Abstract
Bacterial genomes encode hundreds to thousands of enzymes, most of which are specialized for particular functions. However, most enzymes have inefficient promiscuous activities, as well, that generally serve no purpose. Promiscuous reactions can be patched together to form multistep metabolic pathways. Mutations that increase expression or activity of enzymes in such serendipitous pathways can elevate flux through the pathway to a physiologically significant level. In this study, we describe the discovery of three serendipitous pathways that allow synthesis of pyridoxal-5′-phosphate (PLP) in a strain of E. coli that lacks 4-phosphoerythronate (4PE) dehydrogenase (PdxB) when one of seven different genes is overexpressed. We have characterized one of these pathways in detail. This pathway diverts material from serine biosynthesis and generates an intermediate in the normal PLP synthesis pathway downstream of the block caused by lack of PdxB. Steps in the pathway are catalyzed by a protein of unknown function, a broad-specificity enzyme whose physiological role is unknown, and a promiscuous activity of an enzyme that normally serves another function. One step in the pathway may be non-enzymatic.
Keywords: metabolic bypass, multicopy suppression, promiscuity, pyridoxal-5′-phosphate, serendipitous pathway
Introduction
Most enzymes have evolved to be highly efficient catalysts for specific chemical reactions. However, many enzymes also have inefficient promiscuous activities as a result of the assemblage of highly reactive catalytic residues and cofactors in active sites. Promiscuous activities are generally orders of magnitude less efficient than well-evolved activities (O’Brien and Herschlag, 1998, 2001; Wang et al, 2003; Taylor Ringia et al, 2004), but may enhance reaction rates by orders of magnitude relative to those of uncatalyzed reactions (O’Brien and Herschlag, 1998, 2001). Thus, promiscuous activities provide a reservoir of novel catalytic activities that can be recruited to serve new functions.
The evolutionary potential of promiscuous enzymes extends beyond the recruitment of single enzymes to serve new functions. Microbes contain hundreds of enzymes—E. coli contains about 1700 (Freilich et al, 2005)—raising the possibility that promiscuous enzymes can be patched together to generate ‘serendipitous’ pathways that are not part of normal metabolism. We distinguish serendipitous pathways from latent or cryptic pathways, which are bona fide pathways involving dedicated enzymes that are produced only under particular environmental circumstances. In contrast, serendipitous pathways are patched together from enzymes that normally serve other functions and are not regulated in a coordinated manner in response to the need to synthesize or degrade a metabolite.
Serendipitous pathways are of interest for several reasons. First, serendipitous pathways patched together from promiscuous activities may provide starting places for evolution of novel pathways (Jensen, 1976) for biodegradation of anthropogenic compounds or synthesis of pharmaceuticals, chemicals and fuels by ‘green’ processes. Second, serendipitous pathways could foster resistance to antibiotics that block metabolic pathways by providing ways around the block. Finally, serendipitous pathways provide an opportunity to explore whether pathways that did not evolve in nature are intrinsically inferior to existing pathways, or were simply pre-empted by prior emergence of something else.
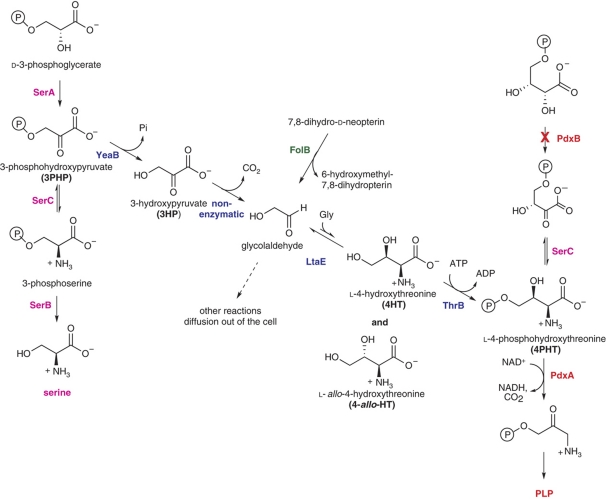
The ASKA collection of E. coli ORFs (Kitagawa et al, 2005) cloned into expression vectors enables a simple approach for identifying promiscuous enzyme activities. Overproduction of an enzyme with a promiscuous activity can generate enough of that activity in favorable cases to replace an enzyme with the corresponding physiological activity. We used this multicopy suppression approach to identify seven genes that restored the ability of an E. coli mutant that lacks 4-phosphoerythronate (4PE) dehydrogenase (PdxB) to grow on M9/glucose. PdxB is required for synthesis of pyridoxal-5′-phosphate (PLP). As E. coli contains at least 60 enzymes that utilize PLP, this cofactor is essential for growth under most conditions. Surprisingly, none of the enzymes encoded by these seven genes has a promiscuous activity that replaces that of PdxB. Rather, overexpression of these genes appears to facilitate serendipitous pathways that generate an intermediate downstream of the block in the PLP synthesis pathway. Genetic complementation experiments allowed us to sort the seven genes into three groups (Figure 1). We have characterized one of these pathways in detail. Reactions in this pathway are catalyzed by a promiscuous activity of an enzyme that serves another function, a broad-specificity enzyme whose physiological role is uncertain, and an enzyme of unknown function. One reaction appears to be non-enzymatic. We emphasize that restoration of growth when this pathway operates is due to a fundamentally different mechanism than those typically responsible for metabolic robustness in microbes. Bacteria that live in changeable environments have evolved metabolic networks that reroute metabolic fluxes by alteration of gene expression and/or enzyme activity to optimize the use of carbon and energy sources in variable environments. These features have been honed by evolution because they increase the fitness of the organism. In contrast, serendipitous pathways—by definition—do not contribute to fitness under normal circumstances. There is no selective pressure to increase the efficiency of such pathways until they become important for fitness, either because of a change in environmental conditions or a mutation that raises the level of flux to a physiologically significant level. In such cases, further genetic changes, including gene duplications, insertions or deletions, and point mutations, can result in evolution of the dedicated, efficient and highly specific enzymes characteristic of well-evolved metabolic pathways.
Figure 1.
The PLP synthesis pathway in E. coli, showing the positions at which serendipitous pathways facilitated by overproduction of the indicated enzymes tie into the normal pathway.
Results
Overexpression of seven different genes restores growth of a strain of E. coli lacking PdxB on M9/glucose
PdxB catalyzes the second step in the PLP synthesis pathway (Figure 1). A strain in which pdxB is deleted cannot grow on minimal medium at 37°C because it cannot synthesize PLP. However, it can grow slowly at 30°C, suggesting that enough PLP can be synthesized by a different route to allow slow growth (Lam and Winkler, 1990). We carried out a multicopy suppression screen with the intention of identifying enzymes for which overproduction provides sufficient promiscuous 4PE dehydrogenase activity to replace PdxB. An expression library containing 3276 ORFs from E. coli but lacking pdxB was assembled using plasmids isolated from clones in the ASKA collection (Kitagawa et al, 2005) and introduced into a ΔpdxB∷kan strain (hereafter referred to as the ΔpdxB strain). Plasmids from 28 colonies that grew on M9/glucose were recovered and the ORF carried by each was identified by sequencing. Remarkably, we found that overexpression of seven different genes restored growth of the ΔpdxB strain on M9/glucose (see Table I, Supplementary Figure 1 and Supplementary Table II). (As three of the seven genes were found only once, it is possible that the population was under-sampled and that additional genes that restore growth remain to be discovered.) Three of the seven genes (hisB, php and yjbQ) enable growth on both liquid and solid medium; the others enable growth only on solid medium.
Table 1. Functions of proteins encoded by genes whose overexpression allows growth of the ΔpdxB strain on glucose.
| Protein | Function |
|---|---|
| aAlso known as NudL. | |
| PdxA | Phosphohydroxythreonine dehydrogenase/decarboxylase |
| AroB | Dehydroquinate synthase |
| ThrB | Homoserine kinase |
| YeaBa | Predicted NUDIX hydrolase |
| HisB | Imidazole-glycerol-phosphate dehydratase/histidinol phosphatase |
| Php | Predicted metallohydrolase |
| YjbQ | Unknown |
Only two of the seven genes that complement the ΔpdxB strain encode enzymes that have dehydrogenase activity. 4-Hydroxy-L-threonine (4HT) dehydrogenase (PdxA) catalyzes a dehydrogenation reaction downstream of the reaction catalyzed by PdxB. Dehydroquinate synthase (AroB) catalyzes a complex reaction in which tightly bound NAD+ is reduced and then later oxidized during the multistep conversion of 3-deoxy-D-arabino-heptulosonic acid 7-phosphate to dehydroquinate (Carpenter et al, 1998). Four of the five remaining enzymes are known or predicted to catalyze dehydration, phosphoryl transfer or hydrolytic reactions. YjbQ exhibits low-level thiamine phosphate synthase activity (Morett et al, 2008), although this is not believed to be its physiological function.
