Abstract
Which transcription factors control the distribution of metabolic fluxes under a given condition? We address this question by systematically quantifying metabolic fluxes in 119 transcription factor deletion mutants of Saccharomyces cerevisiae under five growth conditions. While most knockouts did not affect fluxes, we identified 42 condition-dependent interactions that were mediated by a total of 23 transcription factors that control almost exclusively the cellular decision between respiration and fermentation. This relatively sparse, condition-specific network of active metabolic control contrasts with the much larger gene regulation network inferred from expression and DNA binding data. Based on protein and transcript analyses in key mutants, we identified three enzymes in the tricarboxylic acid cycle as the key targets of this transcriptional control. For the transcription factor Gcn4, we demonstrate that this control is mediated through the PKA and Snf1 signaling cascade. The discrepancy between flux response predictions, based on the known regulatory network architecture and our functional 13C-data, demonstrates the importance of identifying and quantifying the extent to which regulatory effectors alter cellular functions.
Keywords: metabolic flux, omics data, regulatory network, transcription factor, transcriptional regulation
Introduction
Effective control and modulation of cellular behavior is of paramount importance in medicine (Kreeger and Lauffenburger, 2010) and biotechnology (Haynes and Silver, 2009), and requires profound understanding of control mechanisms. In cancer treatment, for example, it would be of great impact to induce apoptosis only in tumor cells but not in healthy ones, while in biotechnology it is important for the cost-effectiveness of a process to minimize the formation of by-products and redirect carbon toward desired compound(s). Learning the mechanisms through which cells regulate their response to changing environments can help in the design of reverse-engineering regulatory circuits to modulate cellular behavior (Csete and Doyle, 2002). To date, regulatory mechanisms are mostly inferred from gene expression, interaction or binding data (Papin et al, 2005; Karlebach and Shamir, 2008; Snyder and Gallagher, 2009). Yet, the ability to predict cellular behavior from such inferred mechanisms is still poor (Bonneau, 2008), owing to the fact that many regulatory events remain hidden. In particular, very little is known about how changes in transcript and protein levels affect metabolic readjustment, and thus, phenotypic behavior (Heinemann and Sauer, 2010).
Transcriptional regulation is arguably at the forefront of a cells's ability to control resource availability, being the first regulatory layer to determine new cellular composition. Over the last decade, transcriptional regulatory networks have been extensively investigated, and the backbone of potential ‘transcription factor–target gene’ interactions has been reconstructed based on genome-wide protein-DNA binding analysis and high-throughput gene expression data (Bonneau, 2008). The first large-scale protein–DNA binding analysis study of the model eukaryote Saccharomyces cerevisiae revealed a highly connected transcription factor network architecture (Lee et al, 2002), whose condition-dependent interaction connectivity was later identified based on protein–DNA binding data from different stress conditions (Harbison et al, 2004). Large-scale genome-wide expression data were used to reconstruct the organization of transcription factor networks by graph theory (Yu and Gerstein, 2006; Hu et al, 2007), probabilistic graphical models (Segal et al, 2003) or clustering algorithms (Ihmels et al, 2002). The integration of protein–DNA binding topology and gene expression data through statistical approaches was used to reconstruct the architecture of the responsive transcriptional regulatory network, unraveling a rewiring of the transcriptional network interactions in response to various stimuli (Luscombe et al, 2004; Balaji et al, 2006; Gitter et al, 2009). An even higher level of integration was achieved by combining protein–DNA binding profiles with genetic perturbations, gene expression data, protein interaction data and systematic phenotyping to reveal causal pathway models that provide global hypotheses of how signaling and transcription are linked (Workman et al, 2006). Despite this extensive knowledge, the link from transcriptional regulation to the functional output is largely missing, because changes in transcript/protein abundance do not necessarily lead to equal (or any) changes in function. Explicitly, if the condition-dependent binding of a transcription factor leads to differential expression of its target gene(s), the consequences of such regulation on cellular operation remains nearly impossible to predict.
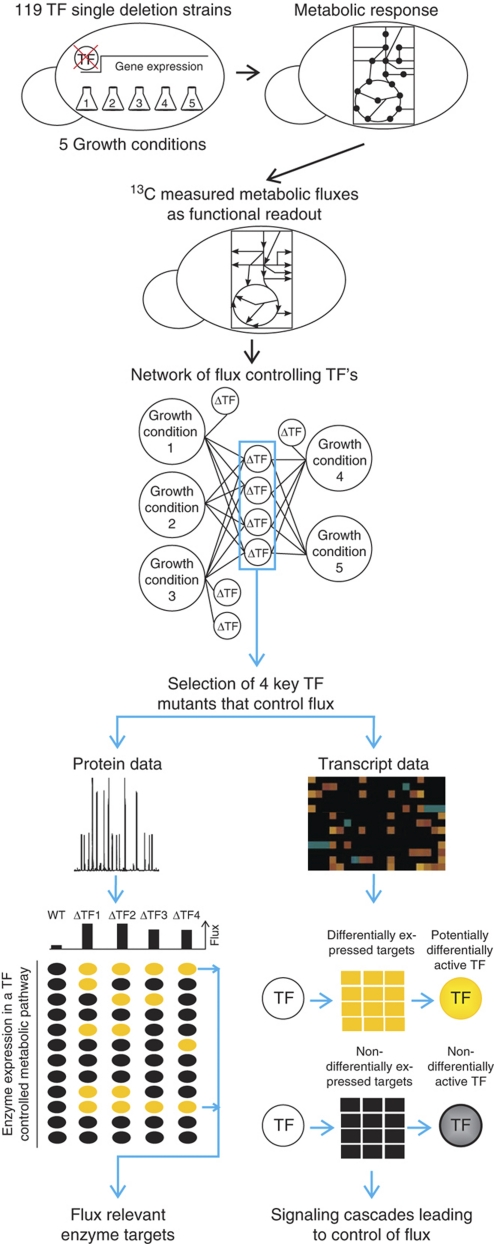
In this study, we aim to elucidate the extent to which transcription factors control the operation of yeast metabolism. As a quantitative readout of metabolic function, we monitored the traffic of small molecules through various pathways of central metabolism by 13C-flux analysis (Sauer, 2006). For a systematic analysis, we quantified the flux distributions (pathway activities) within central carbon metabolism of 119 single deletion strains that lack metabolism-related transcription factors under five different growth conditions. We identified condition-dependent networks of transcription factors that control metabolic pathway activity (Figure 1). Despite their widespread impact on gene expression (Hu et al, 2007), only very few transcription factors affect pathway activity and thus the flux distributions. For transcription factors that affect the flux distribution, we then unraveled flux relevant enzymes based on consistent changes in protein abundances, and further hypothesize on the underlying mechanism leading to the control of metabolic flux distributions based on genome-wide gene expression data (Figure 1).
Figure 1.
Schematic overview on the performed experiments and data analysis. Yellow ellipses and squares indicate altered protein and gene expression in transcription factor mutants compared with the wild type, respectively. Black ellipses and squares indicate no difference between mutant and wild type.
