering the autophagic elimination of depolarized mitochondria by segregating them from the mitochondrial network. We found that upon mitochondrial depolarization, Opa1 cleavage (Ishihara et al., 2006; Griparic et al., 2007) occurs more rapidly than mitofusin degradation (unpublished data), which suggests that the elimination of mitofusins does not cause the initial mitochondrial fragmentation but might delay fusion of mitochondria after uncoupler-induced fragmentation. To determine whether Parkin inhibits fusion of depolarized mitochondria independently of Opa1 cleavage, we examined the rate of recovery of tubular mitochondria after CCCP-induced depolarization and fragmentation of these organelles. Pretreatment with CCCP for 90 min generated depolarized, fragmented, Parkin-positive, and Mfn-negative mitochondria in HeLa: YFP-Parkin cells (Fig. 7, a–d, time 0). We then washed out the CCCP and analyzed the recovery of the tubular mitochondrial networks. After removing the CCCP, the recovery of mitochondrial networks in HeLa: YFP-Parkin cells was slower than in control HeLa cells (Fig. 7, b and d), despite the equal recovery of the longer form of Opa1 detectable both in the presence and absence of YFP-Parkin (Fig. 7 c). To further test the effect of Parkin on mitochondrial connectivity after CCCP washout phenotypes in Fig. 7 b, we applied a FRAP assay (Rizzuto et al., 1998; Karbowski et al., 2007). The fluorescence recovery of mitochondrial matrix-targeted YFP (mito-YFP) was analyzed in HeLa cells transiently expressing mCherry or mCherry-Parkin. The data show that at 90 min after CCCP washout, mCherry-expressing HeLa cells displayed a faster recovery of mito-YFP fluorescence compared with mCherry-Parkin–expressing cells (Fig. 7, e and f [P = 0.0002, n = 30]; and Fig. S5 a). These results suggest that Mfn1/2 degradation promoted by Parkin maintains mitochondria fragmentation after mitochondrial depolarization. Parkin may therefore retain mitochondria in a fragmented state in order to facilitate mitophagy.
Figure 7.
Parkin-promoted mitofusin degradation prevents refusion of damaged mitochondria with healthy mitochondria. (a) Scheme of washout design. HeLa cells or HeLa cells stably transfected with YFP-Parkin (green) were treated with CCCP for 90 min. (b and c) After washing CCCP out and further culture, cells were immunostained (b) and immunoblotted (c) with anti-Tom20, VDAC, Mfn1, Opa1, and Parkin. CCCP, 10 µM for 90 min. Bar, 10 µm. Molecular mass is indicated in kilodaltons next to the gel blots. (d) The recovery rate of the mitochondrial network as in b was plotted. At least 100 cells were counted for the cells with connected networks of mitochondria. (e) After CCCP treatment for 90 min with or without washing out CCCP, HeLa cells transiently expressing Mito-YFP with mCherry empty vector (vector) or mCherry-Parkin (Parkin) were subjected to FRAP assays. The curve represents an average of 30 individual FRAP curves. (f) The mobile fractions of Mito-YFP from e.
Inhibition of mitochondrial fission hinders CCCP- and Parkin-dependent mitophagy
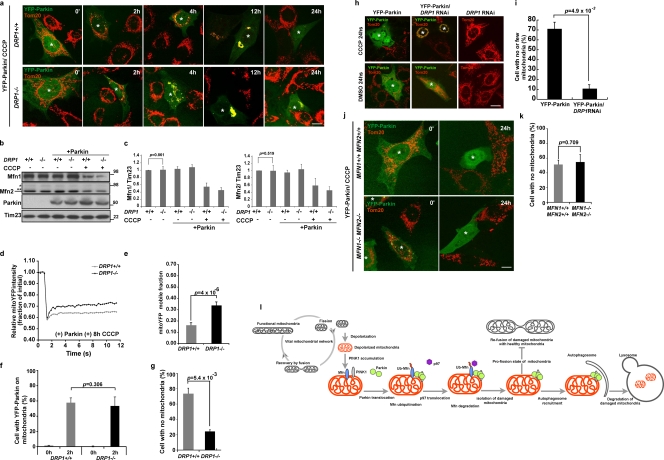
In DRP1−/−MEFs (Ishihara et al., 2009) that display excessively fused mitochondria, we observed a noticeable decrease in Parkin-induced mitophagy compared with identically treated wild-type MEFs (Fig. 8 a, 24 h CCCP; and Fig. 8 g), whereas YFP-Parkin recruitment (Fig. 8 f) and Mfn elimination (Fig. 8, b and c) were identical in these two cell types. To quantify mitochondrial interconnectivity in the DRP1−/−MEFs, we applied FRAP analysis of mito-YFP as described in Fig. 7 (e and f). As expected, mitochondria in DRP1−/− MEFs transiently expressing mCherry-Parkin were more interconnected after CCCP treatment than in similarly treated wild-type MEFs (Fig. 8, d and e). These results indicate that inhibiting mitochondrial fission hinders Parkin-induced mitophagy. We also extended the results in DRP1−/− MEFs to DRP1 knockdown HeLa cells. As in the DRP1−/− MEFs, DRP1 knockdown HeLa cells transiently expressing YFP-Parkin and treated with CCCP displayed clumped mitochondria (Fig. 8 h, CCCP 24 h) and an 80% decrease in mitophagy (Fig. 8 i) relative to control cells. Thus, in both DRP1−/− MEFs and DRP1 knockdown cells, mitophagy is inhibited. This indicates that counteracting mitochondrial fission, which is a normal function of Mfn1/2, inhibits Parkin-mediated mitophagy.
