Abstract
Manual gating of bivariate plots remains the most frequently used data analysis method in flow cytometry. However, gating is operator-dependent and cumbersome, particularly with the increasing complexity of modern multicolor immunophenotyping data. A method that can remove operator bias, enable systematic and thorough analysis of complex high-dimensional data, correlate temporal changes in different subsets and lead to biomaker discovery is needed. Here we apply such a method, called cytometric fingerprinting (CF), to data obtained on peripheral blood B cells from an adult patient with type-1 diabetes who underwent pancreatic islet transplantation. We establish that CF can be used to analyze longitudinal trends in immunophenotypic data, and show that results from CF are comparable to those obtained with traditional gating methods. Both methods reveal the appearance of transitional B cells and subsequent accumulation of more mature B cells following immunosuppression and transplantation. This pattern is consistent with a temporally ordered process of B cell auto-reconstitution. We also show the comparative efficiency of fingerprinting in recognizing relative changes in B cell subsets with respect to time, its ability to couple the data with statistical methods (agglomerative clustering) and its potential to define novel subsets.
Keywords: B lymphocyte, flow cytometry, cytometric fingerprinting, type 1 diabetes, transplantation
1. Introduction
Flow cytometry is a widely used research tool that allows the simultaneous measurement of multiple cellular components in a heterogeneous environment and produces significant biological information. Flow cytometric studies rely on the fact that different cell populations express particular antigens, and that the number of antigens expressed per cell in each population fall within narrow ranges. Thus, conventional flow cytometric data is typically analyzed using pictorial graphs. Here an expert operator, by designing manual “gates” in 1 or 2-dimensional histograms or “dotplots,” is able to isolate and study populations of biological interest. The percentage of events in each gate is then quantified. More complex multiparameter distributions are resolved by applying “gating” procedures in a sequential fashion.
However, “gating” strategies have disadvantages. The process is labor intensive and subjective. There is a limited ability to survey an entire data set systematically, which becomes increasingly problematic with higher levels of dimensionality (more colors). Therefore, there may be informative data in the ungated population, especially in multiple dimensions, that escape detection. Furthermore, some of these drawbacks limit analysis of complex populations, as sequential gating involves imposition of criteria on the data that do not allow for new interpretations of poorly characterized populations. At present analysis via gating remains the most frequently used method, the need for an alternative method that addresses some of these shortcomings is clear. There is also a strong need to automate data analysis to remove operator bias, promote high throughput workflow and, perhaps most importantly, enable systematic and thorough analysis of complex data sets for hypothesis generation and biomarker discovery.
Here, we describe a novel application of Cytometric Fingerprinting (CF), a method for analysis of high-dimensional cytometric data. More details on the analytical aspects of the method and CF software (flow FP) are provided in previous publications (Rogers et al., 2008; Rogers and Holyst, 2009). CF models a multidimensional space via a binning process. The bins are then applied to the data for each time point. Counts of events in bins are flattened into a vector (the "fingerprint"). This representation of the multivariate probability distribution function provides a convenient and efficient form for subsequent application of tools taken from statistical analysis, empirical modeling or machine learning. In this case we are using CF to analyze peripheral blood immunophenotyping data, as a proof of concept for the application of this method to clinical trial data. Clinical trial data are often complex and require quantitative comparisons of the same subject before (e.g., at baseline) vs. at various times after therapy. It is problematic to perform this type of analysis with traditional gating methods, due to their subjective nature. In contrast, CF can be used to generate a model of the aggregated baseline data, enabling rapid and objective longitudinal comparisons.
We apply CF to the analysis of immunophenotyping data obtained on peripheral blood B cells from an adult patient with type 1 diabetes (T1D) who underwent pancreatic islet transplantation. Blood was drawn and studied at two time points while the patient was on transplant waitlist, allowing for the creation of baseline fingerprints. The aggregate of the baseline fingerprints was used as the model (baseline binning model) for the comparison and creation of all of the subsequent time point fingerprints. At the time of transplantation, the patient received T cell targeted induction immunosuppression with thymoglobulin, and following transplantation immunosuppression was maintained with tacrolimus and rapamycin. Peripheral blood was drawn at various time points for over one year following transplant. A total of 10 time points were studied including the two baseline samples. The immunosuppressive regimen, while severely depleting T cells during induction and inactivating their function during maintenance, was only partially effective against B cells, allowing for the analysis of B cell autoreconstitution and the effects of T cell depletion/functional impairment on B cell homeostasis. These phenomena have been described in other settings (for example myeloablative chemotherapy or rituximab treatment (Sanz and Anolik, 2005; Anolik et al., 2007; Sutter et al., 2008)) and provide an established biological context for the analysis of these data. For example, in the case of B cell autoreconstitution, we anticipate the ordered appearance of circulating B cell subsets, starting with the least mature (transitional B cells, CD27−, CD38++), followed by naïve mature (CD27−, CD38+), mature activated (CD27+, CD38+) and finally resting memory (CD27+, CD38dim/−), reflecting normal development (Bearden et al., 2005; Anolik et al., 2007; Liu et al., 2007; Sutter et al., 2008). This longitudinal design permits the use of CF to discover and quantify changing B cell subsets, including potentially novel ones. Finally, it allows for the comparison of CF to traditional flow cytometry data analysis. Herein we highlight how CF can be used to analyze longitudinal immunophenotyping data and demonstrate that CF produces comparable findings in established B cell subsets, with the added benefits of systematic, comprehensive and objective data analysis for high-resolution subset discovery.
2. Methods
2.1 Clinical information
The patient described in the main study is a 49-year-old female with a history of T1D since the age of 15, who fulfilled eligibility requirements for islet cell transplantation and entry into the NIH-sponsored Clinical Islet Transplantation (CIT) consortium protocol CIT-07. Eligibility requirements, exclusion criteria and details about the study protocol are provided at: http://www.isletstudy.org/. In brief, CIT-07 is a phase III, prospective, single-arm, multicenter trial evaluating the safety and efficacy of islet cell transplantation according to standardized islet manufacturing and immunosuppression in patients with long standing T1D complicated by severe hypoglycemia unawareness. The study protocol was approved by the University of Pennsylvania Institutional Review Board, and the patient provided written informed consent to participate. The patient received induction immunotherapy consisting of IV rabbit anti-thymocyte globulin, for a total of 3.75 mg/kg from days −2 to +2, where transplantation occurred on day 0. Maintenance immunotherapy was achieved with oral rapamycin at a dose of 0.1 mg/kg daily adjusted to achieve a 24-hour blood trough level of 10 – 15 ng/ml for the first 3 months and 8 – 12 ng/ml thereafter, and oral tacrolimus at a dose of 0.015 mg/kg twice daily adjusted to achieve a 12-hour blood trough level of 3 – 6 ng/ml. The patient received ABO and flow cross-match compatible islets from a HLA mismatched deceased donor via an intra-portal infusion. Exogenous insulin therapy was tapered off by 64 days post-transplant, and the patient has remained normoglycemic and insulin-independent with full islet graft function by β-cell secretory capacity testing (data not shown). No HLA antibodies have been detected before or after transplantation by Luminex assays. Flow cytometry of peripheral blood lymphocytes was performed at baseline (2 times) and on days 8, 29, 71, 155, 182, 273, 365 and 450 after transplant.
Immunophenotyping data from a second T1D patient (who also fulfilled CIT eligibility criteria) were used to analyze sample variability. This patient had four separate baseline samples (on days 0, 28, 97 and 293). These baseline samples were drawn and analyzed over nearly the same chronological period as the main study subject's post-transplant monitoring. Data from this second patient were also analyzed by CF and include the four baseline samples as well as several post-transplant samples (Supplementary Fig. S4).
