Abstract
Gene expression varies widely between closely related species and strains, yet the genetic basis of most differences is still unknown. Several studies suggested that chromatin regulators have a key role in generating expression diversity, predicting a reduction in the interspecies differences on deletion of genes that influence chromatin structure or modifications. To examine this, we compared the genome-wide expression profiles of two closely related yeast species following the individual deletions of eight chromatin regulators and one transcription factor. In all cases, regulator deletions increased, rather than decreased, the expression differences between the species, revealing hidden genetic variability that was masked in the wild-type backgrounds. This effect was not observed for individual deletions of 11 enzymes involved in central metabolic pathways. The buffered variations were associated with trans differences, as revealed by allele-specific profiling of the interspecific hybrids. Our results support the idea that regulatory proteins serve as capacitors that buffer gene expression against hidden genetic variability.
Keywords: chromatin structure, evolution, gene expression, genetic capacitor
Introduction
Recent studies revealed substantial differences between the gene expression patterns of closely related species (Rifkin et al, 2003; Khaitovich et al, 2006; Tirosh et al, 2009b), but attempts to identify the specific mutations and mechanisms underlying these differences have met with little success. For example, sequence divergence at transcription factor-binding sites accounts for only a small fraction of observed expression differences (Zhang et al, 2004; Tirosh et al, 2008). Several studies suggested that chromatin regulators have a key role in generating expression diversity, either through mutations that directly affect regulator function or abundance, or indirectly, by propagating divergent signals coming from upstream components (Tirosh et al, 2009b). First, S. cerevisiae genes whose expression is sensitive to the deletion of chromatin regulators diverge in expression significantly more than genes whose expression is insensitive to such deletions (Choi and Kim, 2008). Moreover, the nucleosome patterns along the promoters of divergent genes are dynamic, with nucleosomes displaying fuzzy promoter locations and overlap transcription factor-binding sites (Tirosh and Barkai, 2008). In contrast, promoters of genes whose expression is conserved between species appear to be less amendable to regulation, displaying well-positioned nucleosomes and a nucleosome-free region that allows easy access to cis-regulatory elements.
An additional link between chromatin regulators and gene expression divergence came from linkage studies. Kruglyak and colleagues (Brem et al, 2002) mapped the genotypes and the expression profiles of two wild-type (WT) yeast parental strains together with dozens of their progenies. Subsequent analysis of this data linked a large fraction of expression differences to a small number of markers associated with chromatin regulators, suggesting that a significant part of the expression divergence in yeast arises from the evolution of these regulators (Lee et al, 2006).
In a recent study, we directly compared the divergence of nucleosome positioning and that of gene expression between the two closely related yeast species, Saccharomyces cerevisiae and S. paradoxus (Tirosh et al, 2010). While widespread differences were detected between the positioning of nucleosomes in the two species, these differences were excluded from regulatory elements and were not correlated with interspecies expression divergence. Although these results argue against a major role of chromatin structure in the evolution of gene expression, nucleosome positioning is only one aspect of chromatin structure and other aspects, such as histone modifications and higher-order folding, may still have an important role in generating expression divergence.
If chromatin regulators have a major role in generating gene expression divergence, then deletion of such regulators will reduce interspecies differences in gene expression. In contrast, several authors proposed that regulators of gene expression would acquire the ability to buffer genetic differences, predicting that deletion of such regulators will reveal, rather than conceal, phenotypic differences (Siegal and Bergman, 2002; Bergman and Siegal, 2003; Levy and Siegal, 2008). The best-studied example of a protein with buffering capacity is the heat-shock protein, Hsp90 (Rutherford and Lindquist, 1998; Queitsch et al, 2002). Reduced activity of Hsp90 in Drosophila or Arabidopsis out-bred lines generated a wide range of dramatically variable phenotypes. These phenotypes were heritable, consistent with the idea that they result from hidden genetic variability that was revealed on HSP90 inhibition. Hsp90 was thus termed as a genetic capacitor, as it may allow organisms to accumulate hidden genetic variability that could be potentially unleashed on genetic or environmental perturbations. This buffering capacity of Hsp90 was linked to its chaperone activity in promoting the correct folding of proteins in the face of various destabilizing mutations (Tokuriki and Tawfik, 2009), although recent work has questioned this interpretation and instead suggested that inhibition of Hsp90 may result in increased transposon activity (Specchia et al, 2010).
Earlier studies identified other genes, besides HSP90, that reveal hidden genetic variability when mutated. In fact, the classical concept of ‘canalization’, coined by Waddington over 60 years ago, refers to the ‘very general observation… that the wild type of an organism, that is to say, the form which occurs in nature under the influence of natural selection, is much less variable in appearance than the majority of the mutant races’ (Waddington, 1942). Theoretical studies proposed that in complex networks, stabilizing selection by itself might be sufficient to render regulatory genes as capacitors (Stearns, 2002; Siegal and Bergman, 2002; Bergman and Siegal, 2003; Hermisson and Wagner, 2004; Ciliberti et al, 2007). Analysis of morphological variability between individual yeast cells further indicated that cell-to-cell variability increases on the deletion of hundreds of genes, suggesting that these genes function as capacitors of microenvironmental variations (Levy and Siegal, 2008). Importantly, the genes identified as potential environmental capacitors were enriched with chromatin regulators, raising the possibility that these genes will also function as capacitors of genetic variability (Meiklejohn and Hartl, 2002; Lehner, 2010).