Figure 8.
Mitochondrial fission is required for Parkin-mediated mitophagy. (a) MEFs from DRP1+/+ or DRP1−/− mice were transiently transfected with YFP-Parkin. Cells were treated with CCCP and then immunostained with anti-Tom20. Cells expressing YFP-Parkin are indicated with asterisks. CCCP, 20 µM. Bar, 10 µm. (b) Whole cell lysates from DRP1+/+ or DRP1−/− MEFs with or without transient expression of Parkin were subjected to SDS-PAGE. Molecular mass is indicated in kilodaltons next to the gel blots. (c) The average of the Mfn1 or Mfn2 level normalized to the level of Tim23 is presented with the standard deviation indicated by the error bars. Each graph was generated from three independent experiments from b. The protein levels in control (nontreated, nontransfected) cells were set as 1. (d) Clumping mitochondria in DRP1+/+ or DRP1−/− MEFs transiently expressing mCherry-Parkin and Mito-YFP were subjected to the FRAP assay. The curve represents an average of 30 individual FRAP curves. CCCP, 20 µM for 8 h. (e) The mobile fractions of Mito-YFP in d. (f and g) YFP-Parkin translocation (f) and YFP-Parkin–induced mitophagy (g) in DRP1+/+ or DRP1−/− MEFs treated with CCCP as in panel a were quantified (n > 50). CCCP, 20 µM for 24 h. Data represent the mean ± SD with P values of at least three replicates. (h) HeLa cells treated with DRP1 shRNA or control HeLa cells were transiently transfected with YFP-Parkin and treated with or without CCCP. Mitochondria were immunostained with anti-Tom20. Cells expressing YFP-Parkin are indicated with asterisks. CCCP, 10 µM for 24 h. Bar, 10 µm. (i) YFP-Parkin–induced mitophagy in HeLa cells with or without DRP1 RNAi as in h was quantified (n > 50). CCCP, 10 µM for 24 h. Data represent the mean ± SD with P values of at least three replicates. (j and k) YFP-Parkin–induced mitophagy in MFN1+/+2+/+ MEFs or MFN1−/−2−/− MEFs was observed (j) and quantified (k) (n > 50). CCCP, 20 µM for 24 h. Data represent the mean ± SD with P values of at least three replicates. Cells expressing YFP-Parkin are indicated with asterisks. (l) A model for Parkin-mediated Mfn degradation and mitophagy. Depolarized mitochondria (orange) that accumulated PINK1 (gray) are sensed by Parkin (green). After Parkin recruitment to the damaged mitochondria, mitochondrial Parkin induces mitofusin (blue) ubiquitination and degradation. Degradation of mitofusin is regulated by p97 (purple) and is important to prevent impaired mitochondria from fusing with the vital mitochondrial network. Damaged mitochondria isolated by Parkin are engaged with autophagosomes and eliminated by auto-lysosomes.
Collectively, we hypothesize that mitofusin degradation by mitochondria-associated Parkin inhibits the fusion of damaged mitochondria with healthy mitochondria, thereby segregating the impaired mitochondria from the network of fully functional mitochondria to facilitate their selective elimination by autophagy.
Discussion
Parkin, an E3 Ub ligase, and PINK1, a mitochondrial kinase, are mutated in certain familial forms of Parkinson’s disease (Kitada et al., 1998; Valente et al., 2004). Recent studies in Drosophila by several groups put these two gene products in the same molecular pathway (Clark et al., 2006; Park et al., 2006; Yang et al., 2006) and link Parkin and PINK1 to mitochondrial fission pathways (Deng et al., 2008; Poole et al., 2008; Yang et al., 2008; Park et al., 2009). Loss of mitofusin function or mitofusin knockdown partially compensates for PINK1 or Parkin deficiencies, which suggests that PINK1 and Parkin may normally function in a pathway linked to mitochondrial fission. Here we show that Parkin can mediate the ubiquitination and proteasomal degradation of Mfn1 and Mfn2. Mfn1 appears to be more rapidly degraded than Mfn2 via Parkin, and this may relate to differential activity of the two proteins in mitochondrial fusion (Cipolat et al., 2004). These results suggest how Parkin might intersect the mitochondrial fission and fusion pathways consistent with a normal role in promoting mitochondrial fission. Our finding that the uncoupling of mitochondria, a condition that causes Parkin translocation to mitochondria, stimulates mitofusin degradation also points to a vital role for mitofusin degradation in Parkin-mediated mitophagy. Parkin can initiate mitophagy in MFN1/MFN2 double knockout cells, which indicates that mitofusin ubiquitination and proteasomal degradation are not essential to activate mitophagy (Fig. 8, j and k). This is consistent with genetic studies in flies that loss of mitofusin (Marf, a fly Mfn orthologue) compensates for loss of Parkin. Therefore we propose that mitofusin degradation does not serve as a signal for mitophagy activation, but Mfn1 and Mfn2 removal might be important for the promotion of mitochondrial fragmentation that might facilitate subsequent mitophagy.