Assays for 4PE dehydrogenase activity in the seven different enzymes that, when overproduced, restore growth of the ΔpdxB strain on glucose
Generation of a high level of an enzyme with promiscuous 4PE dehydrogenase activity is the most obvious mechanism by which overexpression of a gene might complement the ΔpdxB strain. However, ThrB, YeaB, HisB and Php showed no oxidation of 4PE even after prolonged incubation. YjbQ could not be assayed because it was difficult to obtain soluble protein.
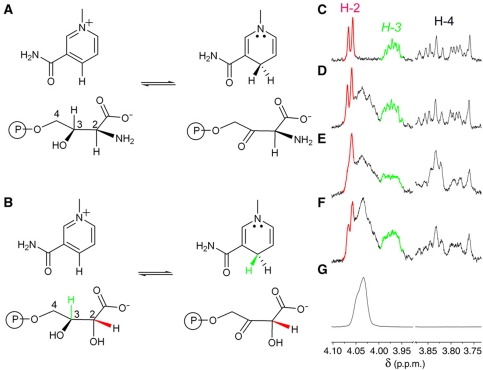
PdxA oxidizes 4PE slowly, with a kcat of 0.020±0.002 s−1, a KM of 150±13 μM and a kcat/KM of 138±19 M−1 s−1, as measured by following reduction of NAD+. PdxB oxidizes 4PE at C-2. We expected that PdxA would oxidize 4PE at C-3, rather than C-2, based on a comparison of the structures of 4PE and 4-phosphohydroxythreonine (4PHT), the normal substrate for PdxA (Figure 2A and B). As the reaction is thermodynamically uphill and the product does not accumulate in sufficient concentrations for characterization by NMR, we determined the site of oxidation indirectly by equilibrating 4PE with NAD+ in the presence of PdxA and excess [4R-2H]NADH or [4S-2H]NADH. The appearance of deuterium in 4PE due to transfer of deuteride to the product in the reverse reaction (see Scheme 1) allowed identification of the site of oxidation. After incubation of 4PE with [4R-2H]NADH and PdxA, the signal due to the C-3 proton of 4PE diminishes, and the signals due to the C-2 and C-4 protons are altered because they are no longer split by a proton on C-3 (Figure 2E). None of the signals is altered in the presence of NADH (Figure 2D) or [4S-2H]NADH (Figure 2F). Thus, oxidation of 4PE occurs at C-3 rather than at C-2, and therefore this activity of PdxA cannot substitute for the activity of PdxB.
Figure 2.
PdxA oxidizes 4PE at C-3. (A) Reaction of PdxA with 4PHT; (B) reaction of PdxA with 4PE; (C–F) 1H-NMR spectra of 4PE (1 mM) incubated with PdxA (30 μM) in the presence of (C) 0.2 mM NAD+; (D) 0.2 mM NAD+ and 1.5 mM NADH; (E) 0.2 mM NAD+ and 1.5 mM [4R-2H]NADH; and (F) 0.2 mM NAD+ and 1.5 mM [4S-2H]NADH; (G) 1H-NMR spectrum of NADH. NAD+ has no interfering peaks in the region shown here. Signals due to the protons at C-2 and C-3 of 4PE are highlighted in red and green, respectively.
Scheme 1.
Equilibration of 4PE with NAD+ and 4-2H-NADH in the presence of an enzyme with 4PE dehydrogenase activity results in incorporation of 2H into 4PE at the position of oxidation by the enzyme.
AroB also oxidizes 4PE, but carries out only a single turnover (data not shown). This is not surprising; during the normal catalytic cycle of AroB, tightly bound NAD+ is reduced to NADH. After elimination of phosphate from the oxidized intermediate, the NADH transfers hydride back to the substrate, regenerating NAD+ at the active site (Carpenter et al, 1998). Oxidation of 4PE would reduce the tightly bound NAD+, but would not afford an opportunity for reoxidation, and would inactivate the enzyme. We cannot determine the position at which 4PE is oxidized by AroB using the approach described above because NADH is not released from the enzyme and cannot be replaced with NAD2H. However, this activity is probably not physiologically relevant; genetic experiments to be described below suggest that complementation of the ΔpdxB strain by overexpression of aroB is not due to its ability to oxidize 4PE.
These results suggest that the restoration of growth in the ΔpdxB strain when ThrB, YeaB, HisB, Php or PdxA (and probably AroB) is overproduced cannot be attributed to replacement of the activity of PdxB. Consequently, we explored the possibility that overproduction of these enzymes facilitates one or more serendipitous pathways that allow PLP to be synthesized in the absence of PdxB.
Three serendipitous pathways are revealed by efforts to complement strains lacking other genes in the PLP synthesis pathway
Multicopy suppression experiments using strains lacking other genes in the PLP synthesis pathway were carried out to identify where the serendipitous pathways facilitated by the enzymes listed in Table I feed into the normal PLP synthesis pathway. In general, strains lacking enzymes upstream of the intersection point should grow when a serendipitous pathway is operating, but strains lacking enzymes downstream of the intersection point should not. These experiments were straightforward for most of the strains, but were complicated by the growth requirements of the ΔserC∷kan strain (hereafter referred to as the ΔserC strain). SerC is required for synthesis of both PLP and serine. Furthermore, deletion of serC reduces the expression of aroA, which is encoded by the same operon (Duncan and Coggins, 1986). Consequently, the ΔserC strain requires addition of serine, pyridoxine and products of the shikimate pathway (Lam and Winkler, 1990) to grow on minimal medium. To examine the ability of serendipitous pathways to produce PLP in the ΔserC strain, we added all of these supplements except pyridoxine. As a control, we examined the effect of these supplements on complementation of the ΔpdxB strain. The supplements have no effect on complementation of ΔpdxB by pdxB, hisB, yjbQ and php (Table II). hisB, php and yjbQ fail to complement strains lacking serC, pdxA, pdxJ or pdxH (Table II), suggesting that pathway 2 forms 2-oxo-3-hydroxy-4-phosphobutanoate (OHPB) (Figure 1). However, the supplements inhibit the ability of thrB, yeaB, pdxA and aroB to complement the ΔpdxB strain. This effect was found to be due solely to the presence of serine (Supplementary Table III). A possible explanation is that the pathways facilitated by overexpression of these genes begin with intermediates in serine biosynthesis that are depleted in the presence of serine due to allosteric inhibition of SerA, which catalyzes the first step in serine biosynthesis (Grant et al, 1996). However, addition of serine has pleiotropic effects (Hama et al, 1990, 1991; Grant et al, 1996), so at this point we choose to use this phenotype simply as a characteristic that distinguishes between different serendipitous pathways.
Table 2. Growth on plates containing M9/glucose at 37°C of strains lacking various enzymes in the PLP synthesis pathway when the indicated genes are overexpressed from pTrcHisB.
| Gene | ΔpdxB (JU283) | ΔpdxB (JU283)+suppl.a | ΔserC (JWK0890)+suppl.a | ΔpdxA (JWK0051) | ΔpdxJ (JWK2548) | ΔpdxH (JWK1630) |
|---|---|---|---|---|---|---|
| Strain designations are defined in Supplementary Table I. | ||||||
| aL-Ser (1.0 mM), L-Phe (0.1 mM), L-Tyr (0.1 mM), L-Trp (0.1 mM), p-hydroxybenzoate (1 μM), p-aminobenzoate (1 μM) and 2,3-dihydroxybenzoate (1 μM). | ||||||
| bColonies reached 1 mm diameter in 1–2 days (++++), 3–5 days (+++) or 6–8 days (++). (+) indicates colonies that were <1 mm after 9 days. | ||||||
| pdxB | ++++ b | ++++ | − | − | − | − |
| hisB | ++++ | ++++ | − | − | − | − |
| yjbQ | +++ | +++ | − | − | − | − |
| php | ++++ | ++++ | − | − | − | − |
| thrB | ++++ | + | − | − | − | − |
| yeaB | +++ | + | − | − | − | − |
| pdxA | +++ | ++ | − | ++++ | − | − |
| aroB | +++ | + | − | ++ | − | − |
yeaB and thrB complement the ΔpdxB strain, but do not complement strains lacking PdxA, PdxJ or PdxH. These data suggest that serendipitous pathway 1 feeds into the normal pathway at the level of either OHPB or 4-phosphohydroxythreonine (4PHT). The ambiguity arises because the serine required for growth of the ΔserC strain interferes with the complementation of the ΔpdxB strain by yeaB and thrB. Thus, we cannot determine from these data whether pathway 1 feeds in before or after the reaction catalyzed by SerC. However, biochemical data (see below) suggest that pathway 1 generates 4PHT.
aroB complements the ΔpdxA strain, but fails to complement strains lacking pdxJ or pdxH. We purified AroB from a strain lacking PdxA and determined that AroB has no detectable 4PHT dehydrogenase activity (data not shown). Therefore, complementation of the ΔpdxA strain by overexpression of aroB is not due simply to replacement of the activity of PdxA. These data suggest that pathway 3 forms 1-amino-propan-2-one-3-phosphate.