Are chromatin regulators generators or capacitors of gene expression variability? To try and distinguish the dominant effect, we deleted chromatin regulators in two closely related yeast species, S. cerevisiae and S. paradoxus, and compared their genome-wide expression profiles. These two yeasts have diverged ∼10 million years ago (Kellis et al, 2003), but maintained practically the same set of genes, display a highly similar physiology and morphology and can readily be mated to produce viable F1 hybrids. Their promoter sequences exhibit substantial divergence (∼82% identity), but their overall gene expression patterns are largely conserved, although, similar to all other species or strains examined, substantial expression differences are readily identified (Tirosh et al, 2009b). We asked whether deletion of chromatin (or transcription) regulators will decrease expression divergence, as expected if the regulators function as generators of variability, or, conversely, will increase expression divergence, as expected if the regulators function primarily as capacitors of expression variability.
Results
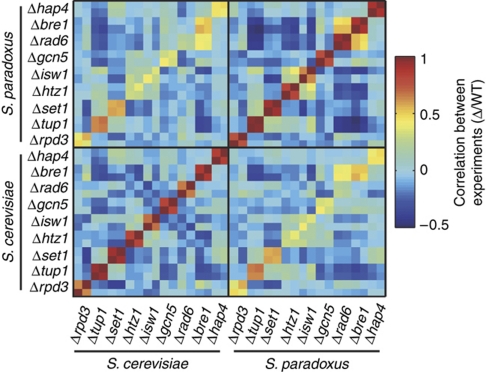
We chose nine chromatin and transcription regulators that are associated with diverse functions: histone modifiers involved in acetylation (Gcn5 and Rpd3), methylation (Set1) and ubiquitination (Bre1 and Rad6) (Kouzarides, 2007), the histone variant H2A.Z (Htz1) (Zhang et al, 2005), the chromatin remodeler Isw1 (Clapier and Cairns, 2009), the general repressor Tup1 (Malave and Dent, 2006) and a central transcription factor involved in respiration (Hap4). We generated S. cerevisiae and S. paradoxus strains deleted (individually) of each of these regulators, and used two-species microarrays to map their gene expression patterns relative to that of the WT strains (Tirosh et al, 2009b; Figure 1). The average correlation for the deletion effect (log-ratio of the mutant versus WT expression levels) between dye-swapped biological repeats was ∼0.85, compared with ∼0 correlation between different mutants and ∼0.4 between strains of the two species deleted of the same regulator.
Figure 1.
Genome-wide expression changes on deletion of chromatin and transcription regulators in S. cerevisiae and S. paradoxus. Expression of mutant strains was measured in dye-swap biological repeats with two-species microarrays (Tirosh et al, 2009b). Shown are the genome-wide correlations between the log2 expression ratios (mutant (Δ)/WT) of all samples, with adjacent samples reflecting biological repeats.
Regulator deletion increases gene expression divergence
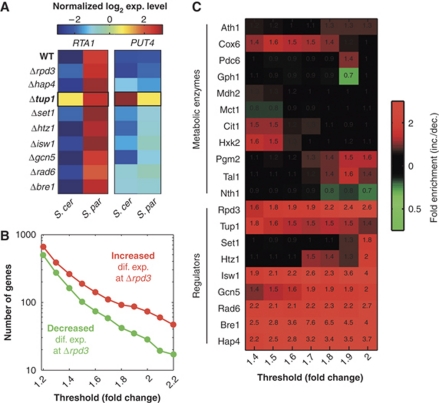
We asked whether the similarity in gene expression patterns of the two species increases or decreases following deletion of the regulators. For example, RTA1 is expressed much more strongly in the WT S. paradoxus than in the WT S. cerevisiae, and this differential expression is also maintained on deletion of most regulators. However, when Tup1 is deleted, RTA1 expression in S. paradoxus becomes more similar to its expression in S. cerevisiae (Figure 2A). Thus, Tup1 is involved in generating the divergent expression of RTA1. An opposite example is provided by PUT4. This gene is expressed at a similar level in the WT S. cerevisiae and S. paradoxus, and also maintains a similar expression level in most deletion strains. However, when Tup1 is deleted, PUT4 expression in S. cerevisiae becomes significantly higher than that found in S. paradoxus (Figure 2A). Thus, in this case, Tup1 buffers the hidden variation that causes variable PUT4 expression in Δtup1.
Figure 2.
Deletion of chromatin and transcription regulators increases the extent of interspecies expression differences. (A) Shown are the color-coded expression levels (log2 of normalized hybridization intensity) for two genes in the wild-type and mutant strains. Upon deletion of a regulator (for example, Dtup1), interspecies expression differences can either decrease (as in RTA1) or increase (as in PUT4) compared with the wild-type strains. (B) Number of genes whose interspecies expression differences are increased (red) or decreased (green) in Δrpd3 compared with wild-type strains. The x axis shows the fold-change thresholds for identifying increases/decreases of expression differences. Similar plots are provided for all nine regulators in Supplementary Figure S2. (C) Enrichment of increased over decreased expression changes for deletions of each of the nine regulators and eleven metabolic enzymes. Shown are ratios of the number of increased divided by decreased expression differences for each deletion strain and at different thresholds. The average (over the different mutants) number of genes whose expression difference increased or decreased by at least 1.2-, 1.5- and 2-fold is 1458, 270 and 101, respectively; the minimal number of genes whose expression difference increased or decreased by the same thresholds is 788, 68 and 18, respectively (all three minimal values are for Set1). Numbers are shown over the heatmap only for values above 1.3 or below 0.7. The corresponding P-values for these enrichments are shown in Supplementary Figure S3. S. cer, S. cerevisiae; S. par, S. paradoxus.