The finding that mitochondrial elongation by DRP1 knockout or knockdown inhibits Parkin-mediated mitophagy is consistent with the model proposed here. We suggest that the inhibition of mitochondrial fusion may isolate damaged mitochondria from healthy mitochondria and thereby promote autophagy of defective organelles. The data shown here indicate that during Parkin-driven mitophagy, mitochondria lose fusion activity through two pathways. First, immediately after depolarization, Opa1 is processed by proteases into a fusion-inactive form; second, after Parkin translocation to depolarized mitochondria, Mfn1 and Mfn2 are degraded in a Parkin-, p97-, and proteasome-dependent manner to prevent mitochondrial refusion. These two pathways may strengthen the isolation of dysfunctional mitochondria from healthy mitochondria within a cell by eliminating fusion of the inner mitochondrial membrane and OMM, respectively. Under low-stress conditions, mitochondrial fusion may aid the rescue of mitochondrial damage, such as the rescue of respiratory chain complex activities or membrane potential, by shuffling materials from healthy to damaged mitochondria (Chen et al., 2005). However, once mitochondria are seriously damaged, they are removed from a cell by autophagy. In this pathway, mitochondrial fission accelerates the isolation of damaged mitochondria, and recent work has shown that fission precedes mitophagy (Nowikovsky et al., 2007; Twig et al., 2008; Kanki et al., 2009). We propose that inhibition of mitochondrial fusion during Parkin-driven mitophagy contributes to an enhanced rate and selectivity of mitophagy, facilitating mitochondrial quality control.
Several recent studies suggest that in mammalian cells, reducing PINK1 activity causes mitochondrial fragmentation (Exner et al., 2007; Dagda et al., 2009; Lutz et al., 2009; Sandebring et al., 2009). As PINK1 functions upstream of Parkin, the model proposed here of Parkin-mediated Mfn degradation would argue the converse: that activation of the PINK1–Parkin pathway would promote fragmentation of mitochondria as suggested by results in fly models. To reconcile the differences described in fly and mammalian systems, we suggest that loss of PINK1 or Parkin function might exacerbate the accumulation of mitochondrial damage due to defective removal of damaged mitochondria, and that this excessive damage results in mitochondrial fragmentation. Obviously, further studies are required to resolve this issue.
Membrane-spanning proteins resident in the ER can be degraded by the proteasome after ubiquitination and retrotranslocation from the ER membrane. The results shown here indicate that Mitofusin degradation by the proteasome is an example of a related process occurring at the OMM. We have shown previously that upon cell cycle arrest, Fzo1 (the yeast Mfn orthologue) is ubiquitinated on the OMM (Neutzner et al., 2007) and degraded by the proteasome (Neutzner and Youle, 2005; Escobar-Henriques et al., 2006; Cohen et al., 2008; Amiott et al., 2009). In analogy to ER-associated degradation (ERAD), we have proposed the term OMMAD for this OMM-associated degradation pathway (Neutzner et al., 2007). In yeast, proteins that may participate in Cullin-mediated ubiquitination—the F-Box protein MDM30 and the Skp1 proteins—have been found to participate in the steady-state proteasomal degradation of Fzo1 during vegetative growth (Dürr et al., 2006; Escobar-Henriques et al., 2006; Cohen et al., 2008), but not during Fzo1 proteasomal degradation upon cell cycle arrest because of α factor (Neutzner and Youle, 2005). Here we find that the E3 Ub ligase Parkin promotes OMMAD of Mfn1 and Mfn2 in mammalian cells in response to mitochondrial uncoupling. Parkin is a RING between RING-type E3 ligase, which has been proposed to participate in Cullin complex–mediated substrate ubiquitination (Staropoli et al., 2003). This suggests that Cullin and mammalian F-box proteins may also be involved in Parkin-mediated mitofusin degradation. Corroborating the idea of an ERAD-like process occurring at the mitochondria, we identified a requirement for p97 activity for proteasomal elimination of the OMM-spanning Mfn1 and Mfn2. In addition, as indicated by the effect of p97QQ on Mfn1 ubiquitination in the absence of Parkin expression, degradation of mitofusins in mammalian cells might be mediated by other E3 Ub ligases during steady-state proteasome-mediated turnover. Furthermore, mitochondrial substrates for Parkin-mediated degradation other than Mfn1 and Mfn2 likely remain to be identified.
We also show that inhibition of p97 activity by the overexpression of dominant-negative p97 mutant prevents the PINK1–Parkin-mediated mitochondrial elimination. Although this role of p97 in mitophagy correlates with its role in Mfn elimination, which suggests that Mfn elimination is required for Parkin-mediated mitophagy, p97 may mediate OMMAD of other mitochondrial substrates or play a role in the maturation of autophagosomes as proposed by Tresse et al. (2010) downstream of Mfn elimination. Current studies in our laboratories address these interesting possibilities.
