Abstract
Eukaryotic cells respond to cellular stresses by the inhibition of translation and the accumulation of mRNAs in cytoplasmic RNA–protein (ribonucleoprotein) granules termed stress granules and P-bodies. An unresolved issue is how different stresses affect formation of messenger RNP (mRNP) granules. In the present study, we examine how sodium azide (NaN3), which inhibits mitochondrial respiration, affects formation of mRNP granules as compared with glucose deprivation in budding yeast. We observed that NaN3 treatment inhibits translation and triggers formation of P-bodies and stress granules. The composition of stress granules induced by NaN3 differs from that of glucose-deprived cells by containing eukaryotic initiation factor (eIF)3, eIF4A/B, eIF5B and eIF1A proteins, and by lacking the heterogeneous nuclear RNP (hnRNP) protein Hrp1. Moreover, in contrast with glucose-deprived stress granules, NaN3-triggered stress granules show different assembly rules, form faster and independently from P-bodies and dock or merge with P-bodies over time. Strikingly, addition of NaN3 and glucose deprivation in combination, regardless of the order, always results in stress granules of a glucose deprivation nature, suggesting that both granules share an mRNP remodeling pathway. These results indicate that stress granule assembly, kinetics and composition in yeast can vary in a stress-specific manner, which we suggest reflects different rate-limiting steps in a common mRNP remodeling pathway.
Keywords: Stress granules, P-bodies, Translation, mRNA
Introduction
The control of translation, stability and subcellular localization of mRNA is a key aspect of the regulation of gene expression in eukaryotic cells. Reflecting such control mechanisms, cytosolic mRNAs are in a dynamic equilibrium between different functional states and subcellular locations. For example, translating mRNAs are found in polysomes throughout the cytoplasm, whereas nontranslating mRNAs often accumulate in cytoplasmic RNA–protein (ribonucleoprotein) granules, such as P-bodies and stress granules. P-bodies are compositionally defined by their bias towards mRNA decay components (Parker and Sheth, 2007), whereas stress granules harbor more factors associated with translation initiation (Anderson and Kedersha, 2009; Buchan and Parker, 2009). However, both granules share specific mRNAs and some proteins and can dock or overlap with each other in a dynamic manner (Kedersha et al., 2005; Hoyle et al., 2007; Buchan et al., 2008). The interaction of P-bodies and stress granules has suggested that mRNPs might be remodeled at their interface and individual mRNAs exchanged between the two granules, although this has not been directly demonstrated (Mollet et al., 2008).
The budding yeast Saccharomyces cerevisiae represents a useful organism to study the formation and function of stress granules and P-bodies. Stress granules have been described in budding yeast during glucose deprivation or severe heat-shock. During glucose deprivation, stress granules form that contain eukaryotic initiation factor (eIF)4E and eIF4G proteins, mRNAs and the poly(A)-binding protein Pab1 (Brengues and Parker, 2007; Hoyle et al., 2007; Buchan et al., 2008); stress granules have also been referred to as EGP bodies because they contain eIF4E, eIF4G and Pab1 (Hoyle et al., 2007). Stress granules or EGP bodies formed during glucose deprivation are seen to first form in conjunction with P-bodies (Hoyle et al., 2007; Buchan et al., 2008), and their formation is reduced in strains deficient in P-body aggregation (Buchan et al., 2008). Stress granules have also been described in budding yeast during severe heat-shock, although those stress granules contain 40S ribosomal subunits and eIF3 factors, which are typically lacking from stress granules formed during glucose deprivation (Grousl et al., 2009; Buchan et al., 2008). Moreover, mutations that impair stress granule assembly under conditions of glucose deprivation have little effect upon assembly of heat-shock stress granules (Buchan et al., 2008; Grousl et al., 2009). These differences are fundamentally similar to observations in mammalian cells indicating that stress granules can have different composition and assembly rules during different types of stress responses (for a review, see Buchan and Parker, 2009), although the mechanisms and functional significance of these differences are unresolved.