Results
In the yeast S. cerevisiae, 275 genes are annotated as ‘transcriptional regulatory active’ in the yeast genome database (Cherry et al, 1998). Out of these, we selected 119 transcription factors related to metabolism or stress responses, covering ∼70% of all transcription factors with target genes in at least one of the two processes. For each of these 119 transcription factors, prototrophic deletion strains were constructed and grown under five conditions: glucose, glucose with high osmolarity, glucose with urea as nitrogen source, glucose with low pH and galactose (Supplementary Table 1). The chosen growth conditions suit the requirements for flux analysis, such as exponential growth on minimal medium (Zamboni et al, 2009). They represent two different regulatory states of reduced (galactose) and maximal carbon source repression (glucose), as well as a different nitrogen metabolism and two common, permanent stress conditions.
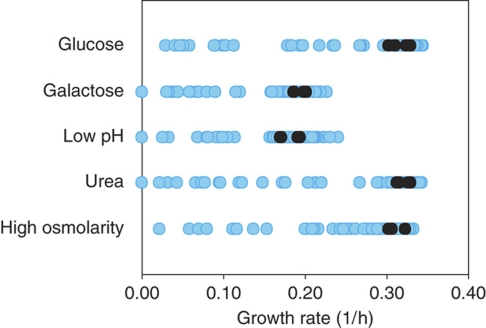
As a general measure for effects of the deleted transcription factors on metabolism, we determined growth rates (Figure 2, Supplementary Table 2). The wild-type grew with a maximum specific growth rate of 0.31–0.33 1/h under three conditions and with a maximum specific growth rate of 0.19–0.20 1/h at low pH or on galactose (Supplementary Table 2). Under all five tested conditions, 13–15% of the investigated mutants exhibited a growth defect >20% and up to six mutants did not grow at all under a given condition. The observed growth defects indicate that the deleted transcription factors were required under the respective growth condition.
Figure 2.
Maximum-specific growth rates of the 119 transcription factor mutants under five growth conditions. Four replicates of the wild type are depicted as black dots. Deletion strains are depicted as blue dots (average of four replicates) (Supplementary Table 2).
Transcription factors that control the distribution of flux
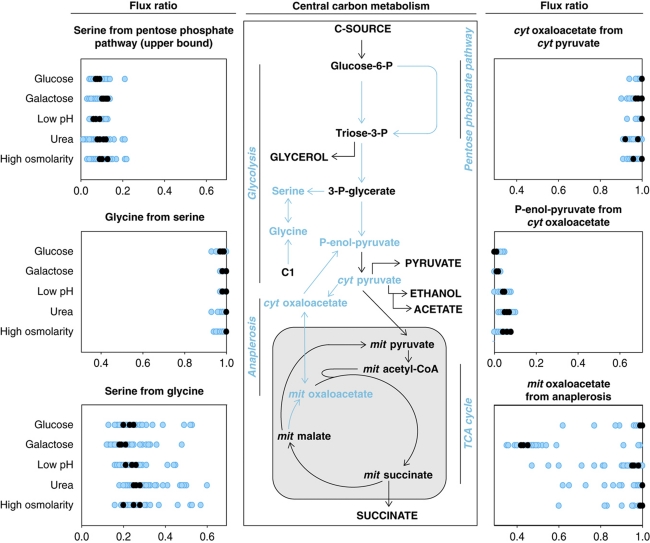
To quantitatively evaluate the effect of single transcription factor deletions on pathway activity and thus on flux distributions, all strains were grown in 20% uniformly 13C-labeled glucose or galactose (Supplementary Table 1). Both substrates enter central metabolism at the level of glucose-6-P, but they lead to primarily fermentative or respiro-fermentative metabolism, respectively (Bro et al, 2005; Küpfer et al, 2005). During fermentative metabolism, ATP is mainly produced through glycolysis with subsequent ethanol formation. During respiro-fermentative metabolism, ATP is simultaneously produced through glycolysis, the tricarboxylic acid (TCA) cycle and respiratory chain with only some formation of ethanol. As glucose and galactose are metabolized to an unequal extent through the alternative pathways of central carbon metabolism, different 13C-labeling patterns emerge that were subsequently determined in protein-bound amino acids by gas chromatography–mass spectrometry (Sauer, 2006; Zamboni et al, 2009). From the determined mass isotopomer abundances in amino acids, we calculated six ratios of converging central metabolic fluxes (Blank and Sauer, 2004; Zamboni et al, 2009), which determine the flux distribution in central carbon metabolism (Figure 3, Supplementary Figure 1, Supplementary Table 1).
Figure 3.
Intracellular ratios of converging fluxes in 119 transcription factor mutants under five growth conditions during steady state. Flux ratios were calculated from uniformly 13C-labeled glucose or galactose experiments. The wild type is depicted as black dots (four replicates) and the deletion strains as blue dots (single measurements) (Supplementary Table 3). Blue metabolites and arrows in the drawing of central carbon metabolism indicate network nodes, for which flux ratios were determined. Capitalized metabolites are extracellular substrates or products. Pathways are depicted in italic.
Depending on the growth condition, between 7 and 13% of the deleted transcription factors altered the determined flux ratios (Figure 3, Supplementary Table 3). Three out of six flux ratios corresponding to gluconeogenesis, glycine production through C1 metabolism and transport of mitochondrial oxaloacetate into the cytosol were never significantly altered in any of the mutants. Thus, these three flux ratios were not controlled by the investigated transcription factors under the tested conditions. The other three potentially transcriptionally controlled flux ratios were the upper bound of ‘serine originating from the pentose phosphate pathway’, which quantifies the relative contribution of glycolysis versus the pentose phosphate pathway; ‘serine originating from glycine’, which quantifies the relative contribution of the backward flux from glycine to serine versus the forward flux from 3-phospho-glycerate to serine; and ‘mitochondrial oxaloacetate derived through anaplerosis’, which quantifies the relative contribution of the respiratory TCA cycle flux versus the replenishment of the biosynthetic precursor. The relative pathway usage of glycolysis and pentose phosphate pathway was altered only in three mutants, whereas the other two flux ratios were altered depending on the growth condition in 1–12% of the transcription factor mutants.
To exclude that the observed alterations in the flux distributions were indirect consequences of altered mutant physiology, we correlated the specific growth rates with the flux ratios by calculating the correlation factor between both (data not shown). If the transcription factors have an indirect effect on flux distributions via reduced growth rates in the deletion mutants, we expect a correlation between mutant growth rates and the determined flux distributions. For transcription factors with a direct effect on metabolism, we expect no such correlation. Relative pathway activity, for the flux distribution between glycolysis and the pentose phosphate pathway, and the convergent ratio of anaplerosis and TCA cycle were not correlated with growth rate (correlation coefficients considering all growth conditions of −0.27 and 0.19, respectively). Thus, they were directly controlled by the deleted transcription factors and not indirectly influenced through altered growth rate. The flux ratio quantifying the backward flux to serine, however, correlated with growth rate (correlation coefficient considering all growth conditions of −0.65), implying that the observed alterations in this flux ratio were indirect consequences of altered growth. As the results were obtained from single experiments in a screening setup, at least triplicate experiments were carried out for all transcription factor deletion mutants with altered flux ratios for the relative pathway activity between glycolysis and the pentose phosphate pathway, and the convergent ratio of anaplerosis and the TCA cycle (Supplementary Table 4). Thereby, 23 out of the 24 originally identified mutants were reconfirmed.