Materials and methods
Cell culture
MEFs, HeLa cells, or HeLa cells stably expressing YFP-Parkin (FlpIn system; Invitrogen) called HeLa: YFP-Parkin were cultured in complete Dulbecco’s minimum essential medium supplemented with 10% heat-inactivated fetal calf serum (Gemini Bio-Products), 100 U/ml penicillin, and 100 µg/ml streptomycin (Invitrogen) in 5% CO2 at 37°C. Stable cell lines were maintained with 250 µg/ml of Zeocin (Invitrogen) or 200 µg/ml of Hygromycin (Sigma-Aldrich). M17 neuroblastoma carrying small hairpin RNA (shRNA) for control or PINK1 (Narendra et al., 2010) were maintained with Opti-MEM, 10% heat-inactivated fetal calf serum (Gemini Bio-Products), and 10 µg/ml Blastidin (Invitrogen). SH-SY5Y neuroblastoma was maintained with Ham’s F12/Dulbecco’s minimum essential medium supplemented with 10% heat-inactivated fetal calf serum (Gemini Bio-Products), 100 U/ml penicillin, and 100 µg/ml streptomycin (Invitrogen). HCT116 cells were maintained in complete McCoy’s 5A medium supplemented with 10% heat-inactivated fetal calf serum (Gemini Bio-Products), 100 U/ml penicillin, and 100 µg/ml streptomycin (Invitrogen) in 5% CO2 at 37°C. HCT116 MARCH5 knockout cells were generously supplied by C. Wang (National Institutes of Health, Bethesda, MD). DRP1 knockout MEFs were supplied by K. Mihara (Kyushu University, Fukuoka, Japan) and N. Ishihara (Kurume University, Fukuoka, Japan). MFN1/2 knockout MEFs were provided by D. Chan (California Institute of Technology, Pasadena, CA).
Antibodies
Anti-Mfn1 and Mfn2 antibodies were generated as described previously (Karbowski et al., 2007). Commercial antibodies were purchased as follows: Opa1 (BD), Fis1 (Enzo Life Sciences, Inc.), Drp1 (BD), VDAC (EMD), Tim23 (BD), Hsp60 (Stressgen), Bip (BD), Tom 20 (BD), Tom40 (Santa Cruz Biotechnology, Inc.), cytochrome c (BD), Ub (P4D1 [Santa Cruz Biotechnology, Inc.] or Fk1 [Enzo Life Sciences, Inc.]), PINK1 (BD), Parkin (PRK8; Santa Cruz Biotechnology, Inc.), and p97 (Thermo Fisher Scientific). Secondary antibodies for the immunostaining were rabbit or mouse Alexa Fluor 488, Alexa Fluor 594, or Alexa Fluor 647 (Invitrogen). Secondary antibodies for immunoblotting were HRP-conjugated anti–rabbit or anti–mouse IgG F (a, b)2 antibodies (GE Healthcare). Western blotting signals were detected by ECL-plus regents (GE Healthcare).
Chemicals
CCCP (Sigma-Aldrich), carbonylcyanide p-trifluoromethoxyphenylhydrazone (FCCP; Sigma-Aldrich), paraquat (Sigma-Aldrich), rotenone (Sigma-Aldrich), CHX (Sigma-Aldrich), Tunicamycin (Sigma-Aldrich), and MG132 (EMD) were prepared from DMSO or water stocks.
DNA and transfection
MBP-Parkin wild type and T415N were a gift from K. Tanaka and N. Matsuda (Tokyo Metropolitan Medical Institute, Tokyo, Japan). Myc-p97 and Myc-p97QQ (E305Q/E578Q) were a gift from S. Fang (University of Maryland, Baltimore, MD). The plasmids carrying shRNA sequences for DRP1, Mito-YFP, mCherry-Parkin, YFP-Parkin (wild type, R275W, C441R), and Mfn2-YFP have been described previously (Karbowski et al., 2007; Narendra et al., 2008, 2010). Transfections were performed with Effectene (QIAGEN) or Fugene 6 HD (Roche). The experiments were performed 16 h after transfections. To select DRP1 knockdown cells, cells were treated with 300 µg/ml of Hygromycin for 3 d, then cultured with 50 µg/ml for 3 d. Selected cells were split again 1 d before assays.
Cell fractionation and IP
Cells were collected in a buffer (220 mM mannitol, 70 mM sucrose, 20 mM Hepes-KOH, pH 7.5, 1 mM EDTA, 0.5 mM PMSF, 2 mg/ml BSA, and protease inhibitor cocktail [Roche]), then homogenized by 20 passages through a 25-G syringe (Tyco) on ice. Homogenates were separated to the postnuclear supernatant (PNS) by 1,000 g for 10 min at 4°C. The PNS was further separated into the mitochondria, and the postmitochondrial fractions were separated by the step-wise centrifugations (3,000 g for 10 min and 16,000 g for 15 min). Both pellets were combined as the mitochondrial fraction. Pellets from each fraction were washed several times with homogenization buffer (250 mM sucrose, 20 mM Hepes-KOH, pH 7.5, and protease inhibitor cocktail [Roche]), to remove the excess of YFP-Parkin proteins. An IP assay with denaturing condition was performed as follows: cell lysates were denatured in TSD buffer (50 mM Tris-HCL, pH 7.5, 1.0% SDS, and 5 mM DTT), then diluted with TNN buffer (20 mM Tris-HCl, pH7.5, 200 mM NaCl, 0.5% NP-40, and protease inhibitor cocktail). The mixtures were subjected to the IP assay with anti-Mfn1 antibody and protein A/G–Sepharose (Santa Cruz Biotechnology, Inc.). Nondenatured IP was performed with IP buffer (50 mM Tris-HCl, pH 7.5, 200 mM NaCl, 2.0% CHAPS, and protease inhibitor cocktail) and protein A/G–Sepharose.