Identification of enzymes that can catalyze reactions in pathway 1
A possible sequence of reactions for pathway 1 is suggested by the observations that addition of 3-hydroxypyruvate (3HP) (Shimizu and Dempsey, 1978), glycolaldehyde (Tani and Dempsey, 1973) or 4HT (Drewke et al, 1993) can support growth of certain E. coli mutants that lack the ability to make PLP. Isotopic labeling experiments confirmed that carbon atoms from glycolaldehyde (Hill and Spenser, 1973; Hill et al, 1977), glycine (Hill et al, 1987) and 4HT (Hill et al, 1996) and nitrogen atoms from glycine (Hill et al, 1987) were incorporated into pyridoxol. For some time, these compounds were considered as likely intermediates in the PLP synthesis pathway (Shimizu and Dempsey, 1978; Duncan and Coggins, 1986), although eventually the pathway shown in Figure 1 was worked out. We confirmed that our ΔpdxB strain grows on M9/glucose at 37°C in the presence of 3HP, glycolaldehyde and 4HT. Our strain also grows in the presence of 3PHP, an intermediate in serine biosynthesis (Supplementary Table IV).
The first step of the proposed pathway 1 is dephosphorylation of 3PHP (Figure 3). E. coli contains 75 known or predicted phosphatases that might catalyze this reaction. An intriguing possibility was suggested by the observation that overproduction of YeaB, a predicted NUDIX hydrolase, restores growth of the ΔpdxB strain. NUDIX hydrolases typically catalyze hydrolysis of organic pyrophosphates (McLennan, 2006) and thus might have low-level phosphatase activity. We expressed YeaB as a fusion with maltose-binding protein (MalE) to obtain soluble protein. The MalE–YeaB fusion protein indeed has 3PHP phosphatase activity (Table III). Because we had a limited supply of 3PHP, we were not able to determine an accurate KM for 3PHP; as the enzyme appears to be saturated at 2 mM 3PHP, we conclude that the KM for 3PHP must be <2 mM. The 3PHP phosphatase activity was due to YeaB rather than MalE, as a MalE–LacZ fusion protein showed no activity. We note that the non-enzymatic rate for conversion of 3PHP to 3HP is 7.2 × 10−10 M−1 s−1. Thus, even though YeaB is an inefficient enzyme, it elevates the rate above the background rate by a factor of 4 × 107.
Figure 3.
Serendipitous pathway 1 converts 3PHP, an intermediate in the serine biosynthesis pathway, to 4PHT, an intermediate in the PLP synthesis pathway.
Table 3. Kinetic parameters for the activities of enzymes that can catalyze reactions in pathway 1.
| Protein | Substrate | kcat (s−1) | KM (mM) | kcat/KM (M−1 s−1) |
|---|---|---|---|---|
| Substrates involved in pathway 1 are highlighted in bold. | ||||
| aYeaB was expressed as a fusion protein with maltose-binding protein (MalE) to improve its solubility. | ||||
| b(Kuzuyama et al, 2000). | ||||
| MalE–YeaBa | 3PHP | 5.7±0.7 × 10−5 | <2 | >0.028±0.003 |
| Dxs | Pyruvate+D-GAPb | 345 | 0.096 (pyruvate) | 3.6 × 106 |
| 0.24 (D-GAP) | 1.4 × 106 | |||
| 3HP | 0.026±0.001 | 0.050±0.008 | 5.2±0.4 × 102 | |
| SucA | 3HP | 0.0139±0.0003 | 0.72±0.07 | 19.3±1.9 |
| LtaE | L-allo-threonine | 3.2±0.2 | 0.052±0.004 | 6.2±0.5 × 104 |
| L-threonine | 1.1±0.1 | 4.0±0.2 | 2.8±0.2 × 102 | |
| 4HT | 1.44±0.03 | 0.027±0.002 | 5.3±0.4 × 104 | |
| ThrB | Homoserine | 46±4 | 0.12±0.02 | 3.8±0.7 × 105 |
| 4HT | 9.7±0.8 | 2.0±0.1 | 4.8±0.6 × 103 | |
Although YeaB is an inefficient catalyst for conversion of 3PHP to 3HP, the flux through this step should be sufficient to supply PLP. The cytoplasmic concentration of MalB–YeaB is ∼45 μM (based upon the production of 4.7 mg of purified protein from 2.2 g of wet cells). YeaB appears to be saturated at 1 mM 3PHP, a reasonable assumption for its cytoplasmic concentration based upon the concentration of its immediate precursor, 3-phosphoglycerate, which is present at 1.5 mM in E. coli growing on glucose (Bennett et al, 2009). Thus, the flux will be approximately kcat [E]=(4.8 × 10−5 s−1) (45 μM)=7.8 μM h−1 (Bennett et al, 2009). In the Supplementary information, we describe a calculation suggesting that the ΔpdxB strain synthesizes PLP at a rate of ∼5.5 μM h−1 when MalB–YeaB is overexpressed. Thus, the rate of conversion of 3PHP to 3HP by YeaB is comparable to the rate of synthesis of PLP.
We note that there is precedence for the ability of such an inefficient enzyme to supply sufficient flux to replace a critical metabolic enzyme. Patrick and Matsumura (2008) have shown that overexpression of a promiscuous enzyme (I198V glutamine 5-phosphoribosyl-1-pyrophosphate amidotransferase (PurF)) substitutes for a lack of phosphoribosylanthranilate isomerase (TrpF), which is required for tryptophan synthesis. The kcat/KM for the promiscuous TrpF activity of I198V PurF is estimated to be 0.012 M−1 s−1 (Patrick and Matsumura, 2008); the kcat/KM for the 3PHP phosphatase activity of YeaB (>0.024 M−1 s−1) is greater than this.
If YeaB is the only enzyme that converts 3PHP to 3HP, a ΔpdxB ΔyeaB strain should not grow on minimal medium supplemented with 3PHP. However, this strain does grow slowly, producing colonies <1 mm in diameter after 9 days, on M9/glucose (Supplementary Table IV). These results suggest that at least one other enzyme can generate 3HP when 3PHP is present at high levels. As the ΔpdxB strain grows after 4 days when YeaB is overproduced, we conclude that overproduction of YeaB is the most efficient way to siphon material from the serine biosynthesis pathway into pathway 1, but YeaB is not the only enzyme that can produce 3HP from 3PHP.
3HP is proposed to be converted to glycolaldehyde in pathway 1. This conversion might occur by either of two routes. Direct decarboxylation of α-keto acids is catalyzed by enzymes that contain thiamine pyrophosphate (TPP). Alternatively, glycolaldehyde might be formed by initial isomerization of 3HP to the β-keto acid tartronate semialdehyde, followed by decarboxylation of tartronate semialdehyde. To search for enzymes in E. coli that catalyze decarboxylation of 3HP by either route, we attempted to measure acceleration of glycolaldehyde production from 3HP by crude extracts of E. coli K-12 BW25113 in the presence of NADH and alcohol dehydrogenase, which converts glycolaldehyde to ethylene glycol (Barngrover et al, 1981). We were unable to detect any acceleration of the conversion of 3HP to glycolaldehyde. As we could not rule out inefficient catalysis that fails to rise significantly above the background rate, we examined the catalytic abilities of specific candidate enzymes that might convert 3HP to glycolaldehyde.
E. coli has 12 enzymes that are known or predicted to utilize TPP. We purified 10 of these: 1-deoxyxylulose-5-phosphate synthase (Dxs), pyruvate oxidase, transketolase A, transketolase B, glyoxalate carboligase and the predicted oxalyl CoA decarboxylase, as well as the TPP-dependent catalytic components of the multisubunit enzymes pyruvate dehydrogenase, α-ketoglutarate dehydrogenase and both isozymes of acetohydroxy acid synthase. YdbK and 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase (MenD) could not be expressed in soluble form. Only Dxs and SucA, the TPP-dependent catalytic component of the α-ketoglutarate dehydrogenase complex, had detectable 3HP decarboxylase activity (Table III).