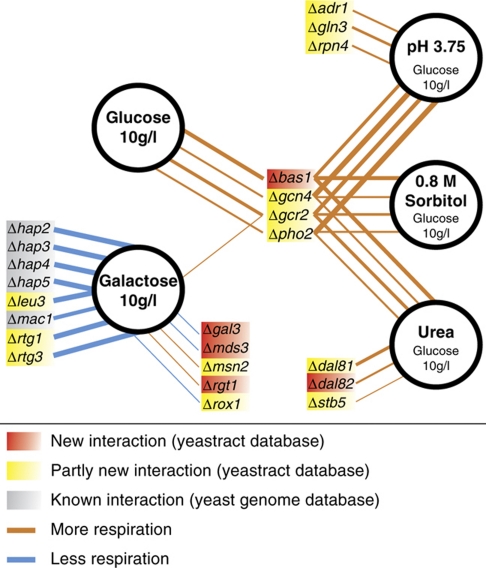
As fluxes and their distribution are a readout for the functional metabolic consequences of a transcript alteration, we conclude that 23 transcription factors control flux distributions under at least one of the tested growth conditions, leading to 42 condition-dependent interactions of transcription factors with metabolic pathway activity. All 23 transcription factors controlled the TCA cycle flux activity. Two of them (Rtg1/3) also controlled the relative activity between glycolysis and the pentose phosphate pathway. Thus, transcriptional control focussed almost exclusively on the TCA cycle and probably also on the functionally connected, but here not observed, respiratory chain, while the remainder of central carbon metabolism was not affected. The control of the TCA cycle flux activity through the 23 identified transcription factors exhibited different magnitudes of alteration (Figure 4): during growth on galactose, the deletion of seven transcription factors led to a completely abolished TCA cycle usage, but none of the deletion mutants with higher relative flux through the TCA cycle actually achieved a TCA cycle flux comparable with that observed on fully respiratory carbon sources (e.g., on pyruvate or ethanol; Fendt and Sauer, 2010).
Figure 4.
Network of transcription factors that controls TCA cycle flux during steady-state growth in batch cultures grown either on glucose or galactose (Supplementary Table 4). Line thickness indicates the magnitude of difference to the wild type.
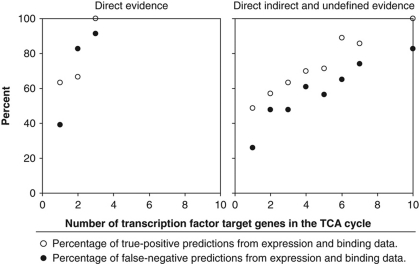
How many transcription factors could have been predicted to affect pathway activity based on their target gene patterns? The Yeastract database lists 71 of the investigated transcription factors with at least one target gene in central metabolism on the basis of literature-curated, direct, indirect or undefined evidence from expression and DNA binding data, which could thus potentially affect pathway activity. Although 55 transcription factors have at least one target in glycolysis or the pentose phosphate pathway, the flux distribution between those two pathways was altered in only two mutants. Of the 35 transcription factors with at least one target in the TCA cycle, in contrast, 23 exerted control under at least one of the tested conditions. To assess the significance of this seemingly better predictive fidelity, we calculated the predictive fidelity of expression and DNA binding data for all TCA cycle genes by considering (i) only direct or (ii) direct, indirect and undefined evidence. While generally predictive fidelity increased with the number of transcription factor target genes in the TCA cycle, there was no particular combination that achieved a high percentage of true-positive predictions at a low false-negative percentage (Figure 5, Supplementary Table 5). Most relevant for a first prediction on potential flux control, where a low false-negative rate is desired, was the combination of all three lines of evidence for transcription factors with one or more target genes in the TCA cycle genes; (26.1% false negatives and 48.6% true positives). Thus, the prediction of flux controlling transcriptional events on the basis of expression and DNA binding data is rather limited for the TCA cycle and not possible for others like the pentose phosphate pathway.
Figure 5.
Predictive fidelity of expression and binding data (Teixeira et al, 2006) for transcription factors that control metabolic fluxes.
In principle, the low predictive power of gene regulation data for functional flux responses could be caused by transcription factor redundancy. To test this hypothesis, we obtained 13C-flux data from the double and triple transcriptional factor mutants Nrg1/2, Msn2/4 and Mig1/2/3 (data not shown). We decided to test these mutants as the deleted transcription factors are main regulators of glucose repression and stress responds (Zaman et al, 2008). Yet, even these multiple deletions did not result in altered TCA cycle flux distributions, indicating that redundancy is not the primary reason for the observed robustness.
Relevant TCA cycle enzymes that enable higher pathway usage
Of the 23 transcription factors that controlled TCA cycle flux distributions under the tested conditions, only Bas1, Gcn4, Gcr2 and Pho2 exerted control under more than one condition (Figure 4). None of these four transcription factors had previously been identified as a key regulator of the TCA cycle. While Gcn4, Gcr2 and Pho2 have known targets in the TCA cycle, our finding is entirely novel for Bas1. Gcn4 is a global regulator of amino-acid biosynthesis and also has five known targets in the TCA cycle (LPD1, CIT3, ACO2, IDH1, IDP1) (Hinnebusch, 2005; Teixeira et al, 2006). Bas1 and Pho2 act together to activate purine and histidine biosynthesis, and only Pho2 has the TCA cycle gene IDH1 as a target (Hannum et al, 2002; Som et al, 2005; Teixeira et al, 2006). Gcr2 is an activator of glycolysis genes, but six TCA cycle genes are also among its known targets (CIT1, CIT3, ACO1, SDH2, SDH3, SDH4) (Chambers et al, 1995; Teixeira et al, 2006). For these four mutants we asked which enzymes are relevant for the higher activity of the TCA cycle.
For this purpose, we determined the abundance of 50 central metabolic enzymes with targeted proteomics in the four mutants during growth on glucose (Figure 6, Supplementary Table 6). In general, glycolytic enzyme abundances were decreased in the GCR2 mutant as expected from the known function of Gcr2, as an activator of glycolysis (Chambers et al, 1995). The GCN4 deletion hardly altered any enzyme abundances, except those of TCA cycle enzymes. The BAS1 and PHO2 mutants exhibited very similar patterns of differentially expressed enzymes; that is, all 11 enzyme alterations observed in the PHO2 mutant were also found in the BAS1 mutant, supporting the view that they act together (Hannum et al, 2002; Som et al, 2005). The consistent increase of several glycolysis enzymes in these two mutants, however, did not lead to an alteration in the relative use of glycolysis and the pentose phosphate pathway.
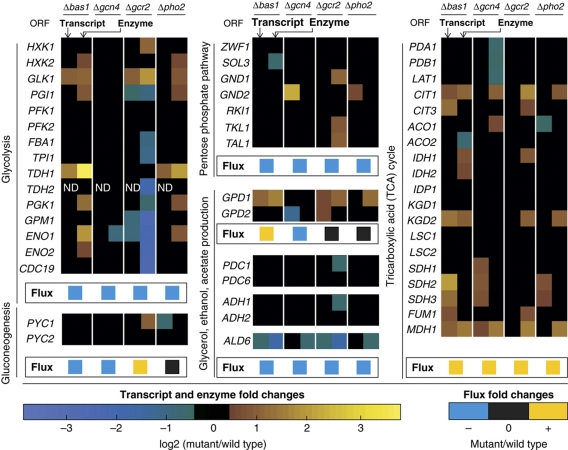
Figure 6.