Immunofluorescence and microscopy
Cells were fixed with 4% PFA/PBS solution for 15 min at 25°C, then permeabilized by 0.5% Triton X-100/PBS solution for 5 min at 25°C. After blocking of samples with 1% BSA/PBS solution for 30 min with gentle shaking, the primary and secondary antibody solutions in 1% BSA/PBS were treated sequentially. Samples were imaged using an inverted microscope (LSM510 Meta; Carl Zeiss, Inc.) with a 63×/1.4 NA oil DIC Plan-Apochromat objective lens at 25°C.
Ubiquitination assays
The recombinant Parkin proteins were synthesized and purified as described previously (Fallon et al., 2006). E1, E2, Ub, and the ATP-regenerating system were purchased from Enzo Life Sciences, Inc. The in vitro ubiquitination assay was performed as described previously (Matsuda et al., 2006), with minor modifications. In brief, isolated mitochondria were generated as described in the cellular fractionation method, and 30 µg of mitochondria fractions were incubated with ubiquitination mixtures at 32°C for 2 h. The reaction was stopped by boiling at 95°C for 10 min with SDS-PAGE sampling buffer (Invitrogen), or subjected to the additional IP assay with denaturing conditions
Measurement of mitochondrial connectivity with FRAP assay
To measure mitochondrial connectivity, FRAP assays were performed as described previously (Karbowski et al., 2006, 2007) with the following modifications. Using cells expressing mito-YFP, a 36 mm × 18 mm rectangle was imaged with a 100× objective lens (zoom 2.5) using a confocal microscope (510; Carl Zeiss, Inc.) before and after a four-iteration photobleaching of the 2.1-µm circle. The circle was randomly placed within the cell, where it encompassed several mitochondria. Using the LSM 510 software, three region of interest (ROIs) were measured: (1) bleach region, (2) nonspecific photo-bleach region, and (3) background fluorescence. Using Excel (Microsoft), the FRAP curves were normalized to the intensity of the ROI from the first image in the series, background subtracted, and corrected for nonspecific bleach according to (Phair and Misteli, 2000). 30 individual FRAP curves were recorded for each sample.
Online supplemental material
Fig. S1 presents the specificities of anti-mitofusin (Mfn1 and Mfn2) antibodies. Fig. S2 shows changes in the level of YFP-Mfn1 upon depolarization of mitochondria. Fig. S3 compares mitofusin degradation in various cell lines upon depolarization of mitochondria. Fig. S4 shows the stability of Mfn1 with p97 expression and MG132 or CHX treatment. Fig. S5 shows examples of the images used for the FRAP assay quantification in Figs. 7 e and 8 d. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.2010071013/DC1.
Acknowledgments
We thank Dr. K. Mihara and Dr. N. Ishihara for kindly sharing DRP1 KO MEFs, Dr. Chunxin Wang for the MARCH5 KO HCT116 cells, Dr. D. Chan for the MFN1/2 KO MEFs, Drs. K. Tanaka and N. Matsuda (Tokyo Metropolitan Medical Institute) for the MBP-Parkin, and Dr. S. Fang for the p97 constructs. We also thank Dr. I. Scott, Dr. A. Neutzner, and the National Institutes of Health fellows Editorial Board for their critical reading of our manuscript, and Dr. C. Smith for help with confocal microscopy.
This work is supported by the National Institute of Neurological Disorders and Stroke intramural program (to R.J. Youle), the National Institute of General Medical Sciences (to M. Karbowski; R01: GM083131), and the Japan Society for the Promotion of Science fellowship (to A. Tanaka).