To determine whether Dxs, SucA, YdbK or MenD is required for operation of pathway 1, we constructed double-knockout strains lacking pdxB and each of the genes encoding these enzymes. We also constructed a ΔpdxB Δdxs ΔsucA strain to determine whether glycolaldehyde formation is due to a combination of the activities of Dxs and SucA. Each of these strains grew on M9/glucose when YeaB or ThrB was overproduced at 37°C (data not shown). These results suggest that TPP-containing enzymes are not responsible for the formation of glycolaldehyde in pathway 1.
The E. coli genome encodes a hydroxypyruvate isomerase (Hyi) that converts 3HP to tartronate semialdehyde. This enzyme is expressed only when cells are grown on glyoxylate (Ashiuchi and Misono, 1999). We ruled out the possibility that Hyi has a role in pathway 1 by constructing a ΔpdxB Δhyi strain. This strain grows on glucose when either yeaB or thrB is overexpressed or when 3HP is supplied in the medium (data not shown).
We next considered the possibility that conversion of 3HP to glycolaldehyde in pathway 1 might be non-enzymatic. We measured the rate of formation of glycolaldehyde from 3HP by 1H-NMR. 3HP is converted to glycolaldehyde, glycolic acid and erythrulose (by reaction with glycolaldehyde, followed by decarboxylation) (Figure 4). A control experiment showed that glycolaldehyde decomposed under these conditions to unidentified products with a first-order rate constant of 0.0062±0.0003 h−1. The kinetic model shown below was used for numerical simulation of the rate constants for appearance of glycolaldehyde (k1=0.0153±0.0003 h−1), erythrulose (k2=0.0092±0.0005 mM−1 h−1) and glycolic acid (k3=0.0069±0.0003 h−1), assuming that k4=0.0062 h−1, using DynaFit (Kuzmič, 1996). Notably, addition of 0.1 mM TPP had no effect on the rate of formation of glycolaldehyde. Thus, glycolaldehyde formation must occur through the initial formation of tartronate semialdehyde. Decarboxylation of β-keto acids such as tartronate semialdehyde can be accelerated by metal ions (Hedrick and Sallach, 1961). However, addition of Mg++ or Ca++ (1 mM) had no effect on the rate of glycolaldehyde formation. Addition of Cu++, Co++, Ni++, Zn++ or Mn++ (1 mM) actually decreased the rate of glycolaldehyde formation, while concomitantly increasing the rate of glycolate formation.
Figure 4.
Non-enzymatic formation of glycolaldehyde from 3HP. (A) Products formed from 3HP. (B) A portion of the 1H-NMR spectrum of the reaction mixture after incubation for 47 h showing peaks used to quantify reaction components. Green, C2-H of glycolate; blue, C4-H of erythrulose; black, C3-H of 3HP-hydrate; red, C2-H of glycolaldehyde-hydrate. (C) Time course showing disappearance of 3HP (•) and appearance of glycolaldehyde ( ), glycolic acid (
), glycolic acid ( ) and erythrulose (
) and erythrulose ( ) during incubation at 37°C in 50 mM potassium phosphate, pH 7.6.
) during incubation at 37°C in 50 mM potassium phosphate, pH 7.6.
 |
The observed rate of conversion of 3HP to glycolaldehyde appears to be sufficient to supply a significant portion of the glycolaldehyde needed for the production of PLP. We estimate that the ΔpdxB strain synthesizes PLP at a rate of 5.5 μM h−1 when yeaB is overexpressed (Supplementary information). We estimated above that the rate of production of 3HP from 3PHP should be ∼8 μM h−1. At steady state, the rates of production and depletion of 3HP are balanced. As formation of erythrulose is negligible unless the glycolaldehyde concentration exceeds 1 mM, which is unlikely, we can calculate the steady state concentration of 3HP by assuming that the rates of formation and disappearance of 3HP are equal at the steady state. Using the relationship 8 μM h−1=(k1+k3) [3HP]ss, we calculate that [3HP]ss is ∼360 μM. If the concentration of 3HP is 360 μM, the rate of formation of glycolaldehyde will be 5.5 μM h−1. Glycolaldehyde will also be formed during conversion of 7,8-dihydro-D-neopterin to 6-hydroxymethyl-7,8-dihydropterin by FolB in the folate biosynthesis pathway. If folate is synthesized at a rate comparable with that of PLP, this reaction could supply glycolaldehyde at ∼5 μM h−1. Although the amount of glycolaldehyde produced by FolB is clearly not sufficient to supply pathway 1 in the ΔpdxB strain, the combined generation of glycolaldehyde from 3HP and 7,8-dihydro-D-neopterin may well be sufficient when yeaB is overexpressed, even if some glycolaldehyde is lost by diffusion through the membrane.
The next step in pathway 1 is formation of 4HT, which is not a recognized metabolite in E. coli. 4HT can be formed by aldol condensation of glycine and glycolaldehyde (Dempsey, 1987). The E. coli genome encodes an enzyme annotated as low-specificity threonine aldolase (LtaE). The physiological function of this enzyme is unknown; it cleaves L-allo-threonine most efficiently among known substrates (Liu et al, 1998), but L-allo-threonine is also not a recognized metabolite in E. coli. We assayed the rate of cleavage of L-allo-threonine, L-threonine and 4HT by LtaE (Table III). (The aldolase reaction is the reverse of the aldol condensation proposed in the latent pathway, but aldol condensation reactions are generally reversible.) Notably, cleavage of 4HT is nearly as efficient as cleavage of L-allo-threonine. We examined the products formed by the LtaE-catalyzed aldol condensation of glycolaldehyde and glycine by NMR (Supplementary Figure 2). The reaction forms 4HT and 4-hydroxy-L-allo-threonine (4-allo-HT) in a ratio of 1.0:0.7. (LtaE does not cleave the D-isomers of threonine and allo-threonine (Liu et al, 1998), so we do not expect formation of 4-hydroxy-D-threonine and 4-hydroxy-D-allo-threonine.) These data show that LtaE catalyzes the aldol condensation of glycine and glycolaldehyde, but the stereochemistry of the reaction is not well controlled, and a substantial amount of the allo-isomer is produced. However, this does not necessarily divert material from pathway 1. As the aldol condensation is reversible and the equilibrium favors the substrates, material sequestered in 4-allo-HT will ultimately be drawn into the pathway as 4HT is converted to downstream products, as long as 4-allo-HT is not converted to any other products. The most obvious enzyme that might utilize 4-allo-HT is ThrB, which phosphorylates 4HT. However, ThrB does not phosphorylate 4-allo-HT (Supplementary Figure 3).
We confirmed that LtaE is responsible for the formation of 4HT by pathway 1 in vivo by constructing a ΔpdxB ΔltaE strain. This strain can grow on M9/glucose in the presence of pyridoxine, but does not grow when intermediates upstream of the proposed aldol condensation (3PHP, 3HP or glycolaldehyde) are supplied (Supplementary Table IV). It can, however, grow when 4HT is supplied. Furthermore, overproduction of YeaB does not complement the ΔpdxB ΔltaE strain (data not shown), confirming that LtaE and YeaB participate in the same pathway.
The last step in the proposed pathway 1 is phosphorylation of 4HT. We suspected that homoserine kinase (ThrB) might catalyze this reaction since it phosphorylates homoserine, which is structurally similar to 4HT. Furthermore, ThrB was previously shown to be required for the growth of a ΔpdxB strain on glucose at 37°C in the presence of glycolaldehyde or 4HT (Zhao and Winkler, 1996a). ThrB does indeed turn over 4HT, but with an efficiency nearly two orders of magnitude lower than that for homoserine (Table III). We confirmed that ThrB is responsible for phosphorylation of 4HT in vivo by constructing a ΔpdxB ΔthrB strain. This strain can grow on glucose in the presence of pyridoxine, but not when intermediates upstream of the proposed phosphorylation reaction are supplied (Supplementary Table IV).
Confirmation that pathway 1 diverts material from serine biosynthesis
We carried out two experiments to test the proposition that YeaB diverts 3PHP from serine biosynthesis into serendipitous pathway 1 in vivo. First, we generated a ΔpdxB ΔserA strain that cannot make 3PHP. Pathway 1 should not operate in this strain even if yeaB is overexpressed. As predicted, the ΔpdxB ΔserA strain cannot grow on M9/glucose supplemented with serine when yeaB is overexpressed, although it can grow when pdxB is overexpressed (see Table IV). Because addition of serine can cause inhibition of ThrA (Hama et al, 1990, 1991) and thus impair synthesis of threonine and methionine, we also assessed the growth of the ΔpdxB ΔserA strain on M9/glucose supplemented with both serine and homoserine. Under these conditions as well, the ΔpdxB ΔserA strain cannot grow when yeaB is overexpressed. These results are consistent with the hypothesis that the lack of growth is due to a lack of 3PHP, but do not rule out an additional unanticipated inhibitory effect of serine on pathway 1. To address this possibility, we constructed a ΔpdxB strain carrying an allele of serA that encodes a serine-insensitive enzyme (designated serA′) (Grant et al, 2005). As SerA′ is insensitive to feedback inhibition by serine, addition of serine does not shut off flux through the serine biosynthesis pathway. Thus, we predict that this strain should grow on M9/glucose supplemented with serine and homoserine when yeaB is overexpressed if there is no other inhibitory effect of serine. Indeed, complementation of the ΔpdxB serA′ strain by yeaB is unaffected by the addition of serine and homoserine (Table IV). Together with the demonstration that YeaB catalyzes conversion of 3PHP to 3HP in vitro, these results provide strong support for the proposition that serendipitous pathway 1 diverts 3PHP from the serine biosynthesis pathway.