To begin to understand the differences in stress granule formation and function, we have previously examined various agents for their ability to trigger stress granule formation in yeast. In this process, we observed that sodium azide (NaN3), which inhibits cytochrome oxidase and thus impairs mitochondrial function (Duncan and Mackler, 1966; Wilson and Chance, 1967; Rikhvanov et al., 2002), caused repression of translation and induced the formation of stress granules and P-bodies. Unlike stress granules formed during glucose deprivation, stress granules induced by NaN3 harbored greater amounts of later-stage initiation factors (e.g. eIF3, eIF1A and eIF5B), could assemble in a manner independent of normal P-body assembly and often exhibited docking behavior with P-bodies. Such features are reminiscent of the behavior of stress granules in mammalian cells. Administration of both NaN3 and glucose deprivation stress in combination revealed that the phenotype of glucose deprivation was dominant, which suggests that the appearance of stress-specific stress granules is due to different rate-limiting steps in mRNP remodeling.
Results
NaN3 reversibly inhibits translation
Numerous stress responses in eukaryotic cells lead to a broad inhibition of protein synthesis (Sonenberg and Hinnebusch, 2009). As a first step in characterizing how the cells respond to NaN3, we examined how treatment of cells with NaN3 affected bulk translation by a polysome analysis, as inhibition of translation initiation typically leads to a loss of polysomes. We observed that treatment of cells with 0.5% NaN3 for 10 minutes led to a reduction in polysomes comparable with the decline in polysomes seen during glucose deprivation (Fig. 1). Consistent with this loss of polysomes, we also observed that NaN3 treatment led to a dramatic decrease in labeling of proteins by [35S]methionine (data not shown), but this is difficult to interpret given that NaN3 can reduce the uptake of amino acids (Kotyk et al., 1971). Removal of the NaN3 led to a restoration of polysomes similar to that seen when glucose is restored to glucose-deprived cultures (Fig. 1). We interpret these observations as demonstrating that NaN3 can strongly and reversibly inhibit protein synthesis. Moreover, because polysomes are lost, it suggests that the inhibition of translation acts upon a step in translation initiation.
Fig. 1.
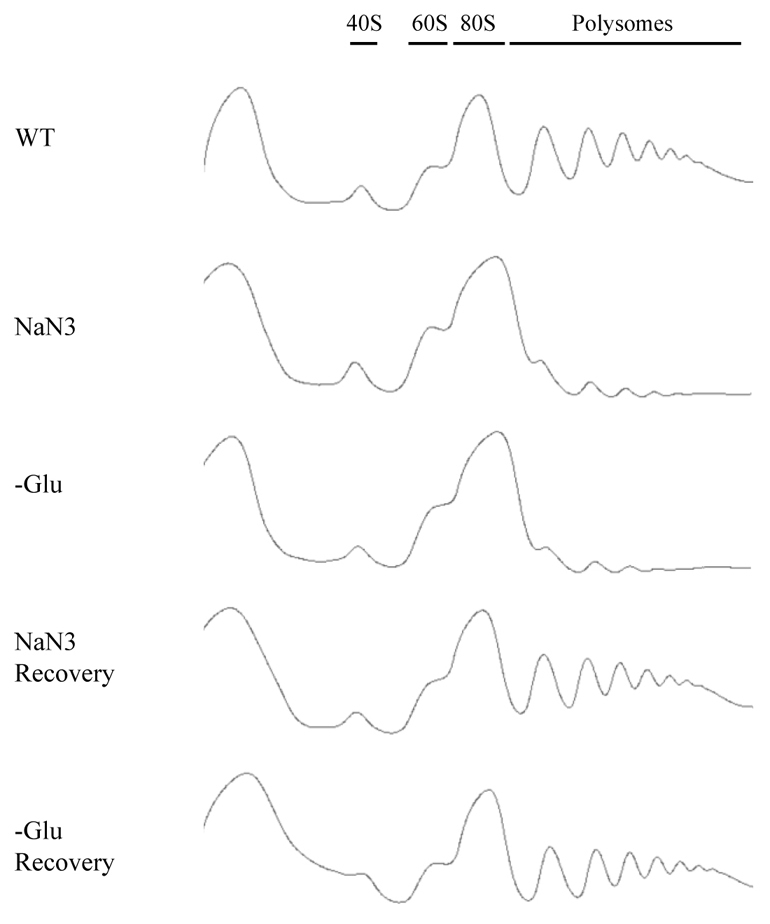
NaN3 stress represses translation. Exponential-phase BY4741 cells were subject to control (WT) or stress conditions for 10 minutes [NaN3 at 0.5% (v/v) or glucose deprivation, –Glu], and stress conditions followed by recovery for 10 minutes in fresh medium. Global translation was monitored by polysome analysis.