Differentially expressed transcripts and enzymes of BAS1, GCN4, GCR2 and PHO2 transcription factor mutants during exponential growth on glucose (fold change ⩾1.3, P-value ⩽0.05). ‘ND’ stands for not determined.
Consistently, increased TCA cycle enzyme abundances in all four mutants was only found for citrate synthase (Cit1) and malate dehydrogenase (Mdh1) (Figure 6, Supplementary Table 6). This suggests that the increase in abundance of these two enzymes is necessary to enable the observed higher activity of the TCA cycle. As the BAS1 and GCR2 mutants displayed the highest TCA cycle activity, we looked for enzymes that were more abundant in these two strains, a criterion that was only fulfilled by isocitrate dehydrogenase (Idh1). In addition, the Idh2 member of the isocitrate dehydrogenase complex was more abundant, but the P-value for the GCR2 mutant was 0.08, a value greater than the chosen cutoff P-value of 0.05. Thus, we concluded that during growth on glucose, increased abundance of Cit1 and Mdh1 is necessary to increase the TCA cycle flux from 0 to 0.01–0.03 mmol/g/h. The additional further flux increase to 0.10–0.13 mmol/g/h then requires an additional increase in abundance of Idh1/2p; as seen in the BAS1 and GCR2 mutants. Although these enzymes are apparently necessary for achieving a higher TCA cycle activity, they are not sufficient as additional components of the respiratory chain must also be expressed at higher levels.
Signaling cascades leading to the control of TCA cycle activity
After identifying relevant enzyme targets for the higher TCA cycle flux, we asked through which signaling cascades Bas1, Gcr2, Gcn4 and Pho2 governed this change in activity. For this purpose we determined genome-wide transcript abundances in the four mutants during growth on glucose. Generally, expression of many genes was altered in all four mutants, including most of the known targets of the four transcription factors (Figure 6, Supplementary Table 7). As already observed for enzyme abundance alterations, the pattern of differentially expressed genes in the BAS1 and PHO2 mutants was very similar, strongly supporting the view that they act together. When comparing protein and transcript abundance alterations in central carbon metabolism, the magnitude of transcript abundance alterations was about half of the abundance alterations in the corresponding enzymes, yet the direction was always consistent (Figure 6).
To identify active signaling cascades leading to the control of TCA cycle activity in the four mutants, we predicted differentially activated transcription factors based on the activity pattern of their target genes (Oliveira et al, 2008). The underlying hypothesis is that a transcription factor is potentially differentially activated in the mutant compared with the wild type when its target genes are differentially expressed. Based on all increased transcripts, that were identified as potentially relevant for the higher TCA cycle flux, we found 47, 53, 31 and 14 transcription factors to be differentially activated in the BAS1, GCN4, GCR2 and PHO2 mutants, respectively (Supplementary Table 8). Not unexpectedly due to the chosen set of investigated transcription factor mutants, the majority of these differentially activated transcription factors were related to metabolism or stress response.
The inferred pattern of differentially activated transcription factors suggests reduced glucose repression in all four mutants (Table I). For the GCR2 mutant, our conclusion is based only on the differential activity of Nrg1, a key transcription factor for maintaining glucose repression (Zhou and Winston, 2001), whereas for the PHO2 mutant it is based on differential activity of the Hap-complex, a global regulator of respiration and a target of glucose repression (Zaman et al, 2008; Turcotte et al, 2009). The evidence is stronger for the BAS1 mutant because both Nrg1 and the Hap-complex appear to be differentially activated. In addition, Adr1, a target of glucose repression through its activating kinase Snf1 (Zaman et al, 2008), was identified as differentially activated. For the GCN4 mutant we have the strongest evidence, as all differentially activated transcription factors described for the other three mutants are also found in GCN4 mutant. Moreover, we also found Mig1, the major transcription factor of the Snf1 repressor complex involved in glucose repression (Turcotte et al, 2009), and Msn2/4 to be differentially activated. Differential activity of Msn2/4, based on the upregulation of its target genes in the GCN4 mutant, indicates less strong signaling of PKA (Zaman et al, 2008), which is one of the two major downstream regulators of glucose repression. Most of the identified differentially activated transcription factors were also tested as single deletions in the primary screen, but did not lead to flux alterations.
Table 1. Potentially differentially activated transcription factors that indicate a reduced glucose repression in the four mutants compared with the wild type.
| Strains | Differentially activated transcription factors |
|---|---|
| Δgcr2 | Nrg1 |
| Δpho2 | Hap-complex |
| Δbas1 | Adr1, Hap-complex, Nrg1 |
| Δgcn4 | Adr1, Hap-complex, Mig1, Msn2/4, Nrg1 |
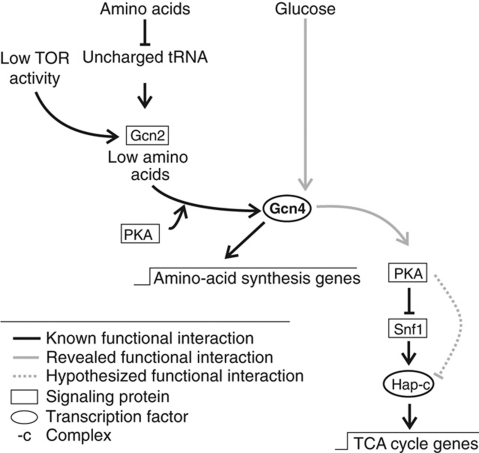
To validate the above hypotheses on glucose repression-related regulation events, we focussed on the GCN4 mutant. The GCN4 mutant has potentially reduced PKA activity, which leads to increased Snf1 activity (Haurie et al, 2004; Hedbacker et al, 2004; Slattery et al, 2008). This in turn leads to the activation of the Hap-complex and its targets (Schüller, 2003; Zaman et al, 2008), resulting in an increased TCA cycle activity (Figure 7). Hence, hyperactivation of PKA should restore TCA cycle activity to wild type levels. Deletion of BCY1 is one possibility to uncouple PKA activity from upstream signals, leading to constitutively active PKA (Zaman et al, 2008). We constructed the BCY1 deletion strain in the GCN4 mutant background, but the double mutant grew very poorly. Nevertheless, the determined TCA cycle activity in cultures that grew to sufficient density was indistinguishable from the activity in wild-type cells, suggesting that the GCN4 mutant phenotype is a result of reduced PKA activity (Table II). Thus, we expected increased Snf1 activity in the GCN4 mutant. Deletion of SNF1 in the GCN4 mutant background reduced TCA cycle activity, but did not fully restore wild-type levels (Table II). These results suggest that although Snf1 activity was higher in the GCN4 mutant, increased Snf1 activity accounted only partially for the observed phenotypes. Hence, PKA regulates TCA cycle activity, at least in part, through a Snf1p-independent mechanism (Figure 7). Finally, simultaneous deletion of GCN4 and HAP4 restored TCA cycle activity to levels and HAP4 overexpression in wild-type significantly increased TCA activity to levels comparable with the GCN4 mutant (Table II). This confirms that the Hap-complex is of crucial importance for Gcn4-dependent regulation of TCA cycle flux. Thus, the GCN4 mutant phenotype of higher TCA cycle flux can be readily explained by decreased activity of PKA and increased activity of Snf1, which impinge on the Hap-complex to regulate flux through the TCA cycle (Figure 7), thereby validating our hypothesis derived from the transcript data analysis. In the biological context, the observed positive feedback loops between PKA and Gcn4 might be advantageous for the cell, due to the interlinkage of the stress response triggered by the lack of amino acids and the substrate response triggered by the highly repressive carbon substrate glucose, as both processes are at least partially dependent on PKA.