Footnotes
Abbreviations used in this paper:
- CCCP
- carbonyl cyanide m-chlorophenylhydrazone
- CHX
- cycloheximide
- IP
- immunoprecipitation
- MBP
- maltose-binding protein
- MEF
- mouse embryonic fibroblast
- OMM
- outer mitochondrial membrane
- PNS
- postnuclear supernatant
- ROI
- region of interest
- shRNA
- small hairpin RNA
- Ub
- ubiquitin
- UPS
- Ub–proteasome system
References
- Amiott E.A., Cohen M.M., Saint-Georges Y., Weissman A.M., Shaw J.M. 2009. A mutation associated with CMT2A neuropathy causes defects in Fzo1 GTP hydrolysis, ubiquitylation, and protein turnover. Mol. Biol. Cell. 20:5026–5035 10.1091/mbc.E09-07-0622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betarbet R., Sherer T.B., MacKenzie G., Garcia-Osuna M., Panov A.V., Greenamyre J.T. 2000. Chronic systemic pesticide exposure reproduces features of Parkinson’s disease. Nat. Neurosci. 3:1301–1306 10.1038/81834 [DOI] [PubMed] [Google Scholar]
- Brooks A.I., Chadwick C.A., Gelbard H.A., Cory-Slechta D.A., Federoff H.J. 1999. Paraquat elicited neurobehavioral syndrome caused by dopaminergic neuron loss. Brain Res. 823:1–10 10.1016/S0006-8993(98)01192-5 [DOI] [PubMed] [Google Scholar]
- Büeler H. 2009. Impaired mitochondrial dynamics and function in the pathogenesis of Parkinson’s disease. Exp. Neurol. 218:235–246 10.1016/j.expneurol.2009.03.006 [DOI] [PubMed] [Google Scholar]
- Chen H., Chomyn A., Chan D.C. 2005. Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J. Biol. Chem. 280:26185–26192 10.1074/jbc.M503062200 [DOI] [PubMed] [Google Scholar]
- Cipolat S., Martins de Brito O., Dal Zilio B., Scorrano L. 2004. OPA1 requires mitofusin 1 to promote mitochondrial fusion. Proc. Natl. Acad. Sci. USA. 101:15927–15932 10.1073/pnas.0407043101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark I.E., Dodson M.W., Jiang C., Cao J.H., Huh J.R., Seol J.H., Yoo S.J., Hay B.A., Guo M. 2006. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 441:1162–1166 10.1038/nature04779 [DOI] [PubMed] [Google Scholar]
- Cochemé H.M., Murphy M.P. 2008. Complex I is the major site of mitochondrial superoxide production by paraquat. J. Biol. Chem. 283:1786–1798 10.1074/jbc.M708597200 [DOI] [PubMed] [Google Scholar]
- Cohen M.M., Leboucher G.P., Livnat-Levanon N., Glickman M.H., Weissman A.M. 2008. Ubiquitin-proteasome-dependent degradation of a mitofusin, a critical regulator of mitochondrial fusion. Mol. Biol. Cell. 19:2457–2464 10.1091/mbc.E08-02-0227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagda R.K., Cherra S.J., III, Kulich S.M., Tandon A., Park D., Chu C.T. 2009. Loss of PINK1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. J. Biol. Chem. 284:13843–13855 10.1074/jbc.M808515200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H., Dodson M.W., Huang H., Guo M. 2008. The Parkinson’s disease genes pink1 and parkin promote mitochondrial fission and/or inhibit fusion in Drosophila. Proc. Natl. Acad. Sci. USA. 105:14503–14508 10.1073/pnas.0803998105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison S.R., Wang F., Becker N.A., Schüle B., Kock N., Phillips L.A., Klein C., Smith D.I. 2003. Alterations in the common fragile site gene Parkin in ovarian and other cancers. Oncogene. 22:8370–8378 10.1038/sj.onc.1207072 [DOI] [PubMed] [Google Scholar]
- Dürr M., Escobar-Henriques M., Merz S., Geimer S., Langer T., Westermann B. 2006. Nonredundant roles of mitochondria-associated F-box proteins Mfb1 and Mdm30 in maintenance of mitochondrial morphology in yeast. Mol. Biol. Cell. 17:3745–3755 10.1091/mbc.E06-01-0053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar-Henriques M., Westermann B., Langer T. 2006. Regulation of mitochondrial fusion by the F-box protein Mdm30 involves proteasome-independent turnover of Fzo1. J. Cell Biol. 173:645–650 10.1083/jcb.200512079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exner N., Treske B., Paquet D., Holmström K., Schiesling C., Gispert S., Carballo-Carbajal I., Berg D., Hoepken H.H., Gasser T., et al. 2007. Loss-of-function of human PINK1 results in mitochondrial pathology and can be rescued by parkin. J. Neurosci. 27:12413–12418 10.1523/JNEUROSCI.0719-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallon L., Bélanger C.M., Corera A.T., Kontogiannea M., Regan-Klapisz E., Moreau F., Voortman J., Haber M., Rouleau G., Thorarinsdottir T., et al. 2006. A regulated interaction with the UIM protein Eps15 implicates parkin in EGF receptor trafficking and PI(3)K-Akt signalling. Nat. Cell Biol. 8:834–842 10.1038/ncb1441 [DOI] [PubMed] [Google Scholar]
- Geisler S., Holmström K.M., Skujat D., Fiesel F.C., Rothfuss O.C., Kahle P.J., Springer W. 2010. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 12:119–131 10.1038/ncb2012 [DOI] [PubMed] [Google Scholar]