NaN3 induces stress granules and P-bodies
Inhibition of translation initiation can often lead to the formation of stress granules and/or P-bodies. To determine whether the addition of NaN3 induced the formation of stress granules and/or P-bodies, we examined the effects of NaN3 on the subcelluar distribution of a range of proteins that concentrate in either P-bodies or stress granules during stress responses (Buchan et al., 2008; Grousl et al., 2009). In each case, we utilized a yeast strain with a C-terminally GFP-tagged protein of interest in the chromosome, transformed with a plasmid expressing either an mCherry (mCh) fusion of a previously identified stress granule [Pub1–mCh, a T-cell-restricted intracellular antigen (TIA)-1 homolog] or P-body (Edc3–mCh) marker (Buchan et al., 2008). These experiments led to the following observations.
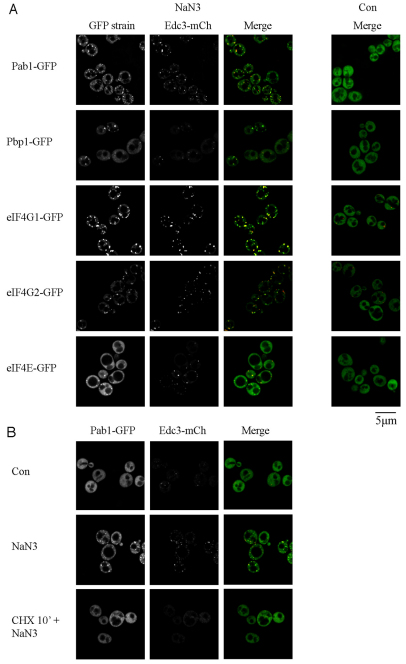
First, we observed that treating cells with 0.5% NaN3 for 30 minutes led to an increase in P-bodies, as judged by increased Edc3 foci (Fig. 2A), as well as increased formation of Dcp2, Dhh1 (a Rck/p54 homolog) and Xrn1 foci, all of which colocalized extremely well with Edc3 (supplementary material Fig. S1A) and partly with the stress granule marker Pub1 (supplementary material Fig. S1B). The same was true at lower concentrations of NaN3 (0.1%), although the increase in P-bodies was not as great (supplementary material Fig. S2A, Table S1). Similar to P-body formation in glucose deprivation (Teixeira et al., 2005), this NaN3-mediated increase in P-bodies was blocked by cycloheximide, which traps mRNAs in polysomes (Fig. 2B), and is consistent with P-bodies forming because of a pool of nontranslating mRNPs.
Fig. 2.
NaN3 induces stress granules and P-bodies that require non-translating mRNA for assembly. (A) Exponential-phase cells expressing chromosomal GFP-tagged proteins and Edc3–mCh (pRP1574) were subjected to 0.5% (v/v) NaN3 for 30 minutes and examined. No GFP foci and only faint P-bodies were observed in glucose-containing control (Con) conditions, hence display of the merge image only. (B) Exponential-phase BY4741 cells expressing pRP1657 were subject to control or NaN3 treatment as above, or treatment with 100 μg/ml cycloheximide (CHX) 10 minutes before NaN3 treatment.
Second, treating cells with 0.5% NaN3 for 30 minutes led to an increase in stress granules, as judged by the accumulation of several proteins previously described to form stress granules during glucose deprivation (Buchan et al., 2008; Hoyle et al., 2007; Brengues and Parker, 2007), including Pab1, Pbp1 (an ataxin-2 homolog), eIF4G1, eIF4G2 and eIF4E, all of which colocalized extremely well with the stress granule marker protein Pub1 (supplementary material Fig. S3). As with P-bodies, 0.1% NaN3 also induced stress granules, but to a lesser degree than 0.5% NaN3 (supplementary material Fig. S2A, Table S2). Similar to stress granules formed during glucose deprivation, NaN3-induced stress granules were sensitive to cycloheximide treatment, suggesting that they also require a pool of free mRNPs for their formation (Fig. 2B). Thus, NaN3 stress induces the formation of stress granules that share numerous components present in stress granules during glucose deprivation.