Figure 7.
Signaling cascades involving Gcn4. Known signaling cascade involving Gcn4 (Schüller, 2003; Slattery et al, 2008; Zaman et al, 2008) and the here revealed Gcn4 signaling cascade of TCA cycle gene expression.
Table 2. Relative TCA cycle flux to mitochondrial oxaloacetate in double deletion strains and HAP4 overexpression strains.
| Strains | Relative TCA cycle flux |
|
|---|---|---|
| Value | Error | |
| Wild type | 0.00 | 0.01 |
| Δgcn4 | 0.13 | 0.03 |
| Δgcn4Δbcy1 | 0.01 | ND |
| Δgcn4Δsnf1 | 0.05 | 0.01 |
| Δgcn4Δhap4 | 0.01 | 0.02 |
| Wild type with empty plasmid | 0.01 | 0.00 |
| Wild type with RPS2-HAP4 | 0.13 | 0.01 |
| Values are determined with 13C-flux analysis as one minus ratio ‘mitochondrial oxaloacetate derived through anaplerosis’ (Blank and Sauer, 2004) (Supplementary Figure 1, Supplementary Table 1). Error ranges were calculated from at least two independent samples. | ||
| ‘ND’ stands for not determined. | ||
Discussion
Starting from the currently largest set of 13C-based flux distributions, we identified networks of individual transcription factors that control metabolic pathway activity. These networks of active metabolic control have the following properties. First, they are highly condition dependent, as at most four transcription factors control the same metabolic flux distribution under more than one growth conditions. Second, they focus almost exclusively on the TCA cycle, thereby controlling the switch between respiratory and fermentative metabolism, which is consistent with more limited transcription factor deletion studies in bacteria (Fischer and Sauer, 2005; Perrenoud and Sauer, 2005; Nanchen et al, 2008). Third, with four to 14 active transcription factors, they are small compared with gene regulation networks that were obtained from expression and DNA binding data.
One of the first large-scale studies to monitor genome-wide gene function with growth rate as a functional readout was performed by Giaever et al (2002). Compared with our results, they found more transcription factors that control cellular function, mainly because growth rate is a more general readout on function than metabolic fluxes, as essentially all cellular processes can affect growth rate. Corroborating our finding of a major discrepancy between the responsive gene regulatory network and the network that actively controls function under a given condition, Giaever et al (2002) found that only 7% of the gene expression-based predicted phenotypes were indeed detected at the level of growth rate. Thus, cellular functions are relatively robust to altered gene expression during steady-state exponential growth.
For the metabolic network studied here, robustness is also apparent from the fact that upregulated TCA cycle fluxes were not sufficient to achieve full respiratory metabolism with absent or low ethanol formation. Several explanations could potentially explain the observed robustness. First, the results might be condition specific, for example, the chosen carbon substrates might require only a small set of transcription factors to control the flux distribution. Second, other regulation mechanisms such as post-transcriptional modifications might actually be the primary flux controlling elements (Heinemann and Sauer, 2010). Third, the redundancy of transcriptional networks (Stelling et al, 2004) might mask the effect of single transcription factor deletions. While we cannot entirely rule out transcription factor redundancy, none of the three tested double and triple transcription factor deletion mutants exhibited a noticeable TCA cycle flux impact; hence, argue against redundancy. Fourth, environmental signals might be transmitted by different signaling pathways to several transcription factors, whose orchestrated action on multiple target genes is necessary to achieve a functional flux response. This latter hypothesis would explain why several transcription factors exert flux effects on the same pathway, but each flux effect is relatively small, as further, coordinated manipulations would be necessary to further increase the respiratory flux. This idea is supported by the observation that combined disruption of two glucose signaling pathways shifts metabolism toward respiration, whereas the single signaling pathway disruptions had no effects (Kuemmel et al, 2010). While likely several reasons contribute to the observed robustness, the fourth hypothesis appears to be the most probable one.
In contrast to the above robustness of fermentative metabolism, S. cerevisiae appears to have no back up for transcription factors that are critical to sustain respiration. During partly respiratory metabolism on galactose, we show that that single transcription factor deletions can essentially abolish the TCA cycle flux, and thus respiration. This fragility of respiration and the preferred fermentative mode of energy production suggest that yeast is a good model for the so-called Warburg effect in many human cancers (Warburg, 1956). Thus, synthetic lethal screens (Costanzo et al, 2010) and large-scale yeast omics data combined with metabolic and hierarchical control analysis (ter Kuile and Westerhoff, 2001; Fell, 2005) or other modeling approaches have the potential to identify key mechanisms and potential drug targets that prevent this metabolic shift during cancer development. Beyond knowledge of regulatory network architecture, our findings demonstrate the importance of identifying and quantifying the extent to which regulatory effectors alter cellular function.
Materials and methods
Strains, medium and cultivation condition
S. cerevisiae wild-type FY4 MATa (Winston et al, 1995) (kindly provided by Fred Winston) was used as wild type. The single deletion strains (Supplementary Table 9) were constructed as whole gene deletion by using a KanMX4 cassette in the prototroph background of FY4 MATa (Winston et al, 1995) (kindly provided by Charlie Boone), which is isogenic to the sequenced S288C strain. All double deletion strains were constructed by crossing the MATa and the MATalpha single deletion strains, except of the Δgcn4Δbcy1 strain. This strain was constructed using a NatMX4 cassette. For overexpression, we used the pRS41H plasmid (Taxis and Knop, 2006) (kindly provided by Eckhard Boles). For overexpression of HAP4, a RPS2 (promotor)–HAP4 (overexpressed gene)–CYC1 (terminator) construct was cloned into the pRS41H plasmid.
Liquid cultivations were carried out in minimal medium batch cultures as described by Blank and Sauer (2004) with 10 g/l glucose or galactose. Adjustments for environmental stress conditions: pH 3.75 was achieved with sulfuric acid (pH 3.75 condition), 0.8 mol/l sorbitol was added to the medium (0.8 M sorbitol condition), ammonium sulfate was substituted with the same amount (mol/mol) of urea (urea condition) and glucose was substituted with the same amount (g/g) of galactose (galactose condition). For the overexpression strain, 300 mg/l hygromycin B (Invitrogen, Basel, Switzerland) was added. Precultures were always grown in glucose minimal medium. Culture aliquots for transcript, enzyme and flux ratio analysis were always harvested during mid-exponential growth phase at an optical density at 600 (OD600) of OD600 0.5–1.2 following a standardized growth curve.