For each of the regulators, we defined two classes of genes: genes that are expressed more similarly between the species upon regulator deletion (factor-generated divergence, exemplified by RTA1) and genes that are expressed more differently upon regulator deletion (factor-buffered divergence, exemplified by PUT4). As shown in Figure 2B, significantly more genes increased in expression divergence in Δrpd3 than those that decreased in expression divergence in Δrpd3, and this result was independent of the threshold used to define changes in the level of divergence. For example, the interspecies differential expression of 58 genes decreased by at least 70% (1.7-fold) in Δrpd3, whereas that of 110 genes increased by at least 1.7-fold in Δrpd3.
Similar analysis with the other deletion strains revealed that deletion of each of the regulators increased the gene expression divergence (Figure 2C and Supplementary Figures S1 and S2). For example, at a threshold of 1.7-fold, the number of genes with increased expression differences was, on average, 2.6-fold higher than the number of genes with decreased expression differences. The enrichments of increased interspecies differences was significant (P<0.05) at multiple thresholds for the nine mutants, with most P-values below 10−4 (binomial test, see Supplementary Figure S3). The only exception was Δset1, in which the difference was not significant as relatively few genes were affected.
Note that, although these thresholds (1.4-fold to twofold) may appear low, they should be considered in the context of expression differences between these closely related species, which is typically in this range of fold changes (Supplementary Figure S4a). Comparison of biological repeats shows that, at these thresholds, increased and decreased expression differences are highly reproducible (Supplementary Figure S4b). Moreover, the enrichment of increased interspecies differences typically became more dramatic as the threshold for defining changes in the level of divergence was raised, eliminating small changes that might be due to technical variations (Figure 2C).
Increased divergence might simply reflect differential fitness effects, for example, if a deletion decreases the fitness of only one of the species and this decrease is accompanied by changes in gene expression. To examine this, we measured the decrease in growth rate of the different deletion strains and found that most deletions have a similar growth-rate effect in the two species (Supplementary Figure S5). The preferential increase of expression differences is not correlated with either the growth-rate effects or with the differential growth-rate effects of the deletions between the two species. Similarly, preferential increase of expression differences is observed regardless of the number of genes whose expression is influenced by each deletion (Supplementary Figure S5).
An increased interspecies expression difference is not expected based on a simple null model of gene deletion (see Materials and methods). As an additional control, we also verified that the increase of interspecies expression differences is not a general property of all deletion mutants. We examined the effects of individual deletions of 11 enzymes involved in various central metabolic pathways (Figure 2C). In contrast to regulators, deletion of different metabolic enzymes did not have a consistent effect. Instead, only 4 of the 11 deletion mutants had a tendency for increased interspecies divergence, although considerably weaker than that of the chromatin and transcription regulators, and the other 7 mutants had either no change or even decreased divergence.
Buffered variations primarily reflect trans differences
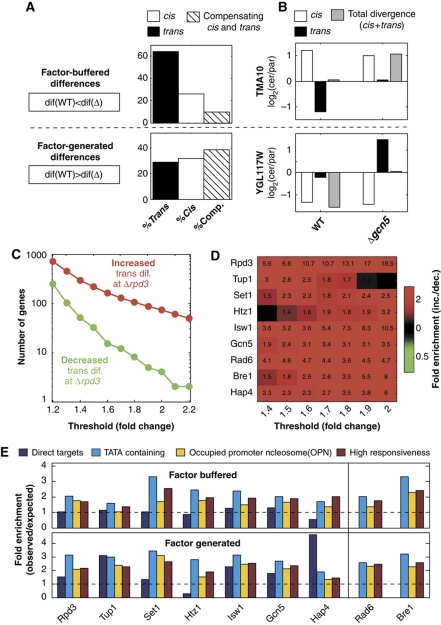
Changes in gene expression are generated by mutations in the DNA sequence that is linked to the gene itself (cis effects), such as mutations in specific binding sites of transcription factors, and by mutations at other genomic regions that impact on the activity or abundance of upstream factors that regulate gene expression (trans effects). We wished to characterize the contribution of cis- and trans effects to the factor-generated and factor-buffered divergence. To this end, we measured the allele-specific expression of the interspecific hybrids formed by mating either the WT S. cerevisiae and S. paradoxus strains or their respective deletion mutants (Tirosh et al, 2009b). In the hybrid backgrounds, both alleles are subject to the same trans environments, so that trans-dependent differences are eliminated, revealing only the cis-dependent part of the variations. This allows us to distinguish the contribution of cis- and trans effects to the interspecies variations in the WT strains (Wittkopp et al, 2004, 2008; Ronald et al, 2005; Springer and Stupar, 2007; Wang et al, 2007; Gagneur et al, 2009; Tirosh et al, 2009b; McManus et al, 2010) and also to the interspecies variations in the different deletion backgrounds. Comparing the two, we sub-divided the genes with factor-dependent divergence into two subgroups, corresponding to divergence that was altered mainly in trans or in cis.
We focused first on the buffered variations and asked which effect (cis or trans) is buffered by the deleted regulators. Notably, most of the buffered variations (64%) were generated in trans (displaying a lower trans difference in the WT), whereas significantly fewer cases (26%) resulted from reduced cis differences (Figure 3A and Supplementary Figure S1). Thus, in the WT background, the regulators mostly buffered trans differences.
Figure 3.