2.2 Flow Cytometry
Immunophenotyping was performed on fresh (<24 hours old) peripheral whole blood anticoagulated with sodium-EDTA. Monoclonal antibodies against CD19-PerCP Cy5.5 (clone SJ25C1, BD-Biosciences), CD27-APC (clone L128, BD-Biosciences), CD38-PE (clone HIT2, BD-Biosciences) and CD10-FITC (clone HI10a, BD-Biosciences) were added directly to 150 µl aliquots of the whole blood and incubated at room temperature for 20 minutes. Red blood cells were lysed using ammonium chloride lysing buffer (BD Pharm Lyse) and the remaining mononuclear cells were washed using FACS buffer (Dulbecco's PBS with 5% fetal calf serum and sodium azide). The cells were fixed in 1% paraformaldehyde (Electron Microscopy Sciences). Stained cells were analyzed on a FACSCalibur (Becton Dickinson, San Jose, CA). CD19+ lymphocyte event counts ranged from 4,500 to 10,000 per time point. Compensation was performed at the time of data acquisition using lymphocytes from the same subject stained with anti-CD8 conjugated to FITC, PE, PerCP-Cy5.5 or APC. Instrument voltage settings were standardized between time points by core facility personnel using rainbow beads (RCP 3-5-7, Spherotech). Three different beads were recorded in each channel. The average coefficient of variation of the mean fluorescence intensity level for the same beads run on the same instrument over the one-year time period of the post-transplant study was 6.6% (range: 4.4–11.5%; n=9). Data were analyzed using FlowJo software (version 8.8.6, Treestar Inc., Ashland, OR). Events were gated on lymphocytes based on forward scatter vs. side scatter and on B cells based on the expression of CD19. Data were acquired in 10-bit mode (1024 channels) except for d273 (acquired in 8-bit mode, 256 channels). Data from d273 were scaled to correspond to the rest of the data for fingerprinting analysis.
2.3. CF analysis
CF analysis was carried out using Bioconductor (Gentleman et al., 2004), which in turn is built upon the R statistical computing framework (R_Development_Core_Team, 2008). The Bioconductor package flowFP was developed to facilitate the comprehensive analysis of flow cytometric data (R_Development_Core_Team, 2008; Rogers and Holyst, 2009). FlowFP is integrated with a broader suite of packages for analysis of flow cytometric data based upon the foundation package flowCore (Hahne et al., 2009).
2.4 Clustering analysis
We applied an agglomerative clustering algorithm, agnes (Kaufman and Rousseeuw, 1990) to cluster the 256 fingerprint bins according to their similarity with respect to the 10 time point observations. We used a manhattan distance metric, and the Unweighted Pair-Group Average (UPGMA) method of linkage for clustering. An intermediate number of clusters was analyzed. If one were to analyze the maximum number of the clusters (corresponding to one cluster for each of the 256 bins) no clear signal would emerge. Conversely, if too few clusters were analyzed separately, then clusters with different temporal signatures would be lumped together, obscuring biologically meaningful temporal correlations.
3. Theory of Cytometric Fingerprinting
3.1 Overview
CF analysis consists of two steps. In the first step, regions (or bins) in multivariate space are determined. In the second step, these bins are used to partition events in individual samples. Event counts in each bin are "flattened" into a list of numbers, which we refer to as a "fingerprint".
3.2 Recursive binning
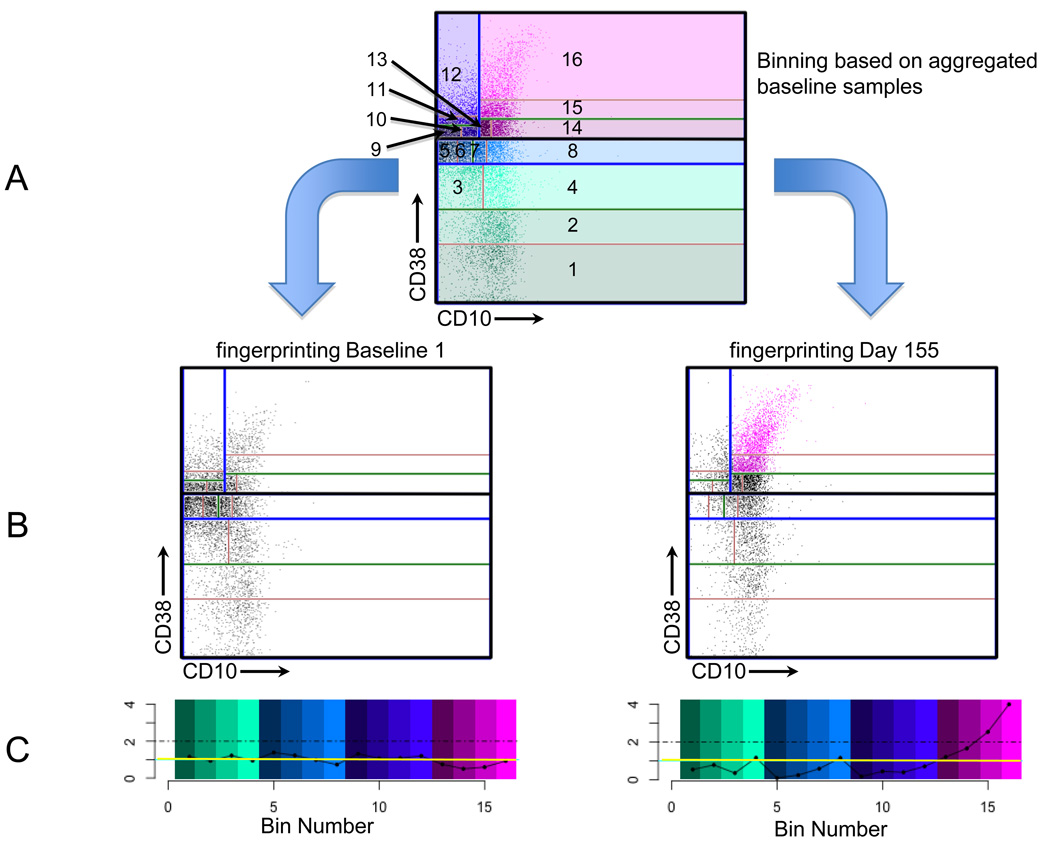
Our binning procedure follows that developed by Roederer and colleagues (Roederer et al., 2001). Bins are first determined by finding the parameter with the largest variance. The rationale is that the parameter values are distributed most broadly on this axis compared to the others, and thus dividing the data into two halves using the median on this axis does the best job of creating uniform distributions. Binning proceeds in a recursive fashion as illustrated in Fig. 1. The complete collection of bins exactly covers the whole space. Moreover, coverage is efficient in that bins have equal event occupancy. By contrast, uniform binning would require a much larger number of bins and would result in many empty bins. The final number of bins in our method is determined by the number of times this recursion is applied, and thus will be a power of 2. As discussed in (Rogers and Holyst, 2009), we chose to use a recursion level of 8, resulting in 256 bins, such that the average number of events per bin was at least 10. This provides a reasonable trade-off between resolution and statistical precision. Binning can be applied to any collection of events. In the present study we chose to use the aggregate of the baseline samples, creating a model against which subsequent time point data can be easily compared. Figures 1A and 1B show a schematic representation of this process and its application to two different time points.
Fig. 1. Schematic representation of CF.
This figure shows the process of binning and fingerprinting. For illustrative purposes, we use only 2 dimensions and 16 bins.
(A) Binning is accomplished recursively, using the aggregate of the two baseline samples for this illustration. In the first step, CD38 is found to have the most variance. The data are split at the CD38 median (thick black horizontal line) into two halves. (The lower half contains bins 1–8 and the upper half contains bins 9–16.) Each of these halves is split again in the direction of maximum variance among the events in each bin (blue lines), and then again (green lines) and once more (red lines). The resulting bins, numbered 1 – 16, are shown. Events are plotted as dots and colored according to their bin membership. The bin and dot coloration match the color scheme provided in (C).
(B) Two samples are compared with the bin boundaries determined in (A). The left panel is one of the two baseline samples and the right panel shows the sample on day 155. The number of events in each of these bins is counted to create the two fingerprints expressing comparison with the baseline aggregate. Events in bins with more than 2-fold increase relative to aggregated baseline are plotted as pink dots. All other events are plotted as black dots.
(C) Fingerprints are plotted on a normalized scale. The left panel corresponds to the baseline sample (as in (B)) and the right panel corresponds to the day 155 sample. The y-axis denotes fold difference relative to the aggregated baseline, where a value of 1 indicates identity and is shown as a horizontal yellow line. The x-axis indicates the bin numbers and the bins are colored as shown in (A). The dot-dashed lines indicate the values of bins with 2-fold increased numbers of events compared to the baseline model. Note that bins 15 and 16 in the right panel exceed the 2-fold threshold.
3.3 Fingerprinting
A fingerprint is computed by counting the number of events in a sample falling into each bin of the model. Thus, a fingerprint is essentially a histogram. The x-axis of the histogram represents a list of bins, and the y-axis represents the number of events in each bin. Fingerprints can be normalized in order to better represent shifts in B cell subsets. Fig. 1C shows the normalized events in each bin relative to the aggregated baseline. Fingerprints represent multidimensional data in a form that lends itself to detailed comparison of changes in distributions.