FY4 was freshly plated from a glycerol stock on a YPD (1% (w/v) yeast extract, 2% (w/v) peptone and 2% (w/v) glucose) plate (2% agar), and the deletion strains were freshly plated from a glycerol stock on a YPD plate containing 300 μg/ml geneticin (G418) (Gibco, Paisley, UK). Liquid precultures were inoculated from YPD plates. Cultivations were performed in 500-ml shake flasks with a culture volume of 50 ml, at 30°C and 300 r.p.m. in a shaker with 50-mm shaking amplitude (proteome measurement), or in 96-deep-well plates (Duetz et al, 2000) (Kuehner AG, Birsfeld, Switzerland) with a culture volume of 1.2 ml, at 30°C and 300 r.p.m. in a shaker with 50-mm shaking amplitude (transcriptome, flux ratio and physiology measurement). To improve mixing, a single 4-mm diameter glass bead (Sigma-Aldrich, Buchs, Switzerland) was added to each well.
Specific growth rates were determined from at least three independent cultures and at least six OD600 data points during the exponential growth phase per culture, measured with a spectra-photometer (Molecular Devices, Sunnyvale, USA).
The minimal medium for the flux experiments contained a mixture of 20% [U-13C]-labeled glucose (13C enrichment ⩾99%, Cambridge Isotope Laboratories, Andover, USA) or galactose (13C enrichment ⩾98%, Omnicron Biochemicals, South Bend, USA) and 80% naturally labeled glucose or galactose, respectively (or 100% [C1-13C]-labeled glucose (13C enrichment ⩾99%, Cambridge Isotope Laboratories)).
Flux ratio analysis
Flux analysis was performed as described by Blank and Sauer (2004). 13C-labeled cultures (20% U-13C) were harvested during mid-exponential growth (OD600 0.5–1.2). The cells were washed three times with ddH2O and stored at −20°C for a gas chromatography-mass spectrometry analysis. Samples for gas chromatography-mass spectrometry analysis were prepared as followed: the frozen cell pellet was hydrolyzed with 6 mol/l HCI for 12 h at 105°C. The samples were dried at 95 °C under a constant air stream. They were derivatized using 20 μl of the solvent DMF (Sigma-Aldrich) and 20 μl of the derivatization agent N-(tert-butyldimethylsilyl)-N-methyl-trifluoroacetamide with 1% tert-butyldimethylchlorosilane (Sigma-Aldrich) for 1 h at 85°C. The mass isotopomer distributions of the protein-bound amino acids were measured with a 6890N GC system (Agilent Technologies, Santa Clara, USA) combined with a 5973 Inert XL MS system (Agilent Technologies). Flux ratios were determined from the mass isotopomer distribution of the protein-bound amino acids with the software FiatFlux (Zamboni et al, 2005), using the analytical equations developed by Blank and Sauer (2004). For the ‘mit oxaloacetate from anaplerosis’ flux ratio, Equation (3) from Blank and Sauer (2004), implemented in the software FiatFlux (Zamboni et al, 2005), was applied (Supplementary Figure 2):
where 2-oxoglutarate25 is the C2–C5 fragment of 2-oxoglutarate; glc1U is one carbon glucose fragments; glc2U are two carbon glucose fragments.
Flux ratio significance cutoff was >10%. The exceptions were ‘mit oxaloacetate from anaplerosis’ and ‘P-enol-pyruvate from cyt oxaloacetate’, where a difference from the wild type >5% was considered as significant, as technical accuracy for these ratios are very good (1.6% instead of >3.0%).
Transcriptome data
For transcriptome analysis, harvesting, extraction and DNase digestion of mRNA aliquotes were performed by the mechanical disruption protocol of the RNAesy Mini Kit (50) (Qiagen, Rapperswil, Switzerland). RNA samples were reverse-transcribed into double-stranded cDNA with One-Cycle cDNA Synthesis Kit (Affymetrix Inc., P/N 900431, Santa Clara, CA). The double-stranded cDNA was purified using a Sample Cleanup Module (Affymetrix Inc., P/N 900371). The purified double-stranded cDNA was in vitro transcribed in the presence of biotin-labeled nucleotides using a IVT Labeling Kit (Affymetrix Inc., P/N 900449). The biotinylated cRNA was purified using a Sample Cleanup Module (Affymetrix Inc., P/N 900371), and its quality and quantity were determined using NanoDrop ND 1000 and Bioanalyzer 2100, respectively. Biotin-labeled cRNA samples were fragmented randomly to 35–200 bp at 94°C in fragmentation buffer (Affymetrix Inc., P/N 900371) and were suspended in 100 μl of hybridization mix (Affymetrix Inc., P/N 900720) containing a hybridization control and control oligonucleotide B2 (Affymetrix Inc., P/N 900454). Samples were hybridized to GeneChip Yeast Genome 2.0 arrays for 16 h at 45°C. Arrays were then washed using an Affymetrix Fluidics Station 450 FS450 0003 protocol. An Affymetrix GeneChip Scanner 3000 (Affymetrix Inc.) was used to determine the fluorescent intensity emitted by the labeled target. Raw data are stored in GEO (GSE19569, and GSE24057).
Proteome data
For proteome analysis, the targeted proteomics protocol as described by Picotti et al (2008, 2009) was applied. Cultures were harvested at mid-exponential growth and washed twice with 4°C cold washing buffer (20 mmol/l Hepes, 2 mmol/l EDTA, pH 7.5). The samples were shock-frozen in liquid nitrogen and stored at −80°C. Cell pellets were disrupted mechanically by vortexing in the presence of glass beads, and proteins were precipitated with −20°C cold acetone. The protein concentration was determined with a Bradford assay (Biorad, Munich, Germany). Fifty μg proteins were mixed with 50 μg 100% [15N]-labeled protein as internal standard (Picotti et al, 2009). Sulfur bridges were reduced with dithiothreitol, blocked with iodoacetamide and proteins were digested with trypsin (Promega, Madison, USA) (1 μg trypsin per 100 μg protein). The resulting peptides were cleaned with a Sep-Pak tC18 (50 mg) reverse-phase cartridge (Waters, Milford, USA). The desalting solution was 0.1% (v/v) formic acid water mixture, and the peptides were eluted from the cartridge with 80% (v/v) acetonitrile water mixture. They were dried under vacuum and resuspended in 0.1% formic acid. Proteins were quantified on a nano-LC-MS/MS system consisting of a Tempo nano LC system (Applied Biosystems, Foster City, USA) and a 4000Qtrap (MSD—Sciex, Applied Biosystems), operated in MS/MS mode. Raw tandem mass spectrometry data have been deposited in the publicly accessible repository of proteomics data PeptideAtlas (www.mrmatlas.org, Picotti et al (2008)), and can be browsed using the yeast genome database (Cherry et al, 1998) accession names.