The cis and trans contribution to expression differences and their buffering. (A) The cis and trans effects were quantified in the wild-type and mutant strains by analyzing the respective interspecific hybrids. Factor-buffered (top) and factor-generated variations (bottom) were classified into changes in the extent of a cis effect, trans effect or compensation between cis and trans: genes in which the change in a cis and/or trans effect is opposite to the change in overall expression difference due to cis–trans compensation (Comp.). Shown are the percentages of these classes, averaged over the nine different mutants. (B) Examples of cis–trans compensation for factor-buffered (top, TMA10) and factor-generated differences (bottom, YGL117W). (C, D) Enrichment of increased over decreased interspecies expression differences for Δrpd3 (C) and for all deletion strains (D). The same analysis was performed as in Figure 2, but restricted to the trans component of interspecies expression differences. Numbers are shown over the heatmap only for values above 1.3 or below 0.7. (E) Fold enrichment of factor-buffered (top) and factor-generated (bottom) variations with the direct targets of each factor (blue; unknown direct targets for Bre1 and Rad6), TATA-containing genes (light blue; Basehoar et al, 2004), OPN genes (yellow; Tirosh and Barkai, 2008) and genes with high responsiveness (red; third of the genes with highest responsiveness, as previously defined; Tirosh and Barkai, 2008). Fold enrichment was calculated as the percentage of factor-buffered/generated genes with the corresponding property divided by the percentage of all genes with that property.
Notably, in 10% of the buffered genes, reduced expression difference in the WT is in fact due to increased cis- or trans differences (Figure 3A). In these cases, opposite cis- and trans effects compensate for one another in the WT background, but this compensation is eliminated in the deletion strains (Landry et al, 2005; Tirosh et al, 2009b). For example, in the WT, TMA10 has higher cis effect but lower trans effect in S. cerevisiae, compared with S. paradoxus. However, owing to compensation among these effects, no difference is observed between the overall TMA10 expression levels of the two species. However, deletion of Gcn5 eliminates the trans difference, thereby revealing the cis difference (Figure 3B). Thus, buffering of TMA10 expression differences was due to compensation between cis- and trans effects.
In contrast to the dominance of trans effects in the buffered variation, factor-generated variations were more dependent on cis effects (Figure 3A and Supplementary Figure S1). Thus, many cis mutations lead to differential gene expression that is eliminated upon deletion of a regulator. Strikingly, in ∼40% of the cases of factor-generated variations, the reduced expression difference in the deletion strains was due to compensation between opposing cis- and trans effects. In these cases, a cis- or trans difference in fact increased in the mutant, but this has led to an overall lower divergence because of the presence of a compensating effect (for example, YGL117W, Figure 3B). Thus, although the overall effect in these cases was of factor-generated variations, that factor actually buffered variations of either a cis effect or, more often, a trans effect.
This observation had led us to re-examine the extent of buffering, focusing now only on the trans effects, which are more likely to be factor dependent. Indeed, when repeating the analysis in Figure 2, focusing on the trans effects only, the results become even more striking (Figure 3C and D). For example, in the case of Rpd3 and a 1.7-fold threshold, the overrepresentation of buffered variations increased from approximately twofold (110/58, Figure 2B) to more than 10-fold (128/12) when considering only trans effects (Figure 3C and D). Similarly, the average enrichment of buffered variations at this threshold among the different regulator deletions increased from 2.6 (Figure 2C) to 3.8-fold (Figure 3D). Cis effects also tended to increase in the mutant strains, but this effect is significantly less pronounced (Supplementary Figure S6).
As buffering comes about mostly from factor-dependent modulation of trans differences between the species, we asked whether the buffered variations are enriched with direct targets of these regulators. Previous studies have characterized the direct targets of seven of the deleted regulators based on ChIP-chip analysis: Hap4 (Harbison et al, 2004), Set1 (Ng et al, 2003), Gcn5 and Rpd3 (Robert et al, 2004), Isw1 (Venters and Pugh, 2009), Htz1 (Zhang et al, 2005) and Tup1 (Buck and Lieb, 2006). These predicted targets, however, were not enriched with factor-buffered variations, and were moderately enriched with factor-generated variations (Figure 3E). Instead, we find that both classes are enriched with genes whose promoters contain a TATA box and that have high nucleosome occupancy upstream of the transcription start site (OPN genes; Figure 3E). These promoter features were previously shown to correlate with high expression responsiveness and evolvability (Tirosh et al, 2006, 2009a; Landry et al, 2007; Tirosh and Barkai, 2008), and indeed we find that genes with factor-buffered or factor-generated variations are associated with high responsiveness, as defined by a microarray compendium of more than a thousand genetic and environmental perturbations (Ihmels et al, 2002). These results suggest that some genes are generally more affected by any network perturbation, including the deletions examined here, and therefore that these genes are preferentially identified as having either factor-buffered or factor-generated variations.
Discussion
We report that individual deletions of eight chromatin regulators and one transcription factor leads invariably to an increase in the expression differences between the closely related yeast species S. cerevisiae and S. paradoxus. These results are consistent with the possibility that chromatin regulators evolved to buffer genetic variability. In contrast, others have proposed that chromatin regulators have a role in generating gene expression variability. These two views might be reconciled if diversity is partially driven by mutations that decrease the activity of chromatin regulators, as these mutations could partially mimic the deletion effects and in this way promote interspecies expression differences. Such mutations may segregate in a population and serve as genetic variation for buffering, which is required for canalization to evolve (Landry, 2009).