CF-based comparisons can be graphically represented in various ways. In the following sections we show (a) the development of a CF model based on the aggregated baseline data, (b) the computation of fingerprints for each of the individual time point data sets, (c) the display of fingerprints as histograms that represent differences in the multivariate distributions between each time points and the baseline model and (d) the mapping of temporally correlated bins (revealed either in fingerprints or by agglomerative clustering) to bivariate plots and parallel coordinate plots to determine their relationship to known or novel lymphocyte subsets.
4. Results
4.1 B cell subset analysis using standard gating techniques
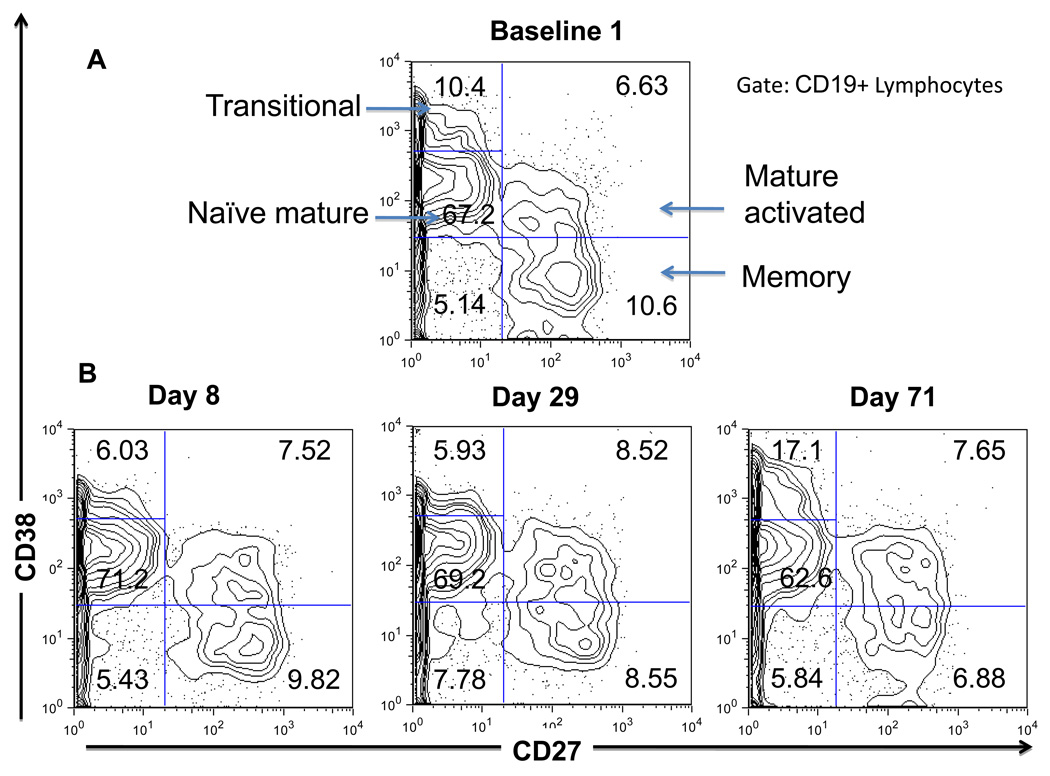
Flow cytometry data from a patient on the transplant waitlist and at various times following pancreatic islet transplantation were analyzed using standard bivariate plots. Through the classical gating and quadrant divisions, circulating B lymphocytes (CD19+) were classified into 5 different well-established subpopulations: transitional (new bone marrow émigrés, CD27−, CD38++), naïve mature (CD27−, CD38+), activated mature (CD27+, CD38+), memory (CD27+, CD38−) and plasmablasts (CD27++, CD38++) (Pascual et al., 1994; Klein et al., 1998; Sims et al., 2005; Abdallah and Prak, 2006). In most samples the plasmablast fraction comprised fewer than 0.5% of the B cells. Thus the analysis was restricted to the four major subpopulations, excluding plasmablasts. Previous experiments with fluorescence minus one controls were used to distinguish positive from negative (background) fluorescence levels (data not shown). However, this method does not distinguish the transitional from the naïve mature B cell subsets. This distinction is made based upon prior knowledge of where the transitional B cell subpopulation typically resides using samples from patients treated with rituximab who are undergoing autoreconstitution in whom nearly all circulating B cells have a transitional phenotype (Sutter et al., 2008). B cells residing in the transitional gate are also known to express additional markers that are characteristic of earlier stage B cells including bright expression of CD24, dim expression of CD10 and subset positivity for CD5 (Sims et al., 2005; Marie-Cardine et al., 2008; Palanichamy et al., 2009; Luning Prak, in-press). This classification scheme is shown for a representative baseline sample in Fig. 2A. The baseline samples (n=2) from this patient show a similar B cell subset distribution expressed as a percentage of total B cells (see Fig. 3, discussed below). Samples from subsequent time points were analyzed by copying the gating settings from the baseline samples and making minor adjustments to accommodate small changes in mean fluorescence intensity. Figure 2B provides examples of this analysis for the first 3 time points post-transplant.
Fig. 2. Standard flow cytometric analysis of peripheral B cell subsets.
(A) CD27 vs. CD38 analysis performed on B cells (CD19+ lymphocytes) at baseline (shown is one of the two baseline time points). Arrows indicate the major B cell subsets: transitional (CD27−CD38++), naïve mature (CD27−CD38+), mature activated (CD27+CD38+) and resting memory (CD27+CD38−). Percentages of B cells are provided within each gated region.
(B) CD27 vs. CD38 analysis performed on B cells at days 8, 29 and 71 post-transplantation. Percentages of B cells are provided within each gated region.
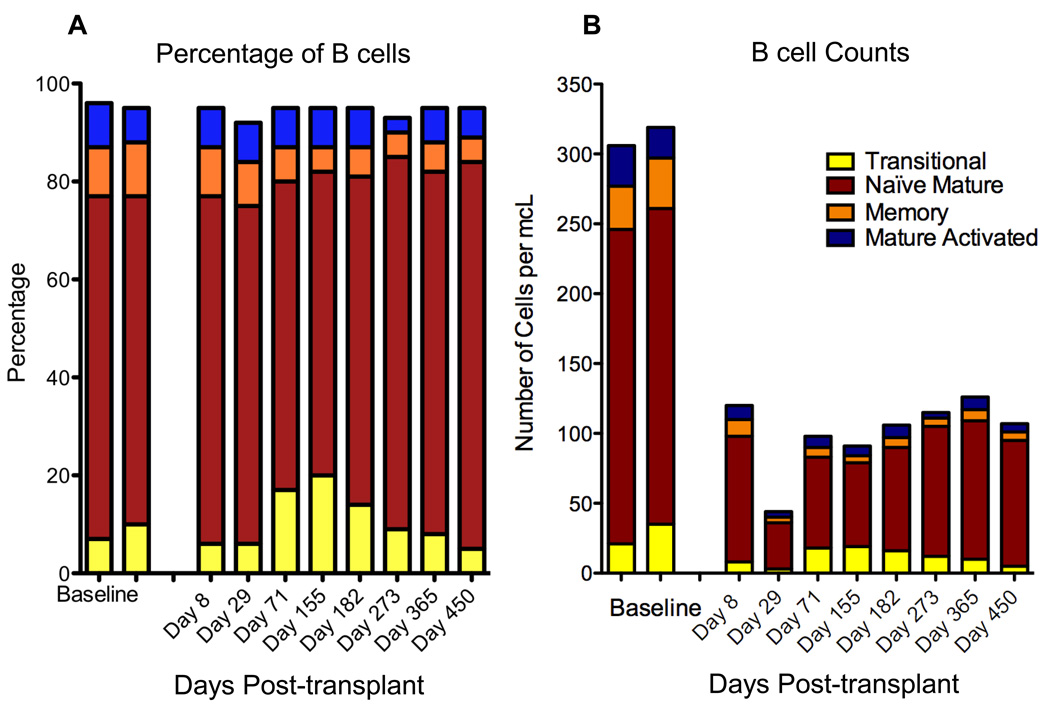
Fig. 3. Changes in B cell subsets over time.
Stacked bars show the progression over time of B cell subsets, analyzed as shown in Fig. 2. Each bar represents a single time point.
(A) Percentage of B cells. The proportion of CD19+ lymphocytes is plotted as a function of the time point post-transplant (in days).
(B) Absolute B cell subsets. The absolute B lymphocyte count (B cells per microliter of whole blood) is plotted as a function of the time point post-transplant (in days). The two leftmost bars in both panels represent baseline data obtained one month apart while the patient was on the transplant waiting list.