Statistical analysis
For transcriptome analysis, the Affymetrix CEL files were processed using R (version 2.8.0; http://www.r-project.org/) and the Bioconductor affy package (Gautier et al, 2004). Probe intensities were normalized for background by using the robust multiarray average method (Irizarry et al, 2003), using only perfect match probes. Normalization was performed using the qsplines algorithm (Workman et al, 2002). Gene expression values were calculated using the Li and Wong (2001) expression index calculation method. The P-values for proteome analysis were calculated with a two-tailed heteroscedastic Student's t-test. Predictive fidelity was calculated based on binding and expression data from the Yeastract database (Teixeira et al, 2006), thereby a transcription factor was counted as potentially flux distribution controlling when it had a target in a certain pathway. For assessing the predictive fidelity, we calculated true-positive predictions (transcription factor that controls flux and has x target gene in the controlled pathway) and false-negative predictions (transcription factor that controls flux but had less than x target gene in the controlled pathway). We varied x between one and the number of target genes that were necessary to achieve 100% true-positive predictions. For prediction of differentially activated transcription factors, the differential gene expression for pairwise comparisons (mutant versus wild type) was assessed using a two-tailed heteroscedastic Student's t-test. The activity of a transcription factor was assessed by using the scoring system described in Oliveira et al (2008). The transcriptional regulatory network derived from Yeastract database (Teixeira et al, 2006) (documented direct only; 20/01/2008) was used as topology for transcription factor—gene interactions. Gene nodes were scored with P-values, whereas information on fold change was used to determine up- or downregulated subnetwork topologies. A transcription factor with z-score ⩾2 is considered to have significantly changed activity.
Supplementary Material
Supplementary Figures S1-2, Supplementary Tables S1, S9
Acknowledgments
We thank Owen Ryan from Charlie Boone's Lab (University of Toronto) for constructing and providing the transcription factor mutants; Fabian Rudolf (ETH Zurich) for the help by constructing the overexpression plasmid; Eckhard Boles (Goethe University of Frankfurt) for providing the pRS41H plasmid; Marian B Carlson (Columbia University) and Hans Ronne (Uppsala University) for providing NRG1/2, MSN2/4 double and MIG1/2/3 triple mutants. For financial support, SMF is grateful to the Competence Center for Systems Physiology and Metabolic Diseases. PP is the recipient of an intra-European Marie Curie Fellowship. This project was funded by the Swiss initiative for systems biology (SystemsX.ch) project YeastX.
Author contributions: SMF designed the study, performed flux, transcript, proteom experiments, data analysis and drafted the paper. APO performed statistical microarray analysis and helped drafting the paper. SC performed flux experiments of double and triple deletion mutants. PP measured proteom samples. RCD constructed double deletion mutants. US conceived and supervised the study, and helped drafting the paper. All authors read and approved the final paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Balaji S, Babu MM, Iyer LM, Luscombe NM, Aravind L (2006) Comprehensive analysis of combinatorial regulation using the transcriptional regulatory network of yeast. J Mol Biol 360: 213–227 [DOI] [PubMed] [Google Scholar]
- Blank LM, Sauer U (2004) TCA cycle activity in Saccharomyces cerevisiae is a function of the environmentally determined specific growth and glucose uptake rates. Microbiol 150: 1085–1093 [DOI] [PubMed] [Google Scholar]
- Bonneau R (2008) Learning biological networks: from modules to dynamics. Nat Chem Biol 4: 658–664 [DOI] [PubMed] [Google Scholar]
- Bro C, Knudsen S, Regensberg B, Olsson L, Nielsen J (2005) Improvement of galactose uptake in Saccaromyces cerevisiae through overexpression of phosphoglucomutase: Example of transcript analysis as a tool in inverse metabolic engineering. Appl Environ Microbiol 71: 6465–6472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers A, Packham EA, Graham IR (1995) Control of glycolytic gene expression in budding yeast (Saccharomyces cerevisiae). Curr Genet 29: 1–9 [DOI] [PubMed] [Google Scholar]
- Cherry JM, Adler C, Ball C, Chervitz SA, Dwight SS, Hester ET, Jia Y, Juvik G, Roe T, Schroeder M, Weng S, Botstein D (1998) SGD: Saccharomyces genome database. Nucleic Acids Res 26: 73–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JL, Toufighi K, Mostafavi S, Prinz J, St Onge RP, VanderSluis B, Makhnevych T, Vizeacoumar FJ, Alizadeh S, Bahr S, Brost RL, Chen Y, Cokol M et al. (2010) The genetic landscape of a cell. Science 327: 425–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csete ME, Doyle JC (2002) Reverse engineering of biological complexity. Science 295: 1664–1669 [DOI] [PubMed] [Google Scholar]
- Duetz WA, Rüedi L, Hermann R, O'Conner K, Büchs J, Witholt B (2000) Methods for intense aeration, growth, storage and replication of bacterial strains in microtiter plates. Appl Environ Microbiol 66: 2641–2646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fell DA (2005) Enzymes, metabolites and fluxes. J Exp Bot 56: 267–272 [DOI] [PubMed] [Google Scholar]
- Fendt SM, Sauer U (2010) Transcriptional regulation of respiration in yeast metabolizing differently repressive carbon substrates. BMC Syst Biol 4: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer E, Sauer U (2005) Large-scale in vivo flux analysis shows rigidity and suboptimal performance of Bacillus subtilis metabolism. Nat Genet 37: 636–640 [DOI] [PubMed] [Google Scholar]
- Gautier L, Cope L, Bolstad BM, Irizarry RA (2004) Affy-analysis of Affymetrix Gene Chip data at the probe level. Bioinformatics 20: 307–315 [DOI] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Véronneau S, Dow S, Lucau-Danila A, Anderson K, André B, Arkin AP, Astromoff A, El-Bakkoury M, Bangham R, Benito R, Brachat S, Campanaro S, Curtiss M, Davis K, Deutschbauer A et al. (2002) Functional profiling of the Saccharomyces cerevisiae genome. Nature 418: 387–391 [DOI] [PubMed] [Google Scholar]
- Gitter A, Siegfried Z, Klutstein M, Fornes O, Oliva B, Simon I, Bar-Joseph Z (2009) Backup in gene regulatory networks explains differences between binding and knockout results. Mol Syst Biol 5: 276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannum C, Kulaeva OI, Sun H, Urbanowski JL, Wendus A, Stillman DJ, Rolfes RJ (2002) Functional mapping of Bas2. Identification of activation and Bas1-interaction domains. J Biol Chem 277: 34003–34009 [DOI] [PubMed] [Google Scholar]
- Harbison CT, Gordon DB, Ihn Lee T, Rinaldi N, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, Jennings EG, Zeitlinger J, Pokholok DK, Kellis M, Rolfe PA, Takusagawa KT, Lander ES, Gifford DK, Fraenkel E, Young RA. et al. (2004) Transcriptional regulatory code of a eukaryotic genome. Nature 431: 99–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haurie V, Sagliocco F, Boucherie H (2004) Dissecting regulatory networks by means of two-dimensional gel electrophoresis: application to the study of the diauxic shift in the yeast Saccharomyces cerevisiae. Proteomics 4: 364–373 [DOI] [PubMed] [Google Scholar]