Table 4. The effect of serine and homoserine on complementation of the ΔpdxB, ΔpdxB ΔserA and ΔpdxB serA’ strains by overexpression of pdxB or yeaB.
| Supplementsa | Strain and overexpressed gene |
|||||
|---|---|---|---|---|---|---|
| ΔpdxB (JU283) |
ΔpdxB ΔserA (JU425) |
ΔpdxB serA’ (JU430) |
||||
| pdxB | yeaB | pdxB | yeaB | pdxB | yeaB | |
| Strain designations are defined in Supplementary Table 1. | ||||||
| aL-serine concentration was 1 mM and L-homoserine concentration was 50 μM. | ||||||
| bColonies reached 1 mm diameter in 1–2 days (++++), 3–5 days (+++) or 6–8 days (++). (+) indicates colonies that were <1 mm after 9 days. | ||||||
| None | ++++b | +++ | − | − | ++++ | ++ |
| L-serine | ++++ | + | +++ | − | +++ | − |
| L-serine+L-homoserine | ++++ | + | +++ | − | ++++ | ++ |
Complementation by PdxA depends on pathway 1
Our observation that overproduction of PdxA complements the ΔpdxB strain is curious. As described above, PdxA oxidizes 4PE at C-3 and therefore cannot substitute for PdxB. Overexpression of pdxA does not restore growth of the ΔpdxB ΔltaE strain on M9/glucose (data not shown), suggesting that complementation by pdxA depends on the presence of LtaE and therefore on pathway 1. We suspect that overproduction of PdxA increases the rate of consumption of 4PHT formed by the inefficient serendipitous pathway, effectively pulling material through the pathway by catalyzing the thermodynamically favorable conversion of 4PHT to 1-amino-propan-2-one-3-phosphate.
Pathway 1 can be elevated to physiological significance at 37°C by spontaneous mutations
Pathway 1 appears to operate even without the overproduction of YeaB or ThrB during growth on minimal medium at 30°C. Previous workers reported that a ΔpdxB strain grows slowly on glucose with mucoid morphology at 30°C, but not at 37°C (Lam and Winkler, 1990). Our ΔpdxB strain, which differs somewhat from the strain used by Lam and Winkler (1990), also shows slow growth on M9/glucose at 30°C. As neither the ΔpdxB ΔltaE strain nor the ΔpdxB ΔthrB strain grows at 30°C (data not shown), pathway 1 can apparently generate sufficient PLP for slow growth at 30°C in the absence of PdxB.
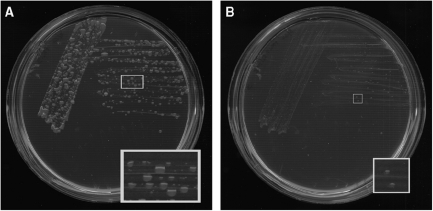
Pseudorevertants of the ΔpdxB strain that grow on M9/glucose at 37°C arise frequently. We have deleted ltaE in one of these pseudorevertants (JK19). Deletion of ltaE slows growth at 37°C substantially; small mucoid colonies can be seen after incubation for 5 days, whereas colonies of the pseudorevertant JK19 are much larger (Figure 5). (The variation in colony size seen in Figure 5A was observed repeatedly, even when plates were streaked from a single colony, suggesting that colony phenotype is influenced by some type of phase variation.) These results suggest that pathway 1 contributes significantly to PLP synthesis in pseudorevertant JK19, although is apparently not the only source of PLP. Efforts to identify the mutations responsible for the newly acquired ability of this pseudorevertant to grow at 37°C on M9/glucose are in progress.
Figure 5.
Growth on M9/glucose at 37°C of (A) JK19, a pseudorevertant of the ΔpdxB strain, and (B) JK125, a derivative of JK19 in which ltaE has been deleted. Pictures were taken after 5 days of incubation. Insets show an expanded view of the designated region from each plate.
Discussion
The experiments described here reveal the existence of three serendipitous pathways that allow synthesis of PLP in the ΔpdxB strain when any one of seven different genes is overexpressed. The number of genes that allow complementation is surprising; most multicopy suppression experiments reveal fewer genes that can complement a strain lacking a metabolic enzyme. For example, Patrick et al (2007) found that 21 of 104 knockout strains of E. coli could be complemented by multicopy suppression using the ASKA library, but in most cases by only one or two genes. One exception, the ΔglyA strain, was complemented by four genes, one of which encodes an antisigma factor. A second unusual case was described by Miller and Raines (2004, 2005), who found that overexpression of four genes encoding glycokinases with promiscuous glucokinase activity complemented a strain lacking glucokinase. Our finding that seven different genes complement the ΔpdxB strain is, to our knowledge, the record. Furthermore, our results are especially interesting because none of the seven genes encodes a protein with a promiscuous 4PE dehydrogenase activity, even though dehydrogenation of alcohols is a very common activity in metabolic enzymes. The results of our multicopy suppression experiments are also notable for the effectiveness of the rescue of a strain that is otherwise unable to grow on M9/glucose. As shown in Table II, it takes 1–2 days for the ΔpdxB strain to grow colonies 1 mm in diameter on M9/glucose when pdxB is overexpressed. Growth takes about the same amount of time when thrB, hisB or php is overexpressed, and takes 3–5 days when yeaB, yjbQ, pdxA or aroB is overexpressed. Thus, overexpression of any of these seven genes enables growth at a rate >20% that of a strain in which PdxB is available. These remarkable findings reveal the potential for innovation inherent within the proteome of E. coli that can be accessed simply by changes in the production of enzymes with catalytically promiscuous activities.
We have identified three enzymes that contribute to one serendipitous pathway. This pathway is patched together using a combination of a promiscuous enzyme (ThrB), a broad-specificity enzyme whose physiological role is unclear (LtaE), and a protein of unknown function (YeaB). Decarboxylation of 3HP appears to be non-enzymatic. We have not yet identified the sequence of reactions in the other two pathways, but these pathways also appear to involve promiscuous activities of enzymes with well-established roles in other pathways (HisB and AroB) and proteins of unknown function (Php and YjbQ).
Our characterization of pathway 1 allows us to rationalize the seemingly disparate observations that growth of the ΔpdxB strain can be restored by the addition of 3HP, glycolaldehyde, or 4HT, or by overexpression of yeaB, thrB or pdxA. Flux through the pathway can apparently be increased to levels sufficient to support growth by increasing the levels of intermediates by supplying them in the medium, or by increasing the levels of certain enzymes in the pathway. Overproduction of YeaB serves to divert flux from the serine biosynthesis pathway toward pathway 1. Overproduction of the downstream enzymes ThrB and PdxA likely increases flux through the pathway by pulling the equilibrium of the aldol condensation of glycolaldehyde and glycine toward 4-HT, which helps to capture glycolaldehyde before it undergoes other reactions or diffuses out of the cell. On the basis of this analysis, we might have expected that overproduction of LtaE would complement the ΔpdxB strain, but it does not. However, overproduction of LtaE causes toxicity even in wild-type E. coli (data not shown), and this likely accounts for the lack of complementation of the ΔpdxB strain when ltaE is overexpressed.
The intermediacy of glycolaldehyde in pathway 1 may provide an explanation for why overexpression of yeaB, thrB or pdxA supports growth on plates but not in liquid medium. Glycolaldehyde is small and uncharged and should easily cross the cell membrane. In liquid culture, loss of glycolaldehyde across the membrane would be essentially irreversible due to enormous dilution in the culture medium. However, glycolaldehyde lost from one cell in a colony on a plate could be taken up by nearby cells and would thus foster growth within the colony. Consistent with this hypothesis is the observation that the ΔpdxB strain can grow in liquid medium when 0.2 mM glycolaldehyde is supplied (Supplementary Figure 4).
The finding that Dxs and SucA, both of which have a respectable promiscuous ability to decarboxylate 3HP, apparently do not contribute to serendipitous pathway 1 in vivo illustrates the important point that a promiscuous activity that is measurable in vitro may not be relevant in vivo. Dxs and SucA catalyze reactions that are essential (for Dxs) or important (for SucA) for growth on glucose. In each case, the substrate(s) for the normal reaction will compete with 3HP for access to the active site. Given that the promiscuous activities are orders of magnitude less efficient than the normal activities, there may be little flux through these promiscuous reactions in vivo.