The percentage and absolute numbers of B cell subsets are shown in Figs. 3A and 3B, respectively. These graphs illustrate the manner and kinetics of lymphocyte auto-reconstitution following transplant in this patient. The pattern of peripheral B cell reconstitution consisted of an initial increased relative proportion of B cells with a transitional phenotype followed by gradual accumulation of naïve mature B cells. This pattern of recovery suggests that the B cell repertoire is in part re-set. These data also reveal the slow kinetics of B cell recovery; the patient exhibited reduced B cell numbers up to and including d450 post transplant. While the relative frequencies of the different B cell subsets appear similar to baseline, the absolute B cell numbers have not yet returned to baseline levels by d450.
4.2 Fingerprinting Analysis: Generation of a Baseline Binning Model and Analysis of Variance
Next, the same raw list mode data (FCS files) were analyzed using CF. Since the aim here was to fingerprint B lymphocytes, a computational “pre-gating” analysis was performed to exclude non-lymphoid populations and include only CD19+ lymphocytes (this analysis is shown in Supplementary Figure S1). The gated B cells were subjected to CF using the following parameters: CD38, CD27, CD19 and CD10.
A baseline binning model was created as described in section 3.2. Fingerprints were then computed and applied in order to compare B cell subset densities in the different samples with respect to this baseline model. Finally, the results were displayed in a stack of fingerprint plots, with each plot corresponding to an individual sample (time point). This representation emphasizes increases in event frequencies relative to the baseline sample, and provides a convenient visual indication of longitudinal population trends. The computing times on a laptop computer with a 1.86 GHz Intel Core 2 Duo processor and 2 GB of memory were as follows: read and scale 10 FCS files (50 sec); compute lymphocyte and CD19+ gates (8 min); bin the baseline samples and compute the fingerprints (1 sec); produce the graphics shown in Figs. 4–7 (5 min).
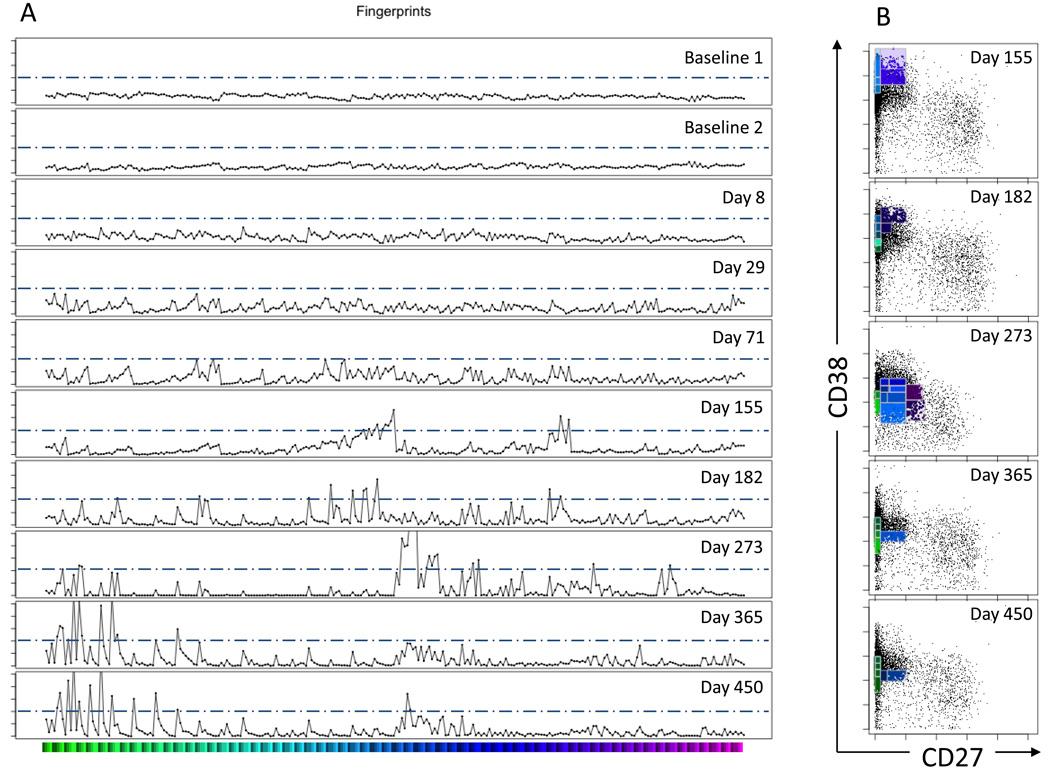
Fig. 4. CF histograms and bivariate plots.
(A) Cytometric Fingerprints. Each row corresponds to the fingerprint of an individual sample relative to the aggregated baseline binning model. The x-axis is the bin number. The y-axis indicates the fold increase relative to the aggregated baseline. The tick marks for the y-axes represent fold intervals of 2. The horizontal bar under the histogram plots provides color-coding for the bin members that are plotted in panel B. The dashed horizontal lines in each fingerprint indicate the 4-fold threshold of increased density relative to the baseline model.
(B) Bivariate plots of highlighted CF histogram bins. At time points d155, d182, d273, d365 and d450 there were fingerprint bins that exceeded the threshold of a 4-fold increase (refer to Panel A). Bivariate plots of CD27 vs. CD38 expression are shown for these time points. The black dots represent all of the events at the indicated time point. The dots highlighted with color correspond to events in the >4-fold increased bins in the corresponding fingerprint. The bivariate plots indicate a progression from less mature (CD27−, CD38++) to more mature (CD27−, CD38+) phenotypes as a function of time post-transplant.
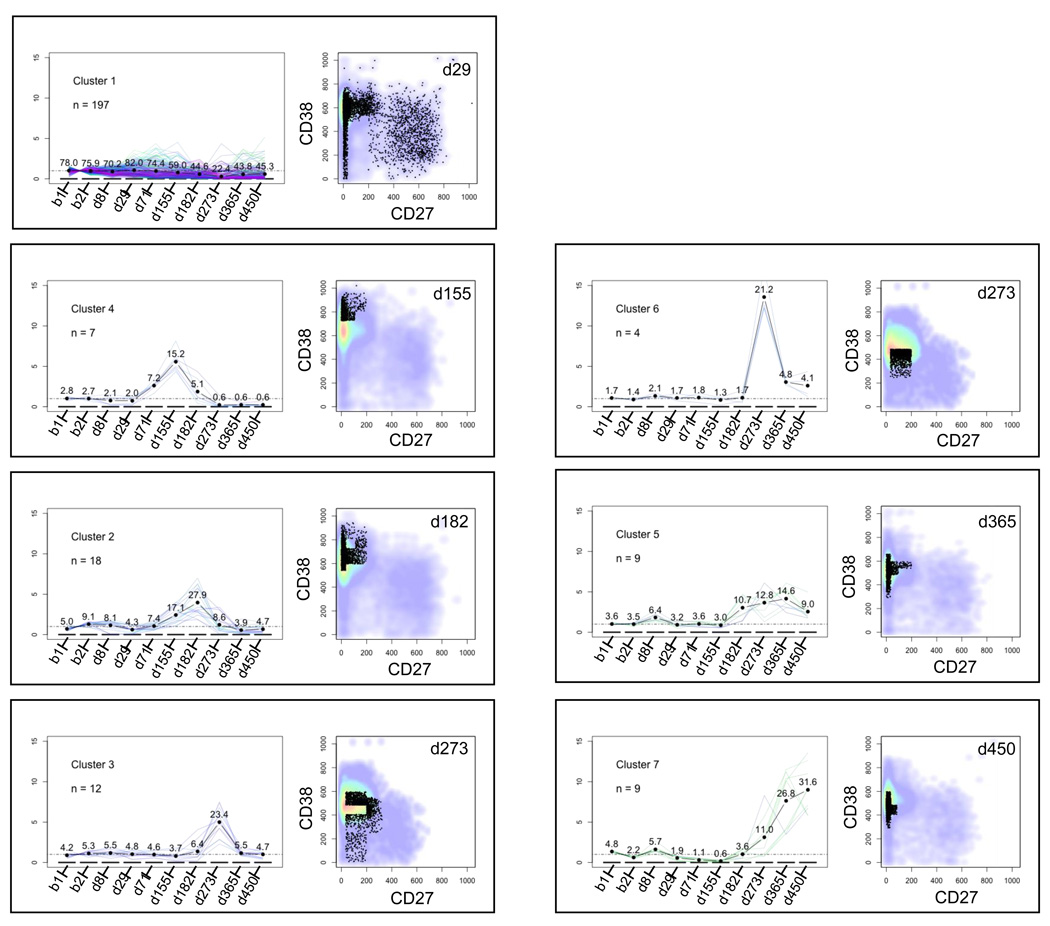
Fig. 7. Analysis of temporally correlated bin clusters.