Buffered expression variations are not enriched among the direct targets of the deleted factors, suggesting that they reflect indirect effects of the deletions. Notably, the buffered variations are correlated among genes that have a similar response to environmental changes (Supplementary Figure S7). This may indicate that these variations originate from divergence of upstream components involved in sensing the environment that cascade through multiple regulatory mechanisms before influencing the expression of target genes (Tirosh et al, 2009b). Deletion of chromatin or transcription factors may modulate these signal transduction pathways, thereby influencing the expression variations at downstream target genes. Accordingly, the affected downstream genes are those that are more responsive to network perturbations and that display distinctive features such as a TATA box and high nucleosome occupancy at promoter regulatory regions (Tirosh et al, 2006; Tirosh and Barkai, 2008).
The idea that mutations (for example, gene deletions) would unleash hidden genetic variability goes back to the canalization concept, first proposed for development (Waddington, 1942) but later extended to other cellular processes (Barkai and Shilo, 2007), which states that WT organisms are ‘robust’, being able to resist genetic or environmental perturbations significantly more than mutants. However, it remains unclear how general is this phenomenon. In particular, it is not clear whether this effect is specific to a limited set of proteins that evolved to be genetic capacitors, as proposed for Hsp90, or whether it presents a general property common to many genes. Our results demonstrate a consistent buffering effect of nine regulators (and another Escherichia coli regulator, see Supplementary Figure S8), but not of eleven metabolic enzymes, and suggests that the ability to buffer gene expression variations might be a characteristic property of large-scale regulators. Additional work would be needed to examine whether buffering is a typical property of all regulators or whether it is specific to chromatin regulators.
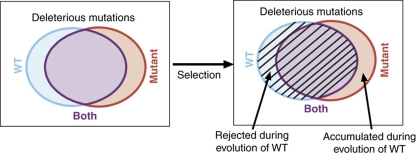
We favor the view that regulators did not evolve to directly buffer gene expression variations, but instead that their buffering emerges naturally during evolution of a complex system, as a consequence of stabilizing selection (Siegal and Bergman, 2002; Bergman and Siegal, 2003; Hermisson and Wagner, 2004; Ciliberti et al, 2007). This possibility can be described by a simple model, as shown in Figure 4: stabilizing selection will eliminate mutations that lead to a deleterious effect in the WT background, but will maintain mutations that are neutral in this background. Such neutral mutations, however, might well be deleterious (change the phenotype) when combined with deletion of an additional gene, if the two mutations are epistatic. Thus, organisms would accumulate conditionally neutral mutations that only have an effect in the mutant background (the hallmark of buffering) while rejecting conditionally neutral mutations that have an effect in the WT background but not in the mutant background (Figure 4; Hermisson and Wagner, 2004). Conditionally neutral mutations are the product of epistatic effects, which are indeed quite common in complex systems (Tong et al, 2004; Gjuvsland et al, 2007), and in the context of gene expression are likely to occur primarily when deleting large-scale regulators such as those analyzed here.
Figure 4.
Model for the buffering of expression variations by large-scale regulators. Squares represent the space of possible mutations and circles represent the fraction of mutations that are (slightly) deleterious for the wild type (WT, blue), mutant (red) or both (purple). A genetic capacitor modulates the effects of mutations in both ways because of epistatic interactions: it buffers the deleterious effects of some mutations while aggravating the effects of other mutations (that is, these mutations are deleterious for the wild type but not for the mutant). As the wild type (and not the deletion mutant) is the product of evolution, natural selection has rejected mutations that are deleterious for the wild type, but accumulated many mutations that are slightly deleterious for the deletion mutant. As a result, deletion of large-scale regulators unleashes the slightly deleterious effects of various mutations and, thus, increases the variability between different strains or species (which accumulated different sets of buffered mutations).
Our study differs from previous experimental analysis of canalization in three ways (Levy and Siegal, 2008). First, we focused directly on buffering genetic rather than environmental variations. Second, we analyzed buffering with respect to genome-wide expression patterns, which can be considered as thousands of different phenotypes. Third, we compared variations between two closely related species rather than within a given species. The two yeast species we studied maintained similar physiology and morphology, but have evolved independently for ∼10 million years. The buffering observed here is consistent with stabilizing selection, and further suggests that stabilizing selection has acted to maintain similar gene expression patterns in these species, despite their divergence.
Materials and methods
Yeast strains and growth conditions
Deletion strains were constructed on the background of S. cerevisiae (BY4741) and S. paradoxus (CBS 432) using standard techniques. The nine regulator deletions were made by introducing G418 and hygromycin B resistance in S. cerevisiae and S. paradoxus (ho∷nat MATα), respectively. The 11 metabolic enzyme deletions were made by introducing hygromycin B resistance on the background of fluorescently labeled strains (BY4741 ho∷Nat:TEF2pro:mCherry, CBS432 ho∷kanMX:TEF2pro:GFP MATα). Mutant hybrids were generated by mating the respective mutants from the two species. All strains were grown to a log-phase growth at 30°C in rich media (YPD medium).
RNA preparation, microarray design and hybridization
All samples were collected at log-phase growth. Starters were grown overnight, diluted to OD(600)=0.1 in 10 ml medium and harvested ∼5 h after dilution when OD(600)∼0.5. Total RNA was extracted using MasterPure™ Yeast RNA Purification Kit (EPICENTRE), amplified with Agilent's Low RNA Input Amplification Kit and hybridized with Agilent's standard protocols and kits to custom two-species microarrays. As previously described (Tirosh et al, 2009b), the microarray contains two blocks, each with ∼105K 60-mer species-specific probes, designed to hybridize to the same positions of most orthologs between S. cerevisiae and S. paradoxus. One to five different probe sequences were designed for each S. cerevisiae gene (and the orthologous positions were used for S. paradoxus) and each probe sequence was placed at three different positions in the microarray to avoid spatial biases. Each gene was therefore assayed, on average, by (3 probe sequences) × (3 positions) × (2 biological repeats)=18 measurements. Arrays were scanned using Agilent microarray scanner and quantified using the Spotreader software (Niles Scientific).
