Not all enzymes are one-reaction wonders. Indeed, secondary capabilities of enzymes can make metabolism a lot more adaptable than previously thought.
Enzymes are usually thought of as specific catalysts of biochemical reactions. Enzyme A acts on substrate B to produce product C. It is, however, becoming increasingly clear that some enzymes are ‘promiscuous', able to act on alternative substrates to catalyze secondary reactions. A recent study by Kim et al (2010) demonstrates that promiscuity of individual enzymes might be just the tip of the iceberg.
Metabolism is a complex network of chemical reactions through which ‘food' is converted into the many compounds necessary for life. Removal of the enzyme catalyzing a given reaction will break the corresponding metabolic link. Sometimes this will be of no consequence—a similar enzyme might catalyze the same reaction or an alternative route to a downstream product may be present among the established reactions. Kim et al (2010) demonstrate a third possibility. New pathways can be constructed by patching together secondary capabilities of enzymes.
Kim et al (2010) start with a bacterium, Escherichia coli, that can produce all of the compounds necessary for growth in a ‘minimal' medium consisting of simple salts and glucose and delete a gene, pdxB, required to produce pyridoxal 5′-phosphate (PLP). Lacking PLP, this deletion strain can no longer grow. From this point, a common next step in microbial genetics is to select for rare mutants that suppress the effect of the deletion and reacquire the ability to grow in minimal medium. Kim et al (2010) performed this step, but with a twist. Rather than selecting a spontaneously arising mutant, they use a genetic library to overexpress many of the normal E. coli genes in the pdxB mutant. Their rationale was that, at high expression, any gene product that had even a weak ability to compensate for the loss of PdxB would allow the mutant to grow.
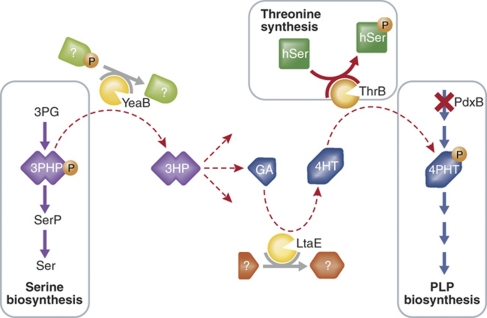
The results were surprising. Overexpression of seven genes could restore growth of the deletion strain. Strikingly, none of these genes encode enzymes that directly substitute for the activity of the PdxB enzyme. Instead, they seem to control key steps in alternative pathways that produce PLP without requiring the reaction catalyzed by PdxB. Kim et al (2010) concentrate on one of these pathways. Through an elegant and comprehensive series of genetic and biochemical analyses, they show that it consists of four separate reactions that provide a bridge diverting material from serine biosynthesis to the production of PLP (Figure 1). At least one of the reactions is controlled by a promiscuous enzyme, catalyzing a reaction different from its primary activity. In another case, a reaction product was not previously recognized as a metabolite in E. coli, suggesting that the enzyme was also functioning promiscuously. Fittingly, Kim et al (2010) term the newly revealed pathway, serendipitous. Although they do not characterize them in detail, at least two other serendipitous pathways appear to be available to produce PLP.
Figure 1.
3PHP from serine biosynthesis can be used to supply 4PHT when the PLP synthesis pathway is blocked. Abbreviations: 3PG, 3-phosphoglycerate; 3PHP, 3-phosphohydroxypyruvate; SerP, 3-phospho-L-serine; Ser, L-serine; 3HP, 3-hydroxypyruvate; GA, glycolaldehyde; 4HT, 4-hydroxy-L-threonine; 4PHT, 4-phosphohydroxy-L-threonine; hSer, L-homoserine; hSerP, O-phospho-L-homoserine; PLP, pyridoxal-5′-phosphate; YeaB (NudL), predicted NUDIX hydrolase; ThrB, homoserine kinase; LtaE, low-specificity threonine aldolase; PdxB, erythronate-4-phosphate dehydrogenase; PdxA.
While Kim et al (2010) clearly demonstrate the action of serendipitous pathways in producing PLP, it is important to note that these pathways required key enzymes to be expressed at high levels before being sufficiently active to support measurable growth. Given that, is it realistic that such pathways may actually have an important role in restoring growth following loss of a necessary enzyme? For at least three reasons, the answer might be yes.
First, although activation of serendipitous pathways that allow growth in PdxB deletion mutants might require overexpression of an enzyme, this might not be a general feature of these pathways. Even at a relatively low metabolic flux, serendipitous pathways may nevertheless be sufficiently active to produce low-demand compounds at levels that support growth. Second, if overexpression of key genes is a common requirement for serendipitous pathways, mutations exist that can do just that. Indeed, several experimental evolution studies have demonstrated fitness benefits from mobile genetic element-mediated increases in gene expression (Chou et al, 2009; Stoebel and Dorman, 2010). Third, a little bit of growth is a powerful thing. Any growth, even at a rate far below that of an unmutated individual, will allow new mutations to occur and rise in frequency, if they confer a benefit—for example, by raising flux through a serendipitous pathway. Indeed, Kim et al (2010) provide some evidence that just this scenario might contribute to growth of secondary mutants of the ΔpdxB strain that can grow in the absence of PLP. It will be interesting to see if any of these or similar spontaneous mechanisms activate beneficial serendipitous pathways in experimental populations. If so, it seems likely that they will also be important in natural populations.
The discovery of the three serendipitous pathways by Kim et al (2010) has major implications for how we think about metabolic networks. How many other pathways might be lying dormant, with the potential to circumvent mutational or therapeutic blocks in metabolism? As rational drug design advances, we will need to consider the possibility that serendipitous pathways might evolve to bypass targeted essential reactions. It will also be interesting to examine whether serendipitous pathways have acted as drivers of biological innovation. With an idea of what to look for, we can begin to develop comparative metabolomic analyses to test whether this has happened.
Footnotes
The author declares that he has no conflict of interest.
References
- Chou HH, Berthet J, Marx CJ (2009) Fast growth increases the selective advantage of a mutation arising recurrently during evolution under metal limitation. PLoS Genet 5: e1000652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kershner JP, Novikov Y, Shoemaker RK, Copley SD (2010) Three serendipitous pathways in E. coli can bypass a block in pyridoxal 5′-phosphate synthesis. Mol Sys Biol 6: 436 10.1038/msb.2010.88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoebel DM, Dorman CJ (2010) The effect of mobile element IS10 on experimental regulatory evolution in Escherichia coli. Mol Biol Evol 27: 2105–2112 [DOI] [PMC free article] [PubMed] [Google Scholar]