Pathway 1 is not known to produce PLP in any organism, and it is interesting to consider whether this pathway is inherently poorly suited for this purpose, or whether it was never discovered because the extant pathway emerged first. The chemical reactions in pathway 1 are not intrinsically difficult or different from typical metabolic reactions. However, glycolaldehyde is small and uncharged and can leak out of cells. Furthermore, 4HT (Shames et al, 1984) and 3HP (data not shown) are toxic. Neither of these factors is likely to be an insurmountable impediment. Many bi-functional enzymes generate highly reactive intermediates at one active site and transfer them through tunnels to a second active site (Weeks et al, 2006); such a mechanism could evolve to transfer glycolaldehyde from the active site at which it is produced to the active site of LtaE, where it would react with glycine to form 4HT. The toxicity of 4HT is probably due to its ability to compete with metabolites such as homoserine and threonine at the active sites of enzymes. Resistance to 4HT toxicity could be achieved by mutations that allow exclusion of 4HT from these active sites. Examples of similar molecular discrimination abound. The mechanism for the toxicity of 3HP is under investigation. Presumably, mechanisms for discrimination against 3HP and 4HT have not evolved because these molecules are not normal metabolites, and therefore there has been no need for macromolecules to discriminate against them.
Although pathway 1 is inefficient, it clearly provides the ΔpdxB strain with the ability to grow under conditions in which survival is otherwise impossible. In general, serendipitous assembly of an inefficient pathway from promiscuous activities of available enzymes will be important whenever the pathway provides increased fitness. This might occur when a critical metabolite is no longer available from the environment, and survival depends on assembly of a new biosynthetic pathway. This is likely the mechanism by which the PLP synthesis pathway in γ-proteobacteria emerged. In fungi, plants and most bacteria, PLP is synthesized from glutamine, ribose-5-phosphate and glyceraldehyde-3-phosphate by the enzyme complex Pdx1/Pdx2, which is absent in γ-proteobacteria. It is believed that the γ-proteobacteria lost the ancestral and more universal Pdx1/Pdx2-dependent pathway at some point, probably during a period in which pyridoxine or a closely related molecule was available in the environment. When the environment changed and the ability to synthesize PLP became important for fitness, the pathway shown in Figure 1 was patched together (Fitzpatrick et al, 2007). Our results suggest that the extant pathway in γ-proteobacteria may have been the one of several possibilities that emerged as the ‘winner’. A second circumstance in which a novel inefficient pathway may improve fitness is the appearance of a novel compound in the environment. A newly assembled and poorly functioning pathway for degradation of pentachlorophenol (PCP), a toxic anthropogenic pesticide introduced in the 1930s, improves fitness of Sphingobium chlorophenolicum by simultaneously allowing detoxification of PCP and access to a novel source of carbon (Copley, 2000). The pathway for degradation of atrazine, another anthropogenic pesticide, enhances microbial fitness by providing a source of ammonia (Shapir et al, 2007). Finally, chemotherapeutic agents that block metabolic pathways in bacteria or cancer cells could provide selective pressure for assembly of serendipitous pathways that allow synthesis of the end product of the blocked pathway and thus a previously unappreciated source of drug resistance. In all of these cases, even an inefficient pathway can provide a selective advantage over other cells in a particular environmental niche, allowing survival and subsequent mutations that elevate the efficiency of the pathway.
The metabolic network of E. coli consists primarily of linear pathways, with a small number of cycles and a rather dense network connecting various sugars in the glycolytic and pentose phosphate pathways. Our work is consistent with the hypothesis that the metabolic network of E. coli is underlain by a denser network of reactions due to promiscuous enzymes that use and generate recognized metabolites, but also unusual metabolites that normally have no physiological role. As the rate of a unimolecular enzymatic reaction is given by v=kcat/KM [E][S] for subsaturating conditions, which will generally be the case for promiscuous activities, flux through a step in the submetabolic network can be elevated either by increasing the concentration of the enzyme or by increasing the concentration of the substrate. (The same applies for bi-molecular reactions, as well.) For the serendipitous pathways reported here, overproduction of particular enzymes raises the flux to a physiologically significant level. For pathway 1, increasing the concentrations of intermediates in the pathway by supplying them in the medium also raises the flux to a physiologically significant level.
The discoveries reported here are particularly important because three serendipitous pathways appear to be able to divert material from normal metabolic pathways to generate an intermediate in the PLP synthesis pathway downstream of the block in the pathway. These findings highlight the abundance of cryptic capabilities in the E. coli proteome that can be drawn on to generate novel pathways. Similar serendipitous pathways might allow inefficient degradation or synthesis of novel compounds. Such pathways could provide a starting place for assembly of more efficient pathways, both in nature and in the hands of metabolic engineers.
Materials and methods
Strains
Strains used are listed in Supplementary Table I. E. coli BW25113 strains in which pdxB, serC, pdxA, pdxJ and pdxH are replaced with a kanamycin resistance gene were obtained from the Keio collection (Nara Institute of Science and Technology) (Baba et al, 2006). Construction of mutant strains lacking pdxB and additional genes is described in the Supplementary information. Primers for PCR amplification of genes and construction of overexpression vectors and knockout strains are listed in Supplementary Tables 5 and 6.
Chemicals
D-1-Deoxyxylulose (Lois et al, 1998), 4HT (Cativiela et al, 1996), 4PHT (Laber et al, 1994), 3PHP (Zhao and Winkler, 1996b), [4R-2H]NADH and [4S-2H]NADH (Viola et al, 1979) were prepared with minor modifications of published procedures. The synthesis of 4PE will be described elsewhere (Y Novikov et al, in preparation). Chemicals synthesized for this project were characterized by 1H-NMR (Supplementary information). All other chemicals were purchased from Sigma-Aldrich (St Louis, MO).
Identification of genes that complement pdxB
The ASKA collection of 3276 clones carrying E. coli BW25113 ORFs in pCA24N was obtained from the Nara Institute of Science and Technology (Kitagawa et al, 2005). A plasmid library lacking pdxB was introduced into the ΔpdxB∷kan strain by electroporation. Transformants were spread on M9/glucose plates containing kanamycin, chloroamphenicol and isopropyl-β-D-thiogalactoside. Plasmids were recovered from colonies that grew on the plates, and the genes responsible for complementation of ΔpdxB were identified by sequencing. The ability of each gene to complement ΔpdxB was confirmed by reintroducing both the corresponding plasmid from the ASKA collection and pTrcHisB carrying the gene that had been amplified from E. coli genomic DNA. The seven genes that were found to complement ΔpdxB were also introduced into strains lacking serC, pdxA, pdxJ or pdxH, and into a ΔpdxB∷kan ΔltaE∷cat strain. Additional details are available in the Supplementary information.
Purification of enzymes
Most enzymes were expressed with an N-terminal His6 tag from pET45b in E. coli BL21(DE3) or from pCA24N in E. coli K12 AG1 and were purified by nickel affinity chromatography. YeaB and LacZ were expressed as fusion proteins with MalE from pMAL-c4x (New England BioLabs Inc.) in E. coli K12 TB1 and purified by amylose affinity chromatography. AroB was expressed with an N-terminal His6 tag from pET45b in a ΔpdxA∷kan strain that had been lysogenized with the λDE3 cassette (Novagen, EMD Chemicals Inc.) (to prevent contamination with PdxA), and purified by nickel affinity chromatography using Ni-Sepharose resin (GE Healthcare Biosciences, Pittsburgh, PA).
Assay of 4PE dehydrogenase activity
4PE dehydrogenase activity was measured by following the reduction of NAD in reaction mixtures containing 2 mM NAD+, variable amounts of 4PE in 100 mM Tris–HCl, pH 7.5, and either 2 μM PdxA at 25°C or 11 μM AroB at 10°C. Prolonged incubation of 4PE and NAD+ with Php (71 μM), HisB (52 μM), ThrB (18 μM) or MalE–YeaB (42 μM) showed no evidence of oxidation of 4PE.
Assay of 4PHT dehydrogenase activity
4PHT dehydrogenase activity was assayed in 50 mM Tris–HCl, pH 7.5, containing 1.2 mM NAD+ and variable amounts of 4PHT with 0.20 μM PdxA or 50 μM AroB at 25°C.
Assay of 3PHP phosphatase activity
3PHP phosphatase activity was measured by following the release of phosphate from 3PHP in reaction mixtures containing 3PHP and MalE–YeaB (5.7 μM) or MalE–LacZ (5.8 μM) in 50 mM Tris–HCl, pH 7.4, at 37°C. The concentration of phosphate in aliquots was determined using the malachite green assay (Van Veldhoven and Mannaerts, 1987).