Microarray data analysis
Expression profiles of all mutant strains were generated by hybridization to two-species microarrays and normalized, as described previously (Tirosh et al, 2009b). The intensities for each array and each dye were log2 transformed and converted into the same distribution using percentile normalization. Probes were then sorted into 10 bins of similar %GC and the log-intensities of each bin were renormalized to the mean and standard deviation of all probes. Probes with a coefficient of variance >40% in at least one of the species were declared as missing values in both species. The log2 expression level of each gene was then defined by the average of all corresponding probes across all replicate experiments. Mating-type-specific genes were excluded from the analysis. Raw and processed expression data will be available at the GEO database (GSE23866).
The deletion effects of each mutant (Figure 1) were defined as the log2 ratio of deletion mutants divided by WT expression levels. Microarray experiments of the WT were conducted together with the Hap4 deletion strain, but separately from all other strains, and we noticed that this leads to technical differences between the two data sets, such that samples within a data set are typically more correlated than samples from the two different data sets. Therefore, when analyzing the expression changes of all mutant strains, except for Hap4, we replaced the WT expression levels with the median expression level of all these mutant strains. This is in fact a more strict definition of expression changes, as a gene will be identified as affected by a specific deletion only if it is not affected by most of the mutants in a similar way.
Expression levels were compared between the two species and we defined the interspecies expression difference for each gene as ∣log2 (S. cer/S. par)∣, in the WT and in each of the mutant strains. We then compared the interspecies expression differences between the WT and each of the mutant strains to identify genes for which the expression difference increases or decreases as a result of the deletions. Increased/decreased expression differences were defined as genes for which the change between the mutant and WT exceeds a given threshold; we repeated the analysis with seven different thresholds (1.4-fold, 1.5-fold, …, 2-fold), and a threshold of 1.5-fold was used to define factor-buffered and factor-generated variations in analysis of cis- and trans effects (Figure 3). Once again, to avoid technical differences between the data sets, we used the median expression difference of all mutants to define the WT expression differences. To verify that this approach does not have a significant effect on the results, we repeated the analysis with the original WT data and obtained qualitatively similar results (Supplementary Figure S1).
Analysis of metabolic enzyme deletion strains
In addition to the 9 regulators, we also examined the deletion effects of 11 enzymes from various central metabolic pathways: glycolysis (Hxk2), gluconeogenesis and glyoxylate cycle (Mdh2), aerobic respiration (Cox6), TCA cycle (Cit1), fermentation (Pdc6), glycogen catabolism (Gph1), fatty acid biosynthesis (Mct1), pentose phosphate pathway (Tal1), trehalose catabolism (Ath1 and Nth1) and Pgm2, which is involved in multiple pathways of hexose metabolism. As for the regulators, each metabolic enzyme was deleted in both species, and the genome-wide expression levels of the mutants were measured in dye-swapped biological repeats with a two-species microarray. This microarray is similar to that used for analysis of regulators but contains fewer genes (∼3750 genes from each of the species). These deletion effects were analyzed in the same way as the regulator deletions. However, unlike regulators, deletion of different metabolic enzymes does not have a consistent effect, as some mutants had increased expression differences (for example, Cox6), other mutants had decreased expression differences (for example, Nth1) and yet other mutants had only negligible effect on the amount of expression differences (for example, Mdh2).
Classification to cis and trans
For each gene, we compared the extent of expression differences between the two species with that between the corresponding hybrid alleles. The expression log2 ratio of the two parental species reflects the total divergence of that gene that includes both cis- and trans components; the log2 ratio of the corresponding hybrid alleles reflects only the cis component; and their subtraction (log2 ratio between parents minus log2 ratio between hybrid alleles) is used to calculate the trans component. Buffering was classified as cis- or trans-dependent based on the effect that increased the most in the mutant strain. Cases of buffering in which the main difference between the WT and mutant strain was an increase of a cis- or a trans effect (while the total difference decreased) were classified as compensation between cis and trans. We excluded from this analysis buffered genes in which all changes in the cis- and trans components were smaller than 1.25-fold. Similarly, factor-generated differences were classified as cis- or trans-dependent based on the effect that decreased the most in the mutant strain or as compensation if the main change in the mutant was an increase of cis- or trans effect that led to decrease of total divergence.
Note that this classification is based on comparison of the parental haploids with the hybrid diploids and thus might be influenced by differences between haploids and diploids. To avoid this problem, we excluded all genes annotated as haploid- or diploid specific or that differ in expression among mating types or between haploids and diploids (Galitski et al, 1999). Furthermore, our previous work (Tirosh et al, 2009b) suggests that this effect does not have a major influence on estimation of cis- or trans effects (see haploid versus diploid section in the Supplementary Information).
A simple null model for the effect of deletions on interspecies expression differences
We need to consider four possibilities:
Genes that are not differentially regulated by the deleted factor, both directly and indirectly. Differential expression of these genes is not affected by the deletion and thus they are irrelevant.
Genes that are differentially regulated only by the deleted factor (directly or indirectly). Deletion of the regulator will abolish (that is, decrease) their differential expression.
