Abstract
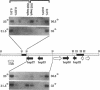
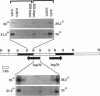
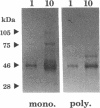
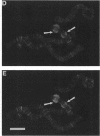
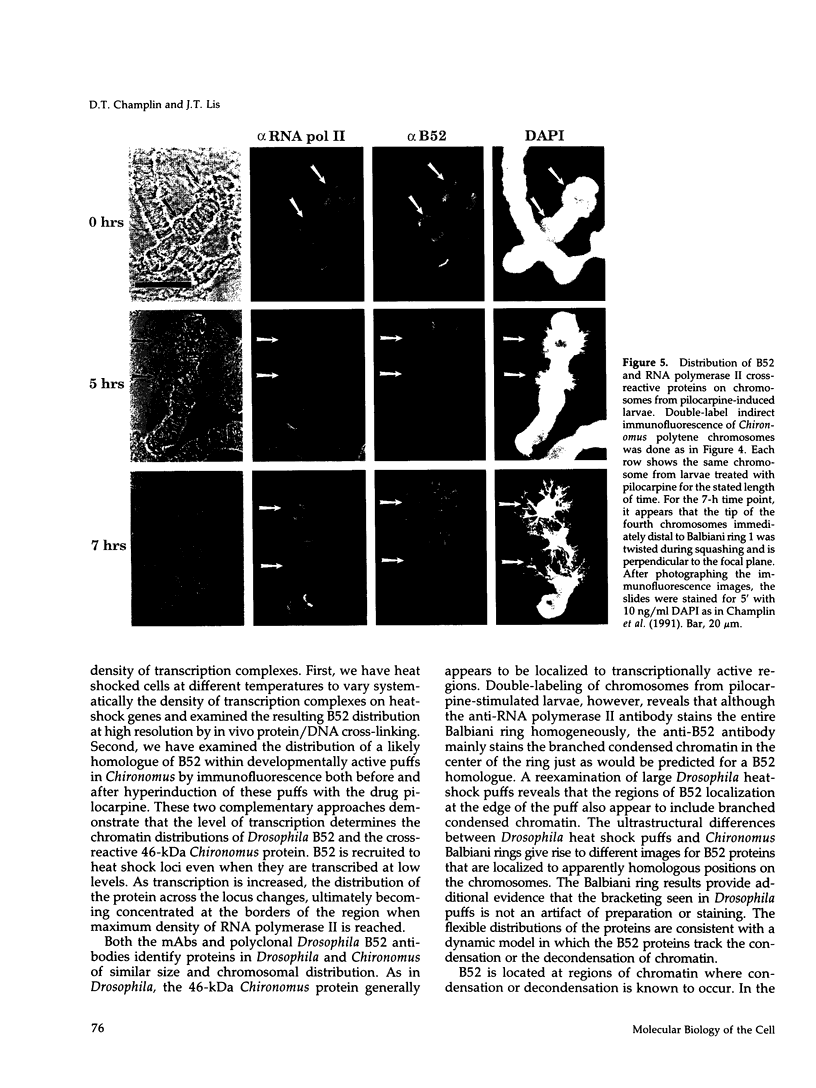
Drosophila B52 protein is a homologue of human ASF/SF2 that functions in vitro as an essential pre-mRNA splicing factor. Immunofluorescence analysis of polytene chromosomes has shown that B52 generally colocalizes with RNA polymerase II; however, in contrast to other splicing factors, B52 brackets RNA polymerase II at highly active heat-shock puffs. Also, UV cross-linking in nonpolytene cells has shown that B52 cross-links in vivo to DNA flanking the highly active transcription units. Here, we find that the distribution of cross-linked B52 at heat-shock loci depends on transcription levels. Heat shocks at low and moderate temperatures, which induce corresponding levels of transcription, recruit B52 both to transcribed DNA and to flanking DNA, whereas a full heat-shock induction concentrates B52 on the DNA that brackets the entire activated region. We have also identified a 46-kDa protein from Chironomus tentans that binds Drosophila B52 antibodies and has a distribution on chromosomes analogous to B52. This protein is found throughout the moderately transcribed Balbiani rings. However, when transcription at these rings is hyperinduced to levels comparable to fully induced Drosophila heat-shock genes, the protein is restricted to the boundaries of highly decondensed chromatin. We suggest that B52 tracks to chromatin fibers that are folding or unfolding, and we discuss this in light of B52's proposed roles in pre-mRNA splicing and control.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beverley S. M., Wilson A. C. Molecular evolution in Drosophila and the higher Diptera II. A time scale for fly evolution. J Mol Evol. 1984;21(1):1–13. doi: 10.1007/BF02100622. [DOI] [PubMed] [Google Scholar]
- Carter K. C., Bowman D., Carrington W., Fogarty K., McNeil J. A., Fay F. S., Lawrence J. B. A three-dimensional view of precursor messenger RNA metabolism within the mammalian nucleus. Science. 1993 Feb 26;259(5099):1330–1335. doi: 10.1126/science.8446902. [DOI] [PubMed] [Google Scholar]