Each panel represents one of the clusters shown in the dendrogram in Fig. 6. The left side of each panel provides a plot of the fold change relative to baseline for the cluster as a function of time. Each thin line in this plot represents one bin in the cluster, while the thick black line represents the average fold change for all of the bins in the cluster. Numbers above the average fold change line denote the percentage of B cells in the cluster at each time point. For example, cluster number 4 at day 155 contains 15.2% of the B cells. The right side of each panel shows a bivariate plot of CD38 vs. CD27 fluorescence intensity (gated on CD19+ lymphocytes). The plots shown correspond to the time points at which the average cluster density is maximal (e.g., d155 for cluster number 4). The black dots represent the B cells in the cluster. The pseudocolor background denotes all of the B cells at the corresponding time point. Panels are ordered from top to bottom and left to right chronologically according to the time at which their average density peaks.
In Fig. 4A, each row corresponds to the fingerprint of an individual sample relative to the baseline binning model. The baseline model was derived from the aggregate of the two baseline samples and the level of variation in the baseline samples was low. Thus, the fingerprints of the individual baseline samples are nearly flat, with minimal deviation. Qualitatively similar fingerprints are obtained if either baseline sample or both baseline samples are used to produce the binning model (Supplementary Fig. S3). However, the aggregated baseline model effectively averages small differences between the baseline samples, potentially permitting the more accurate detection of subtle features in post-transplant samples than would have been possible with only a single baseline sample model.
In contrast to the baseline samples, the fingerprints of the non-baseline samples reveal greater fluctuations (Fig. 4A). Apart from the visual appearance of the fingerprints themselves, one can analyze the level of variability in the fingerprints by calculating the standard deviation of each fingerprint relative to the aggregated baseline. This calculation yields values of 0.30 for each of the baseline samples and values of 0.50, 0.64, 0.84, 1.14, 1.21, 2.21, 1.73 and 1.80 for the subsequent time points (days 8, 29, 71, 155, 182, 273, 365 and 450 post transplant, respectively). All of the standard deviations for the post-transplant samples are higher than the baseline samples. An alternative approach is to measure the maximal fold change relative to aggregated baseline. This analysis yields values of 1.6 and 1.8 for the baseline measurements and values of 2.6, 3.4, 4.0, 7.9, 7.7, 17.5, 11.0 and 13.2 for the subsequent time points (days 8, 29, 71, 155, 182, 273, 365 and 450 post transplant, respectively.) In addition, we performed a separate analysis of a second subject on whom four baseline samples were obtained over a 10-month time period that overlapped with the study for the first subject (Supplementary Fig. S4). It is apparent that the level of variation in the baseline samples from the second subject is also lower than in the post-transplant samples. No baseline sample from either subject has a 4-fold or higher change relative to the aggregated baseline binning model. We therefore chose 4-fold as a cut-off for identifying bins with significantly altered event numbers relative to the baseline binning model.
4.3 Representation of CF data in bivariate and parallel coordinate plots
To determine the corresponding phenotypes of the B cells within the bins containing 4-fold or higher increased numbers relative to the baseline model, the fingerprinting data were represented in conventional bivariate-plots. Shown in Fig. 4B are the bins that meet this 4-fold cut-off. The bins that have changed by 4-fold or higher relative to the baseline binning model are represented in color. The colors correspond to the location of the bin on the x-axis of the fingerprints in Fig. 4A. Only time points in which there were bins with 4-fold or more events relative to the binning model are shown. All of the other events (that have increased by less than 4-fold) are shown as black dots in the bivariate plots. The bivariate plots allow for visualization of changes that are detected by CF, allowing one to relate the events in the altered bins to immunologically defined peripheral B cell subsets.
Fig. 4B shows that CF has identified a relative increase in transitional B cells (CD27−, CD38++) at days 155 and 182, followed by a relative increase in naïve mature B cells (CD27−, CD38+) on days 273, 365 and 450. These results are similar to those obtained using the conventional method described in section 4.1 (see Fig. 3A). With CF, relative increases from the baseline and trends in those increases over time are readily apparent and quantifiable.
To evaluate the expression of all of the markers amongst cells, one can map the altered events onto all possible bivariate plots, as shown in Supplementary Fig. S2. Using the fold changes as cut-offs, events populating bins exceeding the cut-offs can be colored red (≥4-fold increase relative to baseline) or blue (≤0.25-fold relative to baseline), with the remaining events represented in grey. Mapping these quantitative features onto qualitative bivariate plots allows those features to be interpreted in their biological context. Thus, Supplementary Figure S2 allows one to view the B cell subsets of interest in multiple dimensions, to correlate the expression of all of the markers in the tube and thereby to extend the phenotypes of the B cell subsets. For example if one focuses on the red dots, CD10 levels decrease as CD38 levels decrease, accompanying the shift from transitional to naïve mature B cells, consistent with their known marker expression profiles. Thus, CF facilitates temporal and more comprehensive description of B cell subset composition.
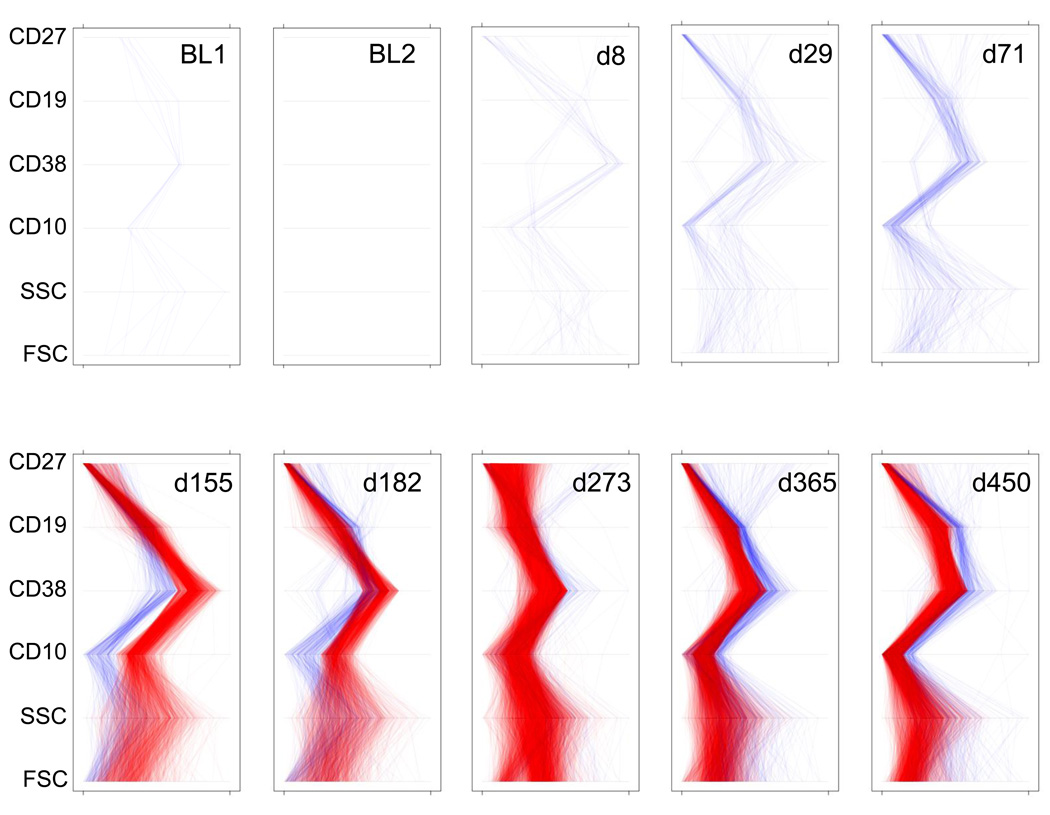
Parallel coordinate plots provide another means of displaying multi-dimensional immunophenotyping data (Inselberg et al., 1987; Streit et al., 2006). An advantage of parallel coordinate views is that, unlike with bivariate plots, all of the markers in an experimental tube can be graphically related in a single plot. Thus, in Fig. 5, each line represents an individual cell. When the lines diverge, it is easy to see shifts in very precisely defined populations with time. For example, on day 155 the transitional cell phenotype (CD19+CD27−CD38++CD10+) is increased compared to baseline. Conversely, the blue lines highlight cells that are decreased relative to the baseline. Fig. 5 compares the expression pattern of these markers and relative density of subsets across time. As was seen before, cells with a transitional phenotype are increased, followed by successive increases in naive mature cells (CD19+, CD27−, CD38+, CD10−) at later time points (for example, at d365).