- Greene J.C., Whitworth A.J., Kuo I., Andrews L.A., Feany M.B., Pallanck L.J. 2003. Mitochondrial pathology and apoptotic muscle degeneration in Drosophila parkin mutants. Proc. Natl. Acad. Sci. USA. 100:4078–4083 10.1073/pnas.0737556100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griparic L., Kanazawa T., van der Bliek A.M. 2007. Regulation of the mitochondrial dynamin-like protein Opa1 by proteolytic cleavage. J. Cell Biol. 178:757–764 10.1083/jcb.200704112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara N., Fujita Y., Oka T., Mihara K. 2006. Regulation of mitochondrial morphology through proteolytic cleavage of OPA1. EMBO J. 25:2966–2977 10.1038/sj.emboj.7601184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara N., Nomura M., Jofuku A., Kato H., Suzuki S.O., Masuda K., Otera H., Nakanishi Y., Nonaka I., Goto Y., et al. 2009. Mitochondrial fission factor Drp1 is essential for embryonic development and synapse formation in mice. Nat. Cell Biol. 11:958–966 10.1038/ncb1907 [DOI] [PubMed] [Google Scholar]
- Kanki T., Wang K., Baba M., Bartholomew C.R., Lynch-Day M.A., Du Z., Geng J., Mao K., Yang Z., Yen W.L., Klionsky D.J. 2009. A genomic screen for yeast mutants defective in selective mitochondria autophagy. Mol. Biol. Cell. 20:4730–4738 10.1091/mbc.E09-03-0225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbowski M., Norris K.L., Cleland M.M., Jeong S.Y., Youle R.J. 2006. Role of Bax and Bak in mitochondrial morphogenesis. Nature. 443:658–662 10.1038/nature05111 [DOI] [PubMed] [Google Scholar]
- Karbowski M., Neutzner A., Youle R.J. 2007. The mitochondrial E3 ubiquitin ligase MARCH5 is required for Drp1 dependent mitochondrial division. J. Cell Biol. 178:71–84 10.1083/jcb.200611064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I., Rodriguez-Enriquez S., Lemasters J.J. 2007. Selective degradation of mitochondria by mitophagy. Arch. Biochem. Biophys. 462:245–253 10.1016/j.abb.2007.03.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitada T., Asakawa S., Hattori N., Matsumine H., Yamamura Y., Minoshima S., Yokochi M., Mizuno Y., Shimizu N. 1998. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 392:605–608 10.1038/33416 [DOI] [PubMed] [Google Scholar]
- Krick R., Bremer S., Welter E., Schlotterhose P., Muehe Y., Eskelinen E.L., Thumm M. 2010. Cdc48/p97 and Shp1/p47 regulate autophagosome biogenesis in concert with ubiquitin-like Atg8. J. Cell Biol. 190:965–973 10.1083/jcb.201002075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.Y., Nagano Y., Taylor J.P., Lim K.L., Yao T.P. 2010. Disease-causing mutations in parkin impair mitochondrial ubiquitination, aggregation, and HDAC6-dependent mitophagy. J. Cell Biol. 189:671–679 10.1083/jcb.201001039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Guo M. 2009. Protein degradation in Parkinson disease revisited: it’s complex. J. Clin. Invest. 119:442–445 10.1172/JCI38619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz A.K., Exner N., Fett M.E., Schlehe J.S., Kloos K., Lämmermann K., Brunner B., Kurz-Drexler A., Vogel F., Reichert A.S., et al. 2009. Loss of parkin or PINK1 function increases Drp1-dependent mitochondrial fragmentation. J. Biol. Chem. 284:22938–22951 10.1074/jbc.M109.035774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda N., Tanaka K. 2010. Does impairment of the ubiquitin-proteasome system or the autophagy-lysosome pathway predispose individuals to neurodegenerative disorders such as Parkinson’s disease? J. Alzheimers Dis. 19:1–9 [DOI] [PubMed] [Google Scholar]
- Matsuda N., Kitami T., Suzuki T., Mizuno Y., Hattori N., Tanaka K. 2006. Diverse effects of pathogenic mutations of Parkin that catalyze multiple monoubiquitylation in vitro. J. Biol. Chem. 281:3204–3209 10.1074/jbc.M510393200 [DOI] [PubMed] [Google Scholar]
- Matsuda N., Sato S., Shiba K., Okatsu K., Saisho K., Gautier C.A., Sou Y.S., Saiki S., Kawajiri S., Sato F., et al. 2010. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J. Cell Biol. 189:211–221 10.1083/jcb.200910140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D., Tanaka A., Suen D.F., Youle R.J. 2008. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 183:795–803 10.1083/jcb.200809125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D.P., Jin S.M., Tanaka A., Suen D.F., Gautier C.A., Shen J., Cookson M.R., Youle R.J. 2010. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 8:e1000298 10.1371/journal.pbio.1000298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neutzner A., Youle R.J. 2005. Instability of the mitofusin Fzo1 regulates mitochondrial morphology during the mating response of the yeast Saccharomyces cerevisiae. J. Biol. Chem. 280:18598–18603 10.1074/jbc.M500807200 [DOI] [PubMed] [Google Scholar]
- Neutzner A., Youle R.J., Karbowski M. 2007. Outer mitochondrial membrane protein degradation by the proteasome. Novartis Found. Symp. 287:4–14, discussion :14–20 10.1002/9780470725207.ch2 [DOI] [PubMed] [Google Scholar]
- Nowikovsky K., Reipert S., Devenish R.J., Schweyen R.J. 2007. Mdm38 protein depletion causes loss of mitochondrial K+/H+ exchange activity, osmotic swelling and mitophagy. Cell Death Differ. 14:1647–1656 10.1038/sj.cdd.4402167 [DOI] [PubMed] [Google Scholar]