- Cartwright I. L., Elgin S. C. Nucleosomal instability and induction of new upstream protein-DNA associations accompany activation of four small heat shock protein genes in Drosophila melanogaster. Mol Cell Biol. 1986 Mar;6(3):779–791. doi: 10.1128/mcb.6.3.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
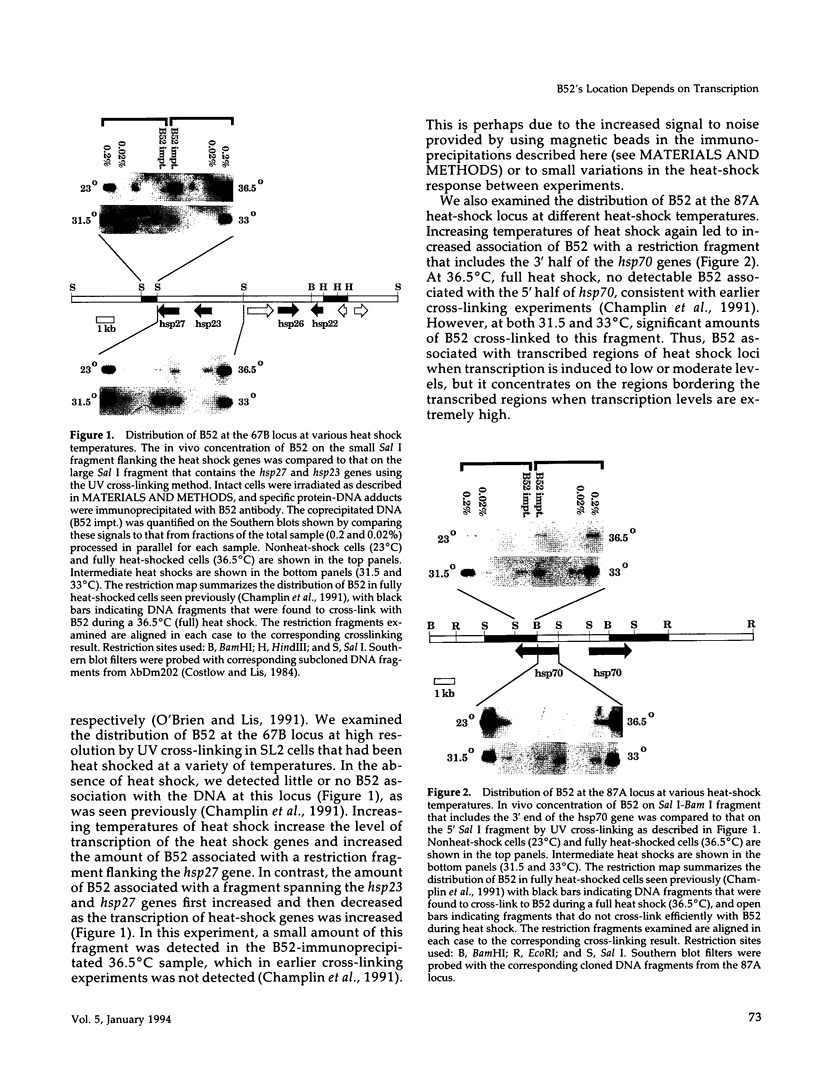
- Champlin D. T., Frasch M., Saumweber H., Lis J. T. Characterization of a Drosophila protein associated with boundaries of transcriptionally active chromatin. Genes Dev. 1991 Sep;5(9):1611–1621. doi: 10.1101/gad.5.9.1611. [DOI] [PubMed] [Google Scholar]
- Christensen M. E., LeStourgeon W. M., Jamrich M., Howard G. C., Serunian L. A., Silver L. M., Elgin S. C. Distribution studies on polytene chromosomes using antibodies directed against hnRNP. J Cell Biol. 1981 Jul;90(1):18–24. doi: 10.1083/jcb.90.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costlow N., Lis J. T. High-resolution mapping of DNase I-hypersensitive sites of Drosophila heat shock genes in Drosophila melanogaster and Saccharomyces cerevisiae. Mol Cell Biol. 1984 Sep;4(9):1853–1863. doi: 10.1128/mcb.4.9.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericsson C., Grossbach U., Björkroth B., Daneholt B. Presence of histone H1 on an active Balbiani ring gene. Cell. 1990 Jan 12;60(1):73–83. doi: 10.1016/0092-8674(90)90717-s. [DOI] [PubMed] [Google Scholar]
- Ericsson C., Mehlin H., Björkroth B., Lamb M. M., Daneholt B. The ultrastructure of upstream and downstream regions of an active Balbiani ring gene. Cell. 1989 Feb 24;56(4):631–639. doi: 10.1016/0092-8674(89)90585-0. [DOI] [PubMed] [Google Scholar]
- Fu X. D., Mayeda A., Maniatis T., Krainer A. R. General splicing factors SF2 and SC35 have equivalent activities in vitro, and both affect alternative 5' and 3' splice site selection. Proc Natl Acad Sci U S A. 1992 Dec 1;89(23):11224–11228. doi: 10.1073/pnas.89.23.11224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge H., Zuo P., Manley J. L. Primary structure of the human splicing factor ASF reveals similarities with Drosophila regulators. Cell. 1991 Jul 26;66(2):373–382. doi: 10.1016/0092-8674(91)90626-a. [DOI] [PubMed] [Google Scholar]