Assay of 3HP decarboxylase activity
3HP decarboxylase activity was measured by coupling the decarboxylation reaction to the reduction of glycolaldehyde by yeast alcohol dehydrogenase. Reaction mixtures contained 0.2 mM TPP, 5 mM MgCl2, 0.5 mM NADH, 500 U ml−1 yeast alcohol dehydrogenase and variable amounts of enzyme and 3HP in 50 mM potassium phosphate, pH 6.8, at 37°C. Oxidation of NADH was monitored at 340 nm. The following concentrations of enzymes were used: TktA (12 μM); TktB (7.8 μM); Gcl (5 μM); PoxB (6.4 μM); IlvB (10 μM); IlvI (1.8 μM); AceE (8.8 μM); Oxc (9.6 μM); Dxs (0.5 μM); and SucA (10 μM). YdbK and MenD were insoluble and not tested. (All enzymes except Dxs were prepared by overexpression from genes cloned into pCA24N. Dxs was prepared by overexpression from dxs cloned into pET45b.) Only the TPP-dependent catalytic components of the multisubunit enzymes (pyruvate dehydrogenase, α-ketoglutarate dehydrogenase and both isozymes of acetohydroxyacid synthase) were assayed.
The formation of glycolaldehyde by decarboxylation of 3HP was confirmed by 1H-NMR analysis of reaction mixtures containing 21 μM Dxs or 91 μM SucA, 0.2 mM TPP, 5 mM MgCl2 and 10 mM 3HP in 50 mM potassium phosphate, pH 6.9. Reaction mixtures were incubated at 37°C and then heated at 90°C for 5 min to stop the reaction at various times. Precipitated proteins were removed by centrifugation at 12 000 × g for 5 min at 4°C. D2O was added to 10% (v/v) to provide a lock signal. 1H-NMR analysis was performed on a Varian Inova 500 MHz NMR spectrometer using the WET solvent suppression technique (Smallcombe et al, 1995).
Assay of LtaE activity
The activity of LtaE was measured by coupling formation of acetaldehyde from L-threonine and L-allo-threonine, or formation of glycolaldehyde from 4HT, to the reduction of the aldehyde product by yeast alcohol dehydrogenase in 0.1 M Tris–HCl, pH 8.0, at 25°C. Assays with L-threonine or L-allo-threonine contained 30 U ml−1 yeast alcohol dehydrogenase. Assays with 4HT contained 500 U ml−1 yeast alcohol dehydrogenase. Assays with L-threonine contained 1.25 μM LtaE. Assays with L-allo-threonine contained 62.5 nM LtaE. Assays with 4HT contained 1 μM LtaE.
Assay of L-homoserine kinase and 4HT kinase activity
The activity of homoserine kinase (ThrB) was measured at 25°C following the method of Burr et al (1976) using 37 nM enzyme to measure activity with L-homoserine and 0.18 μM enzyme to measure activity with 4HT.
Characterization of the product formed by oxidation of 4PE by PdxA
The position of oxidation of 4PE by PdxA was determined by incubating 4PE (1 mM) with NAD+ (0.2 mM), NADH, [4R-2H]NADH or [4S-2H]NADH (1.5 mM), and PdxA (30 μM) in 50 mM sodium phosphate, pH 7.2. After incubation for 27 h at room temperature, reaction mixtures were heated for 5 min at 95°C. Precipitated protein was removed by centrifugation at 12 000 × g for 5 min at 4°C. After lyophilization, samples were dissolved in 1 ml D2O and examined by 1H-NMR using the WET solvent suppression technique (Smallcombe et al, 1995).
Isolation of a pseudorevertant of the ΔpdxB strain that can grow on M9/glucose at 37°C
A single colony of the ΔpdxB strain was inoculated into 10 ml LB medium containing kanamycin and incubated at 37°C with shaking overnight. An aliquot (100 μl) was inoculated into 10 ml LB medium containing kanamycin and grown at 37°C with shaking to mid-log phase. The cells were harvested by centrifugation at 4500 × g for 10 min at 4°C. The cell pellet was washed twice with 10 ml of M9/glucose and then resuspended in 10 ml of M9/glucose. Aliquots (100 μl) were spread onto prewarmed M9/glucose plates containing kanamycin. The plates were incubated at 37°C for 5 days, at which time individual colonies were patched onto fresh plates. Colonies were subsequently streaked onto LB plates containing kanamycin to cleanly separate single colonies. A single pseudorevertant, designated JK19, was used for the studies reported here. JK125 was constructed by knocking out ltaE in JK19 as described above. Single colonies of JK19 or JK125 were resuspended in sterile PBS and washed as described above. Cells were streaked onto M9/glucose agar plates and incubated at 37°C for up to 9 days.
Supplementary Material
Supplementary methods, Supplementary figures S1–4, Supplementary tables SI–VI
Acknowledgments
We thank Dr Johannes Rudolph for helpful suggestions. This work was supported by the US National Institutes of Health Grants GM083285 and GM067749 to SDC.
Author contributions: JK performed the experimental work. JPK isolated and analyzed the pseudorevertant of the ΔpdxB strain. YN synthesized 4HT and 4PE. RS assisted with the NMR analysis. SDC directed the experimental work. JK and SDC wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Ashiuchi M, Misono H (1999) Biochemical evidence that Escherichia coli hyi (orf b0508, gip) gene encodes hydroxypyruvate isomerase. Biochim Biophys Acta 1435: 153–159 [DOI] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H (2006) Construction of Escherichia coli K12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2: 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barngrover DA, Stevens HC, Dills WL Jr (1981) D-Xylulose-1-phosphate: enzymatic assay and production in isolated rat hepatocytes. Biochem Biophys Res Commun 102: 75–80 [DOI] [PubMed] [Google Scholar]
- Bennett BD, Kimball EH, Gao M, Osterhout R, Van Dien SJ, Rabinowitz JD (2009) Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat Chem Biol 5: 593–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burr B, Walker J, Truffa-Bachi P, Cohen GN (1976) Homoserine kinase from Escherichia coli K12. Eur J Biochem 62: 519–526 [DOI] [PubMed] [Google Scholar]
- Carpenter EP, Hawkins AR, Frost JW, Brown KA (1998) Structure of dehydroquinate synthase reveals an active site capable of multistep catalysis. Nature 394: 299–302 [DOI] [PubMed] [Google Scholar]
- Cativiela C, Díaz-de-Villegas MD, Gálvez JA, García JL (1996) Diastereoselective Strecker reaction of D-glyceraldehyde derivatives. A novel route to (2S,3S)- and (2R,3S)-2-amino-3,4-dihydroxybutyric acid. Tetrahedron 52: 9563–9574 [Google Scholar]
- Copley SD (2000) Evolution of a metabolic pathway for degradation of a toxic xenobiotic: the patchwork approach. Trends Biochem Sci 25: 261–265 [DOI] [PubMed] [Google Scholar]
- Dempsey WB (1987) Synthesis of pyridoxal phosphate. In Escherichia coli and Salmonella typhimurium; Cellular and Molecular Biology, Neidhardt FC (ed), Vol. 1, pp 539–543. Washington, D.C.: American Society for Microbiology [Google Scholar]
- Drewke C, Notheis C, Hansen U, Leistner E, Hemscheidt T, Hill RE, Spenser ID (1993) Growth response to 4-hydroxy-L-threonine of Escherichia coli mutants blocked in vitamin B6 biosynthesis. FEBS Lett 318: 125–128 [DOI] [PubMed] [Google Scholar]
- Duncan K, Coggins JR (1986) The serC-aroA operon of Escherichia coli. A mixed function operon encoding enzymes from two different amino acid biosynthetic pathways. Biochem J 234: 49–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick TB, Amrhein N, Kappes B, Macheroux P, Tews I, Raschle T (2007) Two independent routes of de novo vitamin B6 biosynthesis: not that different after all. Biochem J 407: 1–13 [DOI] [PubMed] [Google Scholar]
- Freilich S, Spriggs RV, George RA, Al-Lazikani B, Swindells M, Thornton JM (2005) The complement of enzymatic sets in different species. J Mol Biol 349: 745–763 [DOI] [PubMed] [Google Scholar]