Fig. 5. Parallel Coordinate Plots.
Each panel represents a summary of phenotypic changes at a given time point relative to the aggregated baseline. Each line, from bottom to top of an individual panel, corresponds to a single B cell. Lines are generated by connecting the parameter values for each cell. Red lines indicate cells in bins that are increased by at least 4-fold relative to baseline. Blue lines indicate cells in bins that are decreased at least 4-fold relative to baseline. The y-axis gives the six parameters (FSC, SSC, CD10, CD38, CD19 and CD27). The x-axis for each of the parameters is scaled from the minimum to the maximum observed values for that parameter. BL = baseline. FSC = forward scatter. SSC = side scatter.
4.4. Clustering of fingerprint features
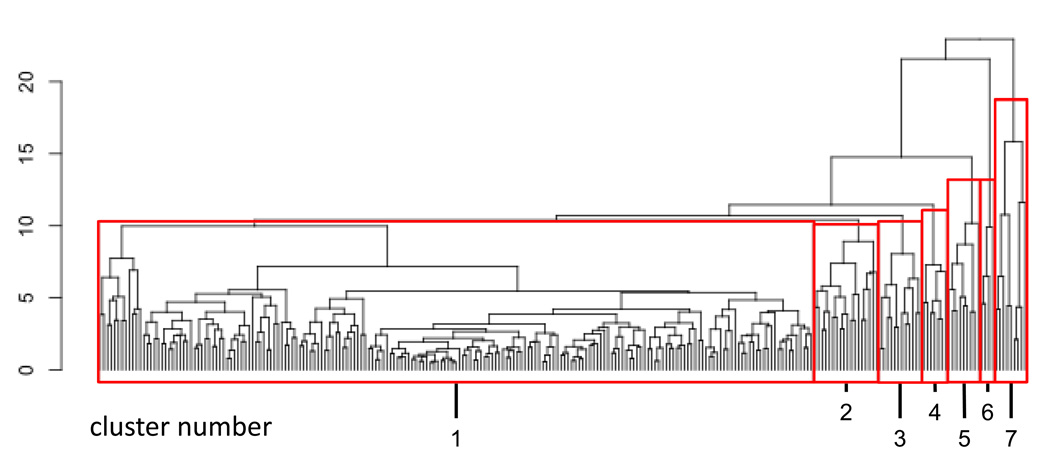
When one visually compares the fingerprints from different time points, it is apparent that multiple bins exhibit correlated shifts that progress over time. Here we wished to determine if CF data could be subjected to clustering analysis to identify temporal progressions in the B cell subsets. This analysis method can be performed without prior knowledge of what the temporal progression is, and thus can be useful for data mining and lymphocyte subset discovery. Thus fingerprint features were clustered according to the similarity of their temporal trajectories (see Section 2.4). Fig. 6 shows the resulting dendrogram. The 256 bins were grouped into 7 clusters as indicated below the dendrogram. Each cluster of bins represents a (possibly disjoint) subregion of the four-dimensional space (CD27, CD38, CD10 and CD19).
Fig. 6. Dendrogram of temporally correlated clusters.
Temporally correlated bins were clustered as described in section 2.4 and plotted as a dendrogram. The y-axis represents the distance (computed as described in the text) at which clusters merge. The dendrogram was parsed into 7 clusters, numbered 1 through 7. The same cluster numbers are used in Fig. 7.
Cluster 1, subtending 215 of the 256 bins, showed little variation over the 10 time-point observations. The other clusters, however, showed pronounced and distinctive variation. Fig. 7 shows the fold change vs. the aggregated baseline for each cluster with respect to time. The accompanying bivariate plots display data from the time point where the maximum fold change vs. baseline occurred for each cluster. The clusters shown in Fig. 7 are ordered chronologically, according to time points at which the maximum change occurs, starting with the earliest. Thus cluster 1 is followed by clusters 4, 2, 3, 6, 5 and 7. (Clusters 3 and 6 have the same time points of maximal increase relative to the aggregated baseline.) The temporal progressions highlighted in bins 2–7 correspond to changes in transitional and naïve mature B cells that were also observed in the bivariate, fold-difference and parallel coordinate analyses (sections 4.2 and 4.3).
There are two types of clusters. In the first type, events have a more heterogeneous pattern of cell surface antigen expression, suggesting the presence of more than one B cell subset that did not change significantly in frequency over time. Cluster 1 exhibits this pattern. In the second type, the events comprising the cluster display a characteristic pattern of cell surface antigen expression, consistent with the presence of a single B cell subset. In keeping with this interpretation, the timing of appearance of some of the clusters corresponds to the known maturation sequence of peripheral B cell subsets. For example, cluster 4, peaking at day 155, corresponds to emerging transitional B cells while cluster 3, peaking at day 273, appears to correspond to a subset of naïve mature B cells.
Finally, the clustering data also appear to discriminate between different groups of cells within known B cell subsets. For example, clusters 4 and 2 appear to be contained within the transitional B cell subset, whereas clusters 3, 6, 5 and 7 appear to be contained within the naïve mature B cell subset. These may represent novel B cell sub-populations.
5. Discussion
5.1. Classical analysis vs. fingerprinting
Here we highlight the use of CF for the analysis of complex, multiparameter flow cytometric data gathered on a single subject over multiple time points. In contrast to traditional immunophenotyping analysis methods (electronic gating followed by visually-guided analysis in bivariate plots), CF utilizes an initial modeling step to establish the baseline binning model, allowing for a statistical analysis of the data. CF is not as reliant upon an underlying assumption of discreteness of populations and allows for a more comprehensive analysis of the data, including potential discovery of new subsets in multiple dimensions. CF also more readily allows for the comparison of temporal shifts in the relative abundance of different lymphocyte subsets. CF can be interfaced with a variety of other data analysis tools including cluster analysis as well as methods for data visualization including bivariate plots and parallel coordinate plots. With these visualization methods significant changes in the B cell subsets relative to baseline can be highlighted by color. In contrast, the use of electronic gating and bivariate plots to analyze flow cytometric data is labor-intensive and there is limited capability to survey the entire data set systematically or to discover or track the temporal progression of populations that are defined in more than two dimensions. Thus, compared to traditional immunophenotyping data analysis, CF is an efficient method for systematically surveying multiparameter data for longitudinal trends in known lymphocyte subsets as well as a tool for novel subset discovery.
There are, however, also some limitations and potential disadvantages to CF. First, CF is more sensitive than traditional gating. Therefore, it is potentially more likely to detect pre-analytical and instrument derived sources of variation. Another limitation of CF, as it is used herein, is that the location of a bin on the x-axis of a cytometric fingerprint is arbitrary and dependent upon the specific multivariate distribution of the data that were used to create the binning model; in other words, it would be meaningless to compare fingerprints that were derived from different models. Therefore, in this example, CF was used for the analysis of intra-individual rather than inter-individual variation. Finally, a direct comparison of CF with conventional bivariate plot data is limited in that there is not a 1:1 correspondence between the probability bins and conventionally defined circulating B cell subsets (because the conventionally defined subsets are only defined in two dimensions after CD19 gating).
CF, like traditional immunophenotyping data analysis methods, measures relative changes in the distribution of events within defined bins. As with conventional bivariate plots, conversion of flow cytometry data from relative events to absolute events can reveal significant biological changes. Also, there are subjective aspects to CF both in terms of establishing the binning model and with respect to data analysis. When establishing the binning model, one has to decide on which samples to use, how many bins there will be, how the bins will be divided and if any pre-gating will be performed (for example, in this study we pre-gated on CD19+ lymphocytes). When analyzing the fingerprinting data, one has to define what constitutes a biologically meaningful cut-off for fold change in a given fingerprint or collection of fingerprints relative to the binning model. Furthermore, if cluster analysis is performed to detect temporal or other correlations in the CF data, one has to decide which type(s) of algorithm to use and define the level of branching.