Genes that are differentially regulated by the deleted factor (directly or indirectly) and by additional mechanisms. Let X be the differential effect of the deleted factor and Y be the effect of all the additional differential mechanisms. Assuming that X and Y are independent (additive), then differential expression in the WT is ∣X+Y∣ and differential expression in the mutant is ∣Y∣. If X and Y act in the same direction (that is, either both increase expression of the S. cerevisiae gene or both decrease expression of the S. cerevisiae gene, compared with its ortholog in S. paradoxus), then ∣X+Y∣>∣Y∣, which means that differential expression will necessarily decrease in the mutant.
Same as no. 3, except that X and Y act in opposite directions (that is, one increases and the other decreases expression of the S. cerevisiae gene, compared with its ortholog in S. paradoxus). In this case, expression divergence could either increase or decrease in the mutant: ∣X+Y∣<∣Y∣ if and only if ∣X∣<2 × ∣Y∣.
Taken together, expression differences would decrease upon deletion in all genes of no. 2, all genes of no. 3 and some of the genes of no. 4; expression differences would increase only in part of the genes of no. 4. If we assume similar proportion of no. 3 and no. 4 (that is, X and Y have equal probability to act in the same or in opposite direction), then the probability that expression difference of a gene would increase upon a gene deletion is clearly <50%, which is the opposite of what we observed. To obtain an enrichment of expression differences that increase on deletion, the proportion of genes in which X and Y act in opposite direction and compensate one another must be much higher than 50%, which would mean that there is hidden variability in the WT. This naive null model clearly fails to capture the complexity of the regulatory network, and we thus examined the deletion effect of 11 metabolic enzymes to obtain a more realistic estimate of what is expected for a typical gene deletion (Supplementary Figure S3).
Supplementary Material
Supplementary Figures S1–8
Acknowledgments
This work was supported by the Helen and Martin Kimmel Award for Innovative Investigations and the European Research Council (Ideas).
Author contributions: IT and NB devised the study. SR, NS and YA performed the experiments. IT performed the computational analysis. IT and NB wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Barkai N, Shilo BZ (2007) Variability and robustness in biomolecular systems. Mol Cell 28: 755–760 [DOI] [PubMed] [Google Scholar]
- Basehoar AD, Zanton SJ, Pugh BF (2004) Identification and distinct regulation of yeast TATA box-containing genes. Cell 116: 699–709 [DOI] [PubMed] [Google Scholar]
- Bergman A, Siegal ML (2003) Evolutionary capacitance as a general feature of complex gene networks. Nature 424: 549–552 [DOI] [PubMed] [Google Scholar]
- Brem RB, Yvert G, Clinton R, Kruglyak L (2002) Genetic dissection of transcriptional regulation in budding yeast. Science 296: 752–755 [DOI] [PubMed] [Google Scholar]
- Buck MJ, Lieb JD (2006) A chromatin-mediated mechanism for specification of conditional transcription factor targets. Nat Genet 38: 1446–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JK, Kim YJ (2008) Epigenetic regulation and the variability of gene expression. Nat Genet 40: 141–147 [DOI] [PubMed] [Google Scholar]
- Ciliberti S, Martin OC, Wagner A (2007) Robustness can evolve gradually in complex regulatory gene networks with varying topology. PLoS Comput Biol 3: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR (2009) The biology of chromatin remodeling complexes. Annu Rev Biochem 78: 273–304 [DOI] [PubMed] [Google Scholar]
- Gagneur J, Sinha H, Perocchi F, Bourgon R, Huber W, Steinmetz LM (2009) Genome-wide allele- and strand-specific expression profiling. Mol Syst Biol 5: 274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galitski T, Saldanha AJ, Styles CA, Lander ES, Fink GR (1999) Ploidy regulation of gene expression. Science 285: 251–254 [DOI] [PubMed] [Google Scholar]
- Gjuvsland AB, Hayes BJ, Omholt SW, Carlborg O (2007) Statistical epistasis is a generic feature of gene regulatory networks. Genetics 175: 411–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, Jennings EG, Zeitlinger J, Pokholok DK, Kellis M, Rolfe PA, Takusagawa KT, Lander ES, Gifford DK, Fraenkel E, Young RA (2004) Transcriptional regulatory code of a eukaryotic genome. Nature 431: 99–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermisson J, Wagner GP (2004) The population genetic theory of hidden variation and genetic robustness. Genetics 168: 2271–2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihmels J, Friedlander G, Bergmann S, Sarig O, Ziv Y, Barkai N (2002) Revealing modular organization in the yeast transcriptional network. Nat Genet 31: 370–377 [DOI] [PubMed] [Google Scholar]
- Kellis M, Patterson N, Endrizzi M, Birren B, Lander ES (2003) Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423: 241–254 [DOI] [PubMed] [Google Scholar]
- Khaitovich P, Enard W, Lachmann M, Paabo S (2006) Evolution of primate gene expression. Nat Rev Genet 7: 693–702 [DOI] [PubMed] [Google Scholar]
- Kouzarides T (2007) Chromatin modifications and their function. Cell 128: 693–705 [DOI] [PubMed] [Google Scholar]
- Landry CR (2009) Systems biology spins off a new model for the study of canalization. Trends Ecol Evol 24: 63–66 [DOI] [PubMed] [Google Scholar]
- Landry CR, Lemos B, Rifkin SA, Dickinson WJ, Hartl DL (2007) Genetic properties influencing the evolvability of gene expression. Science 317: 118–121 [DOI] [PubMed] [Google Scholar]