- Gilmour D. S., Lis J. T. In vivo interactions of RNA polymerase II with genes of Drosophila melanogaster. Mol Cell Biol. 1985 Aug;5(8):2009–2018. doi: 10.1128/mcb.5.8.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour D. S., Lis J. T. RNA polymerase II interacts with the promoter region of the noninduced hsp70 gene in Drosophila melanogaster cells. Mol Cell Biol. 1986 Nov;6(11):3984–3989. doi: 10.1128/mcb.6.11.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour D. S., Rougvie A. E., Lis J. T. Protein-DNA cross-linking as a means to determine the distribution of proteins on DNA in vivo. Methods Cell Biol. 1991;35:369–381. doi: 10.1016/s0091-679x(08)60580-4. [DOI] [PubMed] [Google Scholar]
- Jiménez-García L. F., Spector D. L. In vivo evidence that transcription and splicing are coordinated by a recruiting mechanism. Cell. 1993 Apr 9;73(1):47–59. doi: 10.1016/0092-8674(93)90159-n. [DOI] [PubMed] [Google Scholar]
- Krainer A. R., Mayeda A., Kozak D., Binns G. Functional expression of cloned human splicing factor SF2: homology to RNA-binding proteins, U1 70K, and Drosophila splicing regulators. Cell. 1991 Jul 26;66(2):383–394. doi: 10.1016/0092-8674(91)90627-b. [DOI] [PubMed] [Google Scholar]
- Mayeda A., Zahler A. M., Krainer A. R., Roth M. B. Two members of a conserved family of nuclear phosphoproteins are involved in pre-mRNA splicing. Proc Natl Acad Sci U S A. 1992 Feb 15;89(4):1301–1304. doi: 10.1073/pnas.89.4.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mähr R., Meyer B., Daneholt B., Eppenberger H. M. Activation of Balbiani ring genes in Chironomus tentans after a pilocarpine-induced depletion of the secretory products from the salivary gland lumen. Dev Biol. 1980 Dec;80(2):409–418. doi: 10.1016/0012-1606(80)90415-7. [DOI] [PubMed] [Google Scholar]
- Nacheva G. A., Guschin D. Y., Preobrazhenskaya O. V., Karpov V. L., Ebralidse K. K., Mirzabekov A. D. Change in the pattern of histone binding to DNA upon transcriptional activation. Cell. 1989 Jul 14;58(1):27–36. doi: 10.1016/0092-8674(89)90399-1. [DOI] [PubMed] [Google Scholar]
- O'Brien T., Lis J. T. RNA polymerase II pauses at the 5' end of the transcriptionally induced Drosophila hsp70 gene. Mol Cell Biol. 1991 Oct;11(10):5285–5290. doi: 10.1128/mcb.11.10.5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olins A. L., Olins D. E., Levy H. A., Durfee R. C., Margle S. M., Tinnel E. P. DNA compaction during intense transcription measured by electron microscope tomography. Eur J Cell Biol. 1986 Mar;40(1):105–110. [PubMed] [Google Scholar]