5.2. CF data and B cell subset analysis
The parallel use of CF and traditional flow cytometric data analysis reveals a similar trend in B lymphocyte subset composition using either method in a patient with T1D over time. Using traditional analysis, the two baseline samples exhibit a similar distribution of the major B cell subsets, with a predominance of naïve mature cells. On day 8 following pancreatic islet transplantation, the relative shifts in the B cell subsets are minimal compared to the baseline sample. However, the absolute frequencies of all of the circulating B cell subsets are decreased. By d29 post transplant, the nadir in the absolute B cell count is reached at 47 B cells per microliter of whole blood. We speculate that B cell depletion in this patient was likely due to anti-B cell antibodies within the polyclonal rabbit anti-thymocyte globulin preparation (Zand, 2006). At d71 there is both a relative and an absolute increase in the frequency of transitional B cells, the first B cells to exit from the bone marrow, consistent with autoreconstitution (Sanz and Anolik, 2005). By d155, the transitional B cell fraction reaches its peak. By d182, the relative fraction of transitional B cells starts to decrease while the fraction of naïve mature B cells begins to increase. Naïve mature B cells continue to increase in their relative frequency up to and including day 450 post-transplant. The sequential appearance of transitional followed by naïve mature B cells is consistent with their developmental progression and resembles other examples of autoreconstitution in immunosuppressed patients (Sanz and Anolik, 2005; Anolik et al., 2007; Sutter et al., 2008).
What is remarkable, however, is that the B cells remain depleted up to and including day 450 post transplant. Between days 8 and 450, the patient only received maintenance therapy with tacrolimus and rapamycin. One possible explanation for the sustained B cell depletion in this patient is that T cell depletion therapy indirectly affects B cell maturation or homeostasis. For example, rapamycin inhibits polyclonal B cell activation and differentiation into antibody secreting cells in vitro (Kay et al., 1991; Aagaard-Tillery and Jelinek, 1994). It is also intriguing that the absolute number of transitional B cells does not increase above the baseline level and in fact decreases out to day 450. This pattern of B cell reconstitution differs from that seen in subjects who are depleted only with rituximab (anti-CD20) (Anolik et al., 2007; Sutter et al., 2008). The latter typically experience B cell recovery after 5–6 months and achieve normal or supranormal B cell numbers by 12 months, often with a predominance of transitional B cells. In the T1D transplant patient studied here, the failure of transitional B cells to increase despite the low B cell count could reflect defective or suppressed production of new B cells in the bone marrow. The limited production or maintenance of B cells could be due to an intrinsic defect in the B cell lineage or, perhaps more likely, result from the near absence of T cells. Circulating T cell numbers remained extremely low throughout the post-transplant period, dropping from 1,700 CD3+ lymphocytes per microliter of whole blood to ≤60/µL (range 5–60). Consistent with this hypothesis, previous studies in mice suggest that B cell generation and maintenance are T cell dependent (Chen et al., 1998; Choudhury et al., 2005).
This analysis also highlights how CF facilitates the comparison of multidimensional features of the B cell subsets. For example, in the parallel coordinate plots, the increased transitional cells at day 155 post transplant also express CD10 at higher levels (red lines) than the other circulating B cells, consistent with their early maturational state (Billips et al., 1995; Ghia et al., 1996). Overall, the pattern of B cell recovery following partial depletion, displayed here in a variety of complementary ways through fingerprint analysis, suggests that the B cell repertoire is in part re-set after immunosuppression with a new population of peripheral B cells emerging from the bone marrow.
5.3. The use of CF for lymphocyte subset discovery
The agglomerative clustering analysis highlights the potential for CF to reveal novel subsets or temporal correlations between multiple subsets. For example, the temporal correlation analysis yielded two distinct CD38++, CD27− subpopulations (clusters 4 and 2) that peaked at different time points (days 155 and 182 post transplant, respectively). Recently, transitional subsets have been identified in humans and subdivided on the basis of CD5, CD10 and CD24 expression ((Carsetti et al., 2004; Marie-Cardine et al., 2008; Palanichamy et al., 2009) and our unpublished data). In addition, within the CD38++, CD27− population, there may also be regulatory B cells, which have an extended phenotype of CD5+, CD10+, IgM++ and IgD++ (Blair et al.). Thus, clusters 2 and 4 may represent different transitional subsets or a transitional subset and a regulatory B cell subset.
There is also evidence from the temporal clustering analysis for temporally distinct subpopulations within the CD38+, CD27− region. During reconstitution following B cell depletion or chemotherapy, it has been observed that the fluorescence intensity of CD38 progressively decreases as B cells mature from the transitional fraction into the naïve mature fraction. The progressive decrease of CD38 staining intensity is also observed here. Differences in CD38 intensity may correspond to distinct subsets within the naïve mature B cell population. For example, a population of B cells with intermediate levels of CD38 staining (intermediate between transitional cells and naïve mature cells) has been described. This CD38int, CD27− B cell population is also CD5+ and CD10int and is considered to be "pre-naïve" based upon its immunophenotype and functional activity in vitro (Lee et al., 2009). On the other end of the naïve mature spectrum, there appear to be subsets of CD38dim, CD27− class switched B cells that express CD95 and harbor somatically mutated immunoglobulin genes, suggestive of an antigen-experienced B cell population (Fecteau et al., 2006; Jacobi et al., 2008). Thus, a prediction of the temporal clustering is that cell subsets that fall into temporally correlated bins will have distinctive immunophenotypic characteristics when additional markers are studied either in parallel or in higher dimensional immunophenotyping data sets.
Alternatively or in addition, some of the temporally clustered bins may contain mixed populations of B cells that share a common regulatory mechanism or are affected indirectly by the relative changes in other subsets. For example, cluster 1 contains a mixed population of B cells including naïve mature, resting memory and mature activated B cells. The bivariate plots reveal that the relative frequencies of resting memory, mature activated and subsets of naïve mature B cells appear to decrease following transplantation. This decrease could, in part, be due to a relative increase in earlier B cell subsets including transitional B cells and CD38 brighter naïve mature cells.
5.4. Conclusions and Future Directions
In this paper CF is used to analyze B cell immunophenotyping profiles from a T1D patient at multiple time points following pancreatic islet transplantation. CF permitted the systematic analysis of this multidimensional data set and provided an efficient means for comparing relative changes in B cell subsets with respect to time. Once the baseline binning model is created, it is possible to rapidly construct fingerprints for all of the subsequent samples. Thus, this analysis highlights how CF can be used to screen for temporally correlated B cell subsets and potentially to define novel subsets. Immunophenotyping data can be further analyzed by coupling CF with other data analysis and visualization methods, including agglomerative clustering, parallel coordinate plots and bivariate plots. In addition to longitudinal analysis, a common goal of clinical or translational immunophenotyping is to compare disease states to normal controls. Fingerprinting of the normal controls could also be used to model the multidimensional space and provide a basis for comparison to the disease state. It will also be of interest to further develop the method to compare longitudinal data in different patient groups, but this will require a refinement of the baseline modeling step.
Supplementary Material
Acknowledgments
This research was performed as a project of the Clinical Islet Transplantation Consortium, a collaborative clinical research project headquartered at the NIDDK and NIAID. This work was supported by PHS grants U01-DK070430 (A.N., M.R.R., E.T.L.P., C.L.) and T32-AR-07442-23 (D.R.S.) and a fellowship grant from the Juvenile Diabetes Research Foundation 3-2007-715 (J.S.). We thank the reviewers of this manuscript for their helpful suggestions. We are also indebted to Cornelia Dalton-Bakes, Eileen Markmann, and Maral Palanjian for their coordination of the clinical trial and patient care, to Noah Goodman and Yang-Zhu Du and the Path BioResource Flow Cytometry and Cell Sorting Facility for technical assistance and to the patients for their participation.