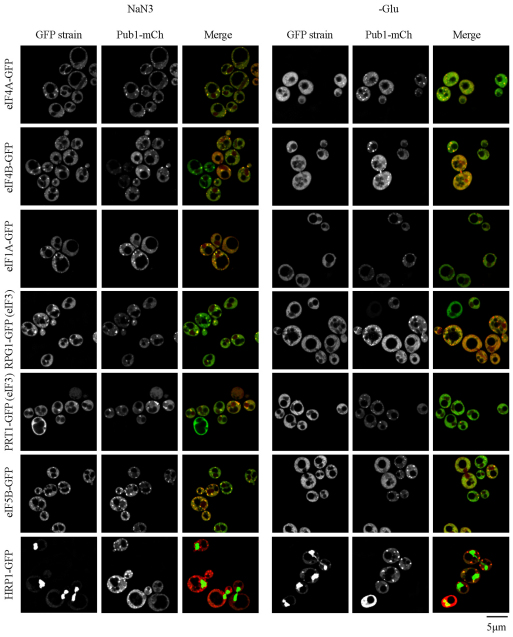
A third significant observation was that stress granules induced by NaN3 contained additional translation factors not typically seen in stress granules induced by glucose deprivation. Specifically, we observed that NaN3 stress caused localization of two core eIF3 subunits, Prt1 and Rpg1, within both Pub1 and some Edc3 foci, whereas glucose-deprived stress granules lacked these factors (Fig. 3). Note that, as Prt1 and Rpg1 foci are not visible in the absence of stress, and always colocalize with Pub1 foci, we consider these to be stress granule factors rather than P-body proteins. However, the intensity of eIF3 foci was weaker than for eIF4F components, such as Pab1 and eIF4G, and indeed Pub1 or Edc3 foci lacking any eIF3 signal were sometimes observed (Fig. 3; supplementary material Fig. S4). By contrast, eIF1A, eIF4A, eIF4B and eIF5B localized robustly to NaN3 stress granules, but only extremely faint signals for eIF4A, eIF4B and eIF5B were evident in some glucose deprivation stress granules (Fig. 3). In summary, in contrast with stress granules formed during glucose deprivation, NaN3-induced stress granules contain additional initiation components typically associated with later stages of initiation. We interpret this as suggesting that NaN3 might inhibit translation at a later step in initiation compared with glucose deprivation (see the Discussion section).
Fig. 3.
NaN3 stress granules harbor additional components associated with later stages of translation initiation. Exponential-phase cells expressing chromosomal GFP-tagged proteins and Pub1–mCh (pRP1661) were subjected to 0.5% (v/v) NaN3 for 30 minutes, or glucose deprivation (–Glu) stress, and examined. Under control conditions (not shown), none of the GFP-tagged factors formed cytoplasmic foci, whereas only faint P-bodies were observed.
A fourth observation from these experiments was that Hrp1, which is a nuclear protein that can accumulate in stress granules during glucose deprivation, was not present in stress granules during NaN3 treatment (Fig. 3). This suggests that there are differences in the shuttling of Hrp1 between the nucleus and cytoplasm during glucose deprivation compared with NaN3 stress (see the Discussion section).
Finally, in order to further determine whether inhibition of mitochondrial function by NaN3 was responsible for induction of stress granules and P-bodies, we exposed yeast to clotrimazole, a known mitochondrial inhibitor (Schuster, 1985; Penso and Beitner, 1998). Although clotrimazole induced stress granules far more modestly than NaN3 (supplementary material Fig. S2A, Table S2), we still observed clear induction compared with non-stress controls and, importantly, saw instances of separated stress granule and P-body juxtaposition that resembled NaN3-induced rather than glucose deprivation foci (see below and the Discussion section). Notably, P-bodies were not induced by clotrimazole (supplementary material Table S1), mimicking previous results in mammalian cells (Kedersha et al., 2005). Taken together, these results are suggestive that NaN3 stimulates stress granule formation in part by inhibiting mitochondrial function, although other effects of NaN3, such as impaired amino acid uptake and/or generation of reactive oxygen species (Kotyk et al., 1971; Grant et al., 1997; Rikhvanov et al., 2001) might also contribute to the induction of stress granules and P-bodies.