- Roth M. B., Zahler A. M., Stolk J. A. A conserved family of nuclear phosphoproteins localized to sites of polymerase II transcription. J Cell Biol. 1991 Nov;115(3):587–596. doi: 10.1083/jcb.115.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sass H., Pederson T. Transcription-dependent localization of U1 and U2 small nuclear ribonucleoproteins at major sites of gene activity in polytene chromosomes. J Mol Biol. 1984 Dec 25;180(4):911–926. doi: 10.1016/0022-2836(84)90263-8. [DOI] [PubMed] [Google Scholar]
- Sass H. RNA polymerase B in polytene chromosomes: immunofluorescent and autoradiographic analysis during stimulated and repressed RNA synthesis. Cell. 1982 Feb;28(2):269–278. doi: 10.1016/0092-8674(82)90345-2. [DOI] [PubMed] [Google Scholar]
- Simon J. A., Sutton C. A., Lobell R. B., Glaser R. L., Lis J. T. Determinants of heat shock-induced chromosome puffing. Cell. 1985 Apr;40(4):805–817. doi: 10.1016/0092-8674(85)90340-x. [DOI] [PubMed] [Google Scholar]
- Solomon M. J., Larsen P. L., Varshavsky A. Mapping protein-DNA interactions in vivo with formaldehyde: evidence that histone H4 is retained on a highly transcribed gene. Cell. 1988 Jun 17;53(6):937–947. doi: 10.1016/s0092-8674(88)90469-2. [DOI] [PubMed] [Google Scholar]
- Spector D. L., Fu X. D., Maniatis T. Associations between distinct pre-mRNA splicing components and the cell nucleus. EMBO J. 1991 Nov;10(11):3467–3481. doi: 10.1002/j.1460-2075.1991.tb04911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wansink D. G., Schul W., van der Kraan I., van Steensel B., van Driel R., de Jong L. Fluorescent labeling of nascent RNA reveals transcription by RNA polymerase II in domains scattered throughout the nucleus. J Cell Biol. 1993 Jul;122(2):283–293. doi: 10.1083/jcb.122.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widmer R. M., Lucchini R., Lezzi M., Meyer B., Sogo J. M., Edström J. E., Koller T. Chromatin structure of a hyperactive secretory protein gene (in Balbiani ring 2) of Chironomus. EMBO J. 1984 Jul;3(7):1635–1641. doi: 10.1002/j.1460-2075.1984.tb02022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Wong Y. C., Elgin S. C. The chromatin structure of specific genes: II. Disruption of chromatin structure during gene activity. Cell. 1979 Apr;16(4):807–814. doi: 10.1016/0092-8674(79)90096-5. [DOI] [PubMed] [Google Scholar]