abbreviations
- CF
cytometric fingerprinting
- T1D
type 1 diabetes
- CIT
clinical islet transplantation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aagaard-Tillery KM, Jelinek DF. Inhibition of human B lymphocyte cell cycle progression and differentiation by rapamycin. Cell Immunol. 1994;156:493–507. doi: 10.1006/cimm.1994.1193. [DOI] [PubMed] [Google Scholar]
- Abdallah KO, Prak ET. B cell monitoring of transplant patients treated with anti-CD20. Clin Transpl. 2006:427–437. [PubMed] [Google Scholar]
- Anolik JH, Friedberg JW, Zheng B, Barnard J, Owen T, Cushing E, Kelly J, Milner EC, Fisher RI, Sanz I. B cell reconstitution after rituximab treatment of lymphoma recapitulates B cell ontogeny. Clin Immunol. 2007;122:139–145. doi: 10.1016/j.clim.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Bearden CM, Agarwal A, Book BK, Vieira CA, Sidner RA, Ochs HD, Young M, Pescovitz MD. Rituximab inhibits the in vivo primary and secondary antibody response to a neoantigen, bacteriophage phiX174. Am J Transplant. 2005;5:50–57. doi: 10.1111/j.1600-6143.2003.00646.x. [DOI] [PubMed] [Google Scholar]
- Billips LG, Nunez CA, Bertrand FE, 3rd, Stankovic AK, Gartland GL, Burrows PD, Cooper MD. Immunoglobulin recombinase gene activity is modulated reciprocally by interleukin 7 and CD19 in B cell progenitors. J Exp Med. 1995;182:973–982. doi: 10.1084/jem.182.4.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair PA, Norena LY, Flores-Borja F, Rawlings DJ, Isenberg DA, Ehrenstein MR, Mauri C. CD19(+)CD24(hi)CD38(hi) B cells exhibit regulatory capacity in healthy individuals but are functionally impaired in systemic Lupus Erythematosus patients. Immunity. 32:129–140. doi: 10.1016/j.immuni.2009.11.009. [DOI] [PubMed] [Google Scholar]
- Carsetti R, Rosado MM, Wardmann H. Peripheral development of B cells in mouse and man. Immunol Rev. 2004;197:179–191. doi: 10.1111/j.0105-2896.2004.0109.x. [DOI] [PubMed] [Google Scholar]
- Chen F, Maldonado MA, Madaio M, Eisenberg RA. The role of host (endogenous) T cells in chronic graft-versus-host autoimmune disease. J Immunol. 1998;161:5880–5885. [PubMed] [Google Scholar]
- Choudhury A, Maldonado MA, Cohen PL, Eisenberg RA. The role of host CD4 T cells in the pathogenesis of the chronic graft-versus-host model of systemic lupus erythematosus. J Immunol. 2005;174:7600–7609. doi: 10.4049/jimmunol.174.12.7600. [DOI] [PubMed] [Google Scholar]
- Fecteau JF, Cote G, Neron S. A new memory CD27-IgG+ B cell population in peripheral blood expressing VH genes with low frequency of somatic mutation. J Immunol. 2006;177:3728–3736. doi: 10.4049/jimmunol.177.6.3728. [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghia P, ten Boekel E, Sanz E, de la Hera A, Rolink A, Melchers F. Ordering of human bone marrow B lymphocyte precursors by single-cell polymerase chain reaction analyses of the rearrangement status of the immunoglobulin H and L chain gene loci. J Exp Med. 1996;184:2217–2229. doi: 10.1084/jem.184.6.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahne F, LeMeur N, Brinkman RR, Ellis B, Haaland P, Sarkar D, Spidlen J, Strain E, Gentleman R. flowCore: a Bioconductor package for high throughput flow cytometry. BMC Bioinformatics. 2009;10:106. doi: 10.1186/1471-2105-10-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inselberg A, Tuval C, Reif M. Convexity algorithms in parallel coordinates. Journal of the ACM. 1987;34:765–801. [Google Scholar]
- Jacobi AM, Reiter K, Mackay M, Aranow C, Hiepe F, Radbruch A, Hansen A, Burmester GR, Diamond B, Lipsky PE, Dorner T. Activated memory B cell subsets correlate with disease activity in systemic lupus erythematosus: delineation by expression of CD27, IgD, and CD95. Arthritis Rheum. 2008;58:1762–1773. doi: 10.1002/art.23498. [DOI] [PubMed] [Google Scholar]
- Kaufman L, Rousseeuw PJ. Finding Groups in Data: An Introduction to Cluster Analysis. New York: Wiley; 1990. [Google Scholar]
- Kay JE, Kromwel L, Doe SE, Denyer M. Inhibition of T and B lymphocyte proliferation by rapamycin. Immunology. 1991;72:544–549. [PMC free article] [PubMed] [Google Scholar]
- Klein U, Rajewsky K, Kuppers R. Human immunoglobulin (Ig)M+IgD+ peripheral blood B cells expressing the CD27 cell surface antigen carry somatically mutated variable region genes: CD27 as a general marker for somatically mutated (memory) B cells. J Exp Med. 1998;188:1679–1689. doi: 10.1084/jem.188.9.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Kuchen S, Fischer R, Chang S, Lipsky PE. Identification and characterization of a human CD5+ pre-naive B cell population. J Immunol. 2009;182:4116–4126. doi: 10.4049/jimmunol.0803391. [DOI] [PubMed] [Google Scholar]
- Liu C, Noorchashm H, Sutter JA, Naji M, Prak EL, Boyer J, Green T, Rickels MR, Tomaszewski JE, Koeberlein B, Wang Z, Paessler ME, Velidedeoglu E, Rostami SY, Yu M, Barker CF, Naji A. B lymphocyte-directed immunotherapy promotes long-term islet allograft survival in nonhuman primates. Nat Med. 2007;13:1295–1298. doi: 10.1038/nm1673. [DOI] [PubMed] [Google Scholar]
- Luning Prak ET, Ross J, Sutter J, Sullivan KE. Age-related trends in pediatric B cell subsets. Pediatric and Developmental Pathology. doi: 10.2350/10-01-0785-OA.1. (in-press) [DOI] [PubMed] [Google Scholar]
- Marie-Cardine A, Divay F, Dutot I, Green A, Perdrix A, Boyer O, Contentin N, Tilly H, Tron F, Vannier JP, Jacquot S. Transitional B cells in humans: characterization and insight from B lymphocyte reconstitution after hematopoietic stem cell transplantation. Clin Immunol. 2008;127:14–25. doi: 10.1016/j.clim.2007.11.013. [DOI] [PubMed] [Google Scholar]
- Palanichamy A, Barnard J, Zheng B, Owen T, Quach T, Wei C, Looney RJ, Sanz I, Anolik JH. Novel human transitional B cell populations revealed by B cell depletion therapy. J Immunol. 2009;182:5982–5993. doi: 10.4049/jimmunol.0801859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual V, Liu YJ, Magalski A, de Bouteiller O, Banchereau J, Capra JD. Analysis of somatic mutation in five B cell subsets of human tonsil. J Exp Med. 1994;180:329–339. doi: 10.1084/jem.180.1.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R_Development_Core_Team. R Foundation for Statistical Computing. Vienna, Austria: 2008. R: A Language and Environment for Statistical Computing. [Google Scholar]
- Roederer M, Moore W, Treister A, Hardy RR, Herzenberg LA. Probability binning comparison: a metric for quantitating multivariate distribution differences. Cytometry. 2001;45:47–55. doi: 10.1002/1097-0320(20010901)45:1<47::aid-cyto1143>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- Rogers WT, Holyst HA. FlowFP: A Bioconductor Package for Fingerprinting Flow Cytometric Data. Adv Bioinformatics. 2009:193947. doi: 10.1155/2009/193947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers WT, Moser AR, Holyst HA, Bantly A, Mohler ER, 3rd, Scangas G, Moore JS. Cytometric fingerprinting: quantitative characterization of multivariate distributions. Cytometry A. 2008;73:430–441. doi: 10.1002/cyto.a.20545. [DOI] [PubMed] [Google Scholar]
- Sanz I, Anolik J. Reconstitution of the adult B cell repertoire after treatment with rituximab. Arthritis Res Ther. 2005;7:175–176. doi: 10.1186/ar1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims GP, Ettinger R, Shirota Y, Yarboro CH, Illei GG, Lipsky PE. Identification and characterization of circulating human transitional B cells. Blood. 2005;105:4390–4398. doi: 10.1182/blood-2004-11-4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streit M, Ecker RC, Osterreicher K, Steiner GE, Bischof H, Bangert C, Kopp T, Rogojanu R. 3D parallel coordinate systems--a new data visualization method in the context of microscopy-based multicolor tissue cytometry. Cytometry A. 2006;69:601–611. doi: 10.1002/cyto.a.20288. [DOI] [PubMed] [Google Scholar]
- Sutter JA, Kwan-Morley J, Dunham J, Du YZ, Kamoun M, Albert D, Eisenberg RA, Luning Prak ET. A longitudinal analysis of SLE patients treated with rituximab (anti-CD20): factors associated with B lymphocyte recovery. Clin Immunol. 2008;126:282–290. doi: 10.1016/j.clim.2007.11.012. [DOI] [PubMed] [Google Scholar]
- Zand MS. B-cell activity of polyclonal antithymocyte globulins. Transplantation. 2006;82:1387–1395. doi: 10.1097/01.tp.0000244063.05338.27. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.