- Park J., Lee S.B., Lee S., Kim Y., Song S., Kim S., Bae E., Kim J., Shong M., Kim J.M., Chung J. 2006. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 441:1157–1161 10.1038/nature04788 [DOI] [PubMed] [Google Scholar]
- Park J., Lee G., Chung J. 2009. The PINK1-Parkin pathway is involved in the regulation of mitochondrial remodeling process. Biochem. Biophys. Res. Commun. 378:518–523 10.1016/j.bbrc.2008.11.086 [DOI] [PubMed] [Google Scholar]
- Pawlyk A.C., Giasson B.I., Sampathu D.M., Perez F.A., Lim K.L., Dawson V.L., Dawson T.M., Palmiter R.D., Trojanowski J.Q., Lee V.M. 2003. Novel monoclonal antibodies demonstrate biochemical variation of brain parkin with age. J. Biol. Chem. 278:48120–48128 10.1074/jbc.M306889200 [DOI] [PubMed] [Google Scholar]
- Phair R.D., Misteli T. 2000. High mobility of proteins in the mammalian cell nucleus. Nature. 404:604–609 10.1038/35007077 [DOI] [PubMed] [Google Scholar]
- Poole A.C., Thomas R.E., Andrews L.A., McBride H.M., Whitworth A.J., Pallanck L.J. 2008. The PINK1/Parkin pathway regulates mitochondrial morphology. Proc. Natl. Acad. Sci. USA. 105:1638–1643 10.1073/pnas.0709336105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole A.C., Thomas R.E., Yu S., Vincow E.S., Pallanck L. 2010. The mitochondrial fusion-promoting factor mitofusin is a substrate of the PINK1/parkin pathway. PLoS One. 5:e10054 10.1371/journal.pone.0010054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovich E., Kerem A., Fröhlich K.U., Diamant N., Bar-Nun S. 2002. AAA-ATPase p97/Cdc48p, a cytosolic chaperone required for endoplasmic reticulum-associated protein degradation. Mol. Cell. Biol. 22:626–634 10.1128/MCB.22.2.626-634.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzuto R., Pinton P., Carrington W., Fay F.S., Fogarty K.E., Lifshitz L.M., Tuft R.A., Pozzan T. 1998. Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science. 280:1763–1766 10.1126/science.280.5370.1763 [DOI] [PubMed] [Google Scholar]
- Sandebring A., Thomas K.J., Beilina A., van der Brug M., Cleland M.M., Ahmad R., Miller D.W., Zambrano I., Cowburn R.F., Behbahani H., et al. 2009. Mitochondrial alterations in PINK1 deficient cells are influenced by calcineurin-dependent dephosphorylation of dynamin-related protein 1. PLoS One. 4:e5701 10.1371/journal.pone.0005701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staropoli J.F., McDermott C., Martinat C., Schulman B., Demireva E., Abeliovich A. 2003. Parkin is a component of an SCF-like ubiquitin ligase complex and protects postmitotic neurons from kainate excitotoxicity. Neuron. 37:735–749 10.1016/S0896-6273(03)00084-9 [DOI] [PubMed] [Google Scholar]
- Tresse E., Salomons F.A., Vesa J., Bott L.C., Kimonis V., Yao T.P., Dantuma N.P., Taylor J.P. 2010. VCP/p97 is essential for maturation of ubiquitin-containing autophagosomes and this function is impaired by mutations that cause IBMPFD. Autophagy. 6:217–227 10.4161/auto.6.2.11014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twig G., Elorza A., Molina A.J., Mohamed H., Wikstrom J.D., Walzer G., Stiles L., Haigh S.E., Katz S., Las G., et al. 2008. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 27:433–446 10.1038/sj.emboj.7601963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente E.M., Abou-Sleiman P.M., Caputo V., Muqit M.M., Harvey K., Gispert S., Ali Z., Del Turco D., Bentivoglio A.R., Healy D.G., et al. 2004. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science. 304:1158–1160 10.1126/science.1096284 [DOI] [PubMed] [Google Scholar]
- Vives-Bauza C., Zhou C., Huang Y., Cui M., de Vries R.L., Kim J., May J., Tocilescu M.A., Liu W., Ko H.S., et al. 2010. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc. Natl. Acad. Sci. USA. 107:378–383 10.1073/pnas.0911187107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Gehrke S., Imai Y., Huang Z., Ouyang Y., Wang J.W., Yang L., Beal M.F., Vogel H., Lu B. 2006. Mitochondrial pathology and muscle and dopaminergic neuron degeneration caused by inactivation of Drosophila Pink1 is rescued by Parkin. Proc. Natl. Acad. Sci. USA. 103:10793–10798 10.1073/pnas.0602493103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Ouyang Y., Yang L., Beal M.F., McQuibban A., Vogel H., Lu B. 2008. Pink1 regulates mitochondrial dynamics through interaction with the fission/fusion machinery. Proc. Natl. Acad. Sci. USA. 105:7070–7075 10.1073/pnas.0711845105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Meyer H.H., Rapoport T.A. 2001. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature. 414:652–656 10.1038/414652a [DOI] [PubMed] [Google Scholar]
- Ye Y., Meyer H.H., Rapoport T.A. 2003. Function of the p97-Ufd1-Npl4 complex in retrotranslocation from the ER to the cytosol: dual recognition of nonubiquitinated polypeptide segments and polyubiquitin chains. J. Cell Biol. 162:71–84 10.1083/jcb.200302169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziviani E., Tao R.N., Whitworth A.J. 2010. Drosophila parkin requires PINK1 for mitochondrial translocation and ubiquitinates mitofusin. Proc. Natl. Acad. Sci. USA. 107:5018–5023 10.1073/pnas.0913485107 [DOI] [PMC free article] [PubMed] [Google Scholar]