- Haynes KA, Silver PA (2009) Eukaryotic systems broaden the scope of synthetic biology. J Cell Biol 187: 589–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedbacker K, Townley R, Carlson M (2004) Cyclic AMP-dependent protein kinase 595 regulates the subcellular localization of Snf1-Sip1 protein kinase. Mol Cell Biol 24: 1836–1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann M, Sauer U (2010) Systems biology of microbial metabolism. Curr Opin Microbiol 13: 337–343 [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG (2005) Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol 59: 407–450 [DOI] [PubMed] [Google Scholar]
- Hu Z, Killon PJ, Iyer VR (2007) Genetic reconstruction of a functional transcriptional regulatory network. Nat Genet 39: 683–687 [DOI] [PubMed] [Google Scholar]
- Ihmels J, Friedlander G, Bergmann S, Sarig O, Ziv Y, Barkai N (2002) Revealing modular organization in the yeast transcriptional network. Nat Genet 31: 370–377 [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264 [DOI] [PubMed] [Google Scholar]
- Karlebach G, Shamir R (2008) Modelling and analysis of gene regulatory networks. Nat Rev Mol Cell Biol 9: 770–780 [DOI] [PubMed] [Google Scholar]
- Küpfer L, Sauer U, Blank LM (2005) Metabolic functions of duplicate genes in Saccharomyces cerevisiae. Genome Res 15: 1421–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreeger PK, Lauffenburger DA (2010) Cancer systems biology: a network modeling perspective. Carcinogenesis 31: 2–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuemmel A, Ewald JC, Fendt SM, Jol SJ, Picotti P, Aebersold R, Sauer U, Zamboni N, Heinemann M (2010) Differential glucose repression in common yeast strains in response to hxk2 deletion. FEMS Yeast Res 10: 322–332 [DOI] [PubMed] [Google Scholar]
- Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannette NM, Harbison CT, Thompson CG, Simon I, Zeitlinger J, Jennings EG, Murray HL, Gordon DB, Ren B, Wyrick JJ, Tagne JB, Volkert TL, Fraenkel E, Gifford DK et al. (2002) Transcriptional regulatory network in Saccharomyces cerevisiae. Science 298: 799–804 [DOI] [PubMed] [Google Scholar]
- Li C, Wong WH (2001) Model-based analysis of ologonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA 98: 31–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luscombe NM, Babu MM, Yu H, Snyder M, Teichmann SA, Gerstein M (2004) Genomic analysis of regulatory network dynamics reveals large topological changes. Nature 431: 308–312 [DOI] [PubMed] [Google Scholar]
- Nanchen A, Schicker A, Revelles O, Sauer U (2008) Cyclic AMP-dependent catabolite repression is the dominant control mechanism of metabolic fluxes under glucose limitation in Escherichia coli. J Bacteriol 190: 2323–2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira AP, Patil KR, Nielsen J (2008) Architecture of transcriptional regulatory circuits is knitted over the topology of bio-molecular interaction networks. BMC Syst Biol 2: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papin JA, Hunter T, Palsson BO, Subremaniam S (2005) Reconstruction of cellular signalling networks and analysis of their properties. Nat Rev Mol Cell Biol 6: 99–111 [DOI] [PubMed] [Google Scholar]
- Perrenoud A, Sauer U (2005) Impact of global transcriptional regulation by ArcA, ArcB, Cra, Crp, Fnr, and Mlc on glucose catabolism in Escherichia coli. J Bacteriol 187: 3171–3179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picotti P, Bodenmiller B, Mueller LN, Domon B, Aebersold R (2009) Full dynamic range proteome analysis of S. cerevisiae by targeted proteomics. Cell 38: 795–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picotti P, Lam H, Campbell D, Deutsch EW, Mirzaei H, Ranish J, Domon B, Aebersold R (2008) A database of mass spectrometric assays for the yeast proteome. Nat Methods 5: 913–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer U (2006) Metabolic networks in motion: 13C-based flux analysis. Mol Sys Biol 2: 62; DOI: 10.1038/msb4100109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüller HJ (2003) Transcriptional control of nonfermentative metabolism in the yeast Saccharomyces cerevisiae. Curr Genet 43: 139–160 [DOI] [PubMed] [Google Scholar]
- Segal E, Shapira M, Regev A, Peer D, Botstein D, Koller D, Friedman N (2003) Module networks: Identifying regulatory modules and their condition-specific regulators from gene expression data. Nat Genet 34: 166–176 [DOI] [PubMed] [Google Scholar]
- Slattery MG, Liko D, Heideman W (2008) Protein kinase A, TOR, and glucose transport control the response to nutrient repletion in Saccharomyces cerevisiae. Eukaryot Cell 7: 538–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder M, Gallagher JE (2009) Systems biology from a yeast omics perspective. FEBS Lett 583: 3895–3899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Som I, Mitsch RN, Urbanowski JL, Rolfes RJ (2005) DNA-bound Bas1 recruits Pho2 to activate ADE genes in Saccharomyces cerevisiae. Eukaryot Cell 4: 1725–1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelling J, Sauer U, Szallasi Z, Doyle FJ, Doyle J (2004) Robustness of cellular functions. Cell 118: 675–685 [DOI] [PubMed] [Google Scholar]
- Taxis C, Knop M (2006) System of centromeric, episomal, and integrative vectors based on drug resistance markers for Saccharomyces cerevisiae. BioTechniques 40: 73–77 [DOI] [PubMed] [Google Scholar]
- Teixeira MC, Monteiro P, Jain P, Tenreiro S, Fernandes AR, Mira NP, Alenquer M, Oliveira AL, Sá-Correira I (2006) The YEASTRACT database: A tool for the analysis of transcription regulatory associations in Saccharomyces cerevisiae. Nucleic Acids Res 34: 446–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Kuile BH, Westerhoff HV (2001) Transcriptome meets metabolome: hierarchical and metabolic regulation of the glycolytic pathway. FEBS Lett 500: 169–171 [DOI] [PubMed] [Google Scholar]
- Turcotte B, Liang XB, Robert F, Soontorngun N (2009) Transcriptional regulation of nonfermentable carbon utilization in budding yeast. FEMS Yeast Res 10: 2–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O (1956) On respiratory impairment in cancer cells. Science 124: 269–270 [PubMed] [Google Scholar]
- Winston F, Dollard C, Ricupero-Hovasse SL (1995) Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 11: 53–55 [DOI] [PubMed] [Google Scholar]
- Workman C, Jensen LJ, Jarmer H, Berka R, Gautier L, Nielser HB, Saxild HH, Nielsen C, Brunak S, Knudsen S (2002) A new non-linear normalization method for reducing variability in DNA microarray experiments. Genome Biol 3: 0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman CT, Mak HC, McCuine S, Tagne JB, M A, Ozier O, Begley TJ, Samson LD, Ideker T (2006) A systems approach to mapping DNA damage response pathways. Science 312: 1054–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Gerstein M (2006) Genomic analysis of the hierarchical structure of regulatory networks. Proc Natl Acad Sci USA 103: 14724–14731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaman S, Lippman SI, Zhao X, Broach JR (2008) How Saccharomyces responds to nutrients. Annu Rev Genet 42: 27–81 [DOI] [PubMed] [Google Scholar]
- Zamboni N, Fendt SM, Ruehl M, Sauer U (2009) 13C-based metabolic flux analysis. Nat Protoc 4: 878–892 [DOI] [PubMed] [Google Scholar]
- Zamboni N, Fischer E, Sauer U (2005) FiatFlux–a software for metabolic flux analysis from 13C—glucose experiments. BMC Bioinformatics 25: 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Winston F (2001) NRG1 is required for glucose repression of the SUC2 and GAL genes of Saccharomyces cerevisiae. BMC Genet 2: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1-2, Supplementary Tables S1, S9