- Grant GA, Hu Z, Xu XL (2005) Identification of amino acid residues contributing to the mechanism of cooperativity in Escherichia coli D-3-phosphoglycerate dehydrogenase. Biochemistry 44: 16844–16852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant GA, Schuller DJ, Banaszak LJ (1996) A model for the regulation of D-3-phosphoglycerate dehydrogenase, a Vmax-type allosteric enzyme. Protein Sci 5: 34–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hama H, Kayahara T, Tsuda M, Tsuchiya T (1991) Inhibition of homoserine dehydrogenase I by L-serine in Escherichia coli. J Biochem 109: 604–608 [DOI] [PubMed] [Google Scholar]
- Hama H, Sumita Y, Kakutani Y, Tsuda M, Tsuchiya T (1990) Target of serine inhibition in Escherichia coli. Biochem Biophys Res Commun 168: 1211–1216 [DOI] [PubMed] [Google Scholar]
- Hedrick JL, Sallach HJ (1961) The metabolism of hydroxypyruvate. I. The nonenzymatic decarboxylation and autoxddation of hydroxypyruvate. J Biol Chem 236: 1867–1871 [PubMed] [Google Scholar]
- Hill RE, Himmeldirk K, Kennedy IA, Pauloski RM, Sayer BG, Wolf E, Spenser ID (1996) The biogenetic anatomy of vitamin B6. A 13C NMR investigation of the biosynthesis of pyridoxol in Escherichia coli. J Biol Chem 271: 30426–30435 [DOI] [PubMed] [Google Scholar]
- Hill RE, Iwanow A, Sayer BG, Wysocka W, Spenser ID (1987) The regiochemistry and stereochemistry of the biosynthesis of vitamin B6 from triose units. J Biol Chem 262: 7463–7471 [PubMed] [Google Scholar]
- Hill RE, Miura I, Spenser ID (1977) Biosynthesis of Vitamin B6. The incorporation of [1,3-13C2]glycero1. J Am Chem Soc 99: 4179–4181 [DOI] [PubMed] [Google Scholar]
- Hill RE, Spenser ID (1973) Biosynthesis of vitamin B6. Incorporation of terminally labelled glucose. Can J Biochem 51: 1412–1416 [DOI] [PubMed] [Google Scholar]
- Jensen RA (1976) Enzyme recruitment in evolution of new function. Annu Rev Microbiol 30: 409–425 [DOI] [PubMed] [Google Scholar]
- Kitagawa M, Ara T, Arifuzzaman M, Ioka-Nakamichi T, Inamoto E, Toyonaga H, Mori H (2005) Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res 12: 291–299 [DOI] [PubMed] [Google Scholar]
- Kuzmič P (1996) Program DYNAFIT for the analysis of enzyme kinetic data: application to HIV proteinase. Anal Biochem 237: 260–273 [DOI] [PubMed] [Google Scholar]
- Kuzuyama T, Takagi M, Takahashi S, Seto H (2000) Cloning and characterization of 1-deoxy-D-xylulose 5-phosphate synthase from Streptomyces sp. Strain CL190, which uses both the mevalonate and nonmevalonate pathways for isopentenyl diphosphate biosynthesis. J Bacteriol 182: 891–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laber B, Gerbling KP, Harde C, Neff KH, Nordhoff E, Pohlenz HD (1994) Mechanisms of interaction of Escherichia coli threonine synthase with substrates and inhibitors. Biochemistry 33: 3413–3423 [DOI] [PubMed] [Google Scholar]
- Lam HM, Winkler ME (1990) Metabolic relationships between pyridoxine (vitamin B6) and serine biosynthesis in Escherichia coli K-12. J Bacteriol 172: 6518–6528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JQ, Dairi T, Itoh N, Kataoka M, Shimizu S, Yamada H (1998) Gene cloning, biochemical characterization and physiological role of a thermostable low-specificity L-threonine aldolase from Escherichia coli. Eur J Biochem 255: 220–226 [DOI] [PubMed] [Google Scholar]
- Lois LM, Campos N, Putra SR, Danielsen K, Rohmer M, Boronat A (1998) Cloning and characterization of a gene from Escherichia coli encoding a transketolase-like enzyme that catalyzes the synthesis of D-1-deoxyxylulose 5-phosphate, a common precursor for isoprenoid, thiamin, and pyridoxol biosynthesis. Proc Natl Acad Sci USA 95: 2105–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLennan AG (2006) The Nudix hydrolase superfamily. Cell Mol Life Sci 63: 123–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller BG, Raines RT (2004) Identifying latent enzyme activities: substrate ambiguity within modern bacterial sugar kinases. Biochemistry 43: 6387–6392 [DOI] [PubMed] [Google Scholar]
- Miller BG, Raines RT (2005) Reconstitution of a defunct glycolytic pathway via recruitment of ambiguous sugar kinases. Biochemistry 44: 10776–10783 [DOI] [PubMed] [Google Scholar]
- Morett E, Saab-Rincon G, Olvera L, Olvera M, Flores H, Grande R (2008) Sensitive genome-wide screen for low secondary enzymatic activities: the YjbQ family shows thiamin phosphate synthase activity. J Mol Biol 376: 839–853 [DOI] [PubMed] [Google Scholar]
- O’Brien PJ, Herschlag D (1998) Sulfatase activity of E. coli alkaline phosphatase demonstrates a functional link to arylsulfatases, an evolutionarily related enzyme family. J Am Chem Soc 120: 12369–12370 [Google Scholar]
- O’Brien PJ, Herschlag D (2001) Functional interrelationships in the alkaline phosphatase superfamily: phosphodiesterase activity of Escherichia coli alkaline phosphatase. Biochemistry 40: 5691–5699 [DOI] [PubMed] [Google Scholar]
- Patrick WM, Matsumura I (2008) A study in molecular contingency: glutamine phosphoribosylpyrophosphate amidotransferase is a promiscuous and evolvable phosphoribosylanthranilate isomerase. J Mol Biol 377: 323–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick WM, Quandt EM, Swartzlander DB, Matsumura I (2007) Multicopy suppression underpins metabolic evolvability. Mol Biol Evol 24: 2716–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shames SL, Ash DE, Wedler FC, Villafranca JJ (1984) Interaction of aspartate and aspartate-derived antimetabolites with the enzymes of the threonine biosynthetic pathway of Escherichia coli. J Biol Chem 259: 15331–15339 [PubMed] [Google Scholar]
- Shapir N, Mongodin EF, Sadowsky MJ, Daugherty SC, Nelson KE, Wackett LP (2007) Evolution of catabolic pathways: genomic insights into microbial s-triazine metabolism. J Bacteriol 189: 674–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu S, Dempsey WB (1978) 3-hydroxypyruvate substitutes for pyridoxine in serC mutants of Escherichia coli K-12. J Bacteriol 134: 944–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smallcombe SH, Patt SL, Keifer PA (1995) WET solvent suppression and its applications to LC NMR and high-resolution NMR spectroscopy. J Magn Reson A 117: 295–303 [Google Scholar]
- Tani Y, Dempsey WB (1973) Glycolaldehyde is a precursor of pyridoxal phosphate in Escherichia coli B. J Bacteriol 116: 341–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor Ringia EA, Garrett JB, Thoden JB, Holden HM, Rayment I, Gerlt JA (2004) Evolution of enzymatic activity in the enolase superfamily: functional studies of the promiscuous o-succinylbenzoate synthase from Amycolatopsis. Biochemistry 43: 224–229 [DOI] [PubMed] [Google Scholar]
- Van Veldhoven PP, Mannaerts GP (1987) Inorganic and organic phosphate measurements in the nanomolar range. Anal Biochem 161: 45–48 [DOI] [PubMed] [Google Scholar]
- Viola RE, Cook PF, Cleland WW (1979) Stereoselective preparation of deuterated reduced nicotinamide adenine nucleotides and substrates by enzymatic synthesis. Anal Biochem 96: 334–340 [DOI] [PubMed] [Google Scholar]
- Wang SC, Johnson WH Jr, Whitman CP (2003) The 4-oxalocrotonate tautomerase- and YwhB-catalyzed hydration of 3E-haloacrylates: implications for the evolution of new enzymatic activities. J Am Chem Soc 125: 14282–14283 [DOI] [PubMed] [Google Scholar]
- Weeks A, Lund L, Raushel FM (2006) Tunneling of intermediates in enzyme-catalyzed reactions. Curr Opin Chem Biol 10: 465–472 [DOI] [PubMed] [Google Scholar]
- Zhao G, Winkler ME (1996a) 4-Phospho-hydroxy-L-threonine is an obligatory intermediate in pyridoxal 5′-phosphate coenzyme biosynthesis in Escherichia coli K-12. FEMS Microbiol Lett 135: 275–280 [DOI] [PubMed] [Google Scholar]
- Zhao G, Winkler ME (1996b) A novel α-ketoglutarate reductase activity of the serA-encoded 3-phosphoglycerate dehydrogenase of Escherichia coli K-12 and its possible implications for human 2-hydroxyglutaric aciduria. J Bacteriol 178: 232–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary methods, Supplementary figures S1–4, Supplementary tables SI–VI