- Landry CR, Wittkopp PJ, Taubes CH, Ranz JM, Clark AG, Hartl DL (2005) Compensatory cis-trans evolution and the dysregulation of gene expression in interspecific hybrids of Drosophila. Genetics 171: 1813–1822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SI, Pe'er D, Dudley AM, Church GM, Koller D (2006) Identifying regulatory mechanisms using individual variation reveals key role for chromatin modification. Proc Natl Acad Sci USA 103: 14062–14067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner B (2010) Genes confer similar robustness to environmental, stochastic, and genetic perturbations in yeast. PLoS One 5: e9035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy SF, Siegal ML (2008) Network hubs buffer environmental variation in Saccharomyces cerevisiae. PLoS Biol 6: e264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malave TM, Dent SY (2006) Transcriptional repression by Tup1-Ssn6. Biochem Cell Biol 84: 437–443 [DOI] [PubMed] [Google Scholar]
- McManus CJ, Coolon JD, Duff MO, Eipper-Mains J, Graveley BR, Wittkopp PJ (2010) Regulatory divergence in Drosophila revealed by mRNA-seq. Genome Res 20: 816–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiklejohn CD, Hartl DL (2002) A single mode of canalization. Trends Ecol Evol 17: 468–473 [Google Scholar]
- Ng HH, Robert F, Young RA, Struhl K (2003) Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell 11: 709–719 [DOI] [PubMed] [Google Scholar]
- Queitsch C, Sangster TA, Lindquist S (2002) Hsp90 as a capacitor of phenotypic variation. Nature 417: 618–624 [DOI] [PubMed] [Google Scholar]
- Rifkin SA, Kim J, White KP (2003) Evolution of gene expression in the Drosophila melanogaster subgroup. Nat Genet 33: 138–144 [DOI] [PubMed] [Google Scholar]
- Robert F, Pokholok DK, Hannett NM, Rinaldi NJ, Chandy M, Rolfe A, Workman JL, Gifford DK, Young RA (2004) Global position and recruitment of HATs and HDACs in the yeast genome. Mol Cell 16: 199–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronald J, Brem RB, Whittle J, Kruglyak L (2005) Local regulatory variation in Saccharomyces cerevisiae. PLoS Genet 1: e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford SL, Lindquist S (1998) Hsp90 as a capacitor for morphological evolution. Nature 396: 336–342 [DOI] [PubMed] [Google Scholar]
- Siegal ML, Bergman A (2002) Waddington's canalization revisited: developmental stability and evolution. Proc Natl Acad Sci USA 99: 10528–10532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Specchia V, Piacentini L, Tritto P, Fanti L, D'Alessandro R, Palumbo G, Pimpinelli S, Bozzetti MP (2010) Hsp90 prevents phenotypic variation by suppressing the mutagenic activity of transposons. Nature 463: 662–665 [DOI] [PubMed] [Google Scholar]
- Springer NM, Stupar RM (2007) Allele-specific expression patterns reveal biases and embryo-specific parent-of-origin effects in hybrid maize. Plant Cell 19: 2391–2402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns SC (2002) Progress on canalization. Proc Natl Acad Sci USA 99: 10229–10230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Barkai N (2008) Two strategies for gene regulation by promoter nucleosomes. Genome Res 18: 1084–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Barkai N, Verstrepen KJ (2009a) Promoter architecture and the evolvability of gene expression. J Biol 8: 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Reikhav S, Levy AA, Barkai N (2009b) A yeast hybrid provides insight into the evolution of gene expression regulation. Science 324: 659–662 [DOI] [PubMed] [Google Scholar]
- Tirosh I, Sigal N, Barkai N (2010) Divergence of nucleosome positioning between two closely related yeast species: genetic basis and functional consequences. Mol Syst Biol 6: 365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Weinberger A, Bezalel D, Kaganovich M, Barkai N (2008) On the relation between promoter divergence and gene expression evolution. Mol Syst Biol 4: 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Weinberger A, Carmi M, Barkai N (2006) A genetic signature of interspecies variations in gene expression. Nat Genet 38: 830–834 [DOI] [PubMed] [Google Scholar]
- Tokuriki N, Tawfik DS (2009) Chaperonin overexpression promotes genetic variation and enzyme evolution. Nature 459: 668–673 [DOI] [PubMed] [Google Scholar]
- Tong AH, Lesage G, Bader GD, Ding H, Xu H, Xin X, Young J, Berriz GF, Brost RL, Chang M, Chen Y, Cheng X, Chua G, Friesen H, Goldberg DS, Haynes J, Humphries C, He G, Hussein S, Ke L et al. (2004) Global mapping of the yeast genetic interaction network. Science 303: 808–813 [DOI] [PubMed] [Google Scholar]
- Venters BJ, Pugh BF (2009) A canonical promoter organization of the transcription machinery and its regulators in the Saccharomyces genome. Genome Res 19: 360–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington CH (1942) Canalization of development and the inheritance of acquired characters. Nature 150: 563–565 [DOI] [PubMed] [Google Scholar]
- Wang D, Sung HM, Wang TY, Huang CJ, Yang P, Chang T, Wang YC, Tseng DL, Wu JP, Lee TC, Shih MC, Li WH (2007) Expression evolution in yeast genes of single-input modules is mainly due to changes in trans-acting factors. Genome Res 17: 1161–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp PJ, Haerum BK, Clark AG (2004) Evolutionary changes in cis and trans gene regulation. Nature 430: 85–88 [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, Haerum BK, Clark AG (2008) Regulatory changes underlying expression differences within and between Drosophila species. Nat Genet 40: 346–350 [DOI] [PubMed] [Google Scholar]
- Zhang H, Roberts DN, Cairns BR (2005) Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell 123: 219–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Gu J, Gu X (2004) How much expression divergence after yeast gene duplication could be explained by regulatory motif evolution? Trends Genet 20: 403–407 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1–8