Kinetics and features of stress granule and P-body assembly differ between glucose deprivation and NaN3 treatment
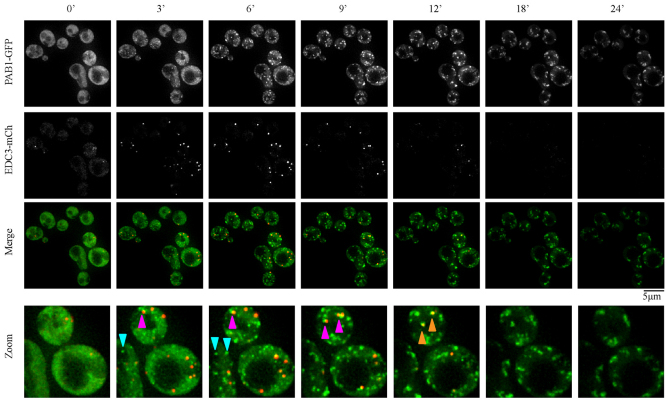
To further characterize the NaN3 stress response, we examined the formation of P-bodies and stress granules during NaN3 treatment and compared that with what has been described above for glucose deprivation. In these experiments, wild-type yeast expressing a plasmid bearing Pab1 (a stress granule marker) fused to green fluorescent protein (GFP) and Edc3–mCh (a P-body marker) were exposed to NaN3, and images were collected every 90 seconds for up to 24 minutes. These results are presented as a supplementary movie (supplementary material Movie 1) and as a series of images recorded at different timepoints (Fig. 4). These experiments revealed the observations described below.
Fig. 4.
Kinetic analysis of NaN3 stress granule and P-body formation. Exponential-phase BY4741, transformed with pRP1657, were subjected to NaN3 stress and immediately spotted onto microscopy slides for examination. Still images are taken from supplementary material Movie 1. Turquoise arrowheads indicate stress granule foci initially forming separately from visible P-bodies, whereas magenta arrowheads indicate P-body-associated stress granule formation. These stress granules often remain docked before, in some cases, fusing more completely with P-bodies (orange arrowheads). Note that, at later timepoints (at 18 and 24 minutes), significant photobleaching of the P-body signal has occurred, giving a false impression of P-body disappearance.
First, P-bodies were induced quickly, forming as early as 3 minutes after NaN3 addition. However, the degree of P-body induction after NaN3 treatment during early timepoints was far less than that seen during glucose deprivation, as verified by the intensity and size measurements of P-bodies (supplementary material Table S3). This suggests that NaN3 gives a less robust accumulation of P-bodies compared with that produced by glucose deprivation. Second, multiple small Pab1 foci formed rapidly and early during the stress timecourse, with substantial Pab1 foci being present at 3 minutes. Although some foci initially appeared docked closely to, or within a P-body, within the first 5 minutes of a typical NaN3 timecourse approximately 80% of Pab1 foci were distinct from P-bodies (supplementary material Movie 1; Fig. 4, zoom panels; supplementary material Table S4). This is in contrast with the formation of stress granules during glucose deprivation (Buchan et al., 2008), which initially formed at a later timepoint, between 5 and 10 minutes, and were almost exclusively observed within P-bodies (95% during this time period, supplementary material Table S4), before going on to form P-body-distinct foci. As time progressed during a NaN3 stress timecourse, Pab1 foci appeared to coalesce, and increased in intensity and size, often as the result of visible fusion events, which ultimately reduced their number compared with earlier stages in the timecourse (supplementary material Movie 1; Fig. 4; supplementary material Table S4). Throughout the timecourse, many stress granules exhibited a partial overlap with P-bodies, appearing to be docked to the Pab1 foci, although complete merging of the two foci tended to increase over time (Fig. 4, zoom panel). This docking behavior is reminiscent of the arsenite-induced assembly of stress granules in mammalian cells (Mollet et al., 2008; Kedersha et al., 2005) and also differs from glucose deprivation where stress granules and P-bodies tend either to be completely distinct or overlap more completely (Buchan et al., 2008; Hoyle et al., 2007). Finally, although it appeared that P-bodies levels declined at later timepoints, control experiments indicated this was due to photobleaching of Edc3–mCh (data not shown).
In summary, the kinetics of granule assembly between different stress conditions varies in yeast, which might reflect the existence of stress-specific pathways of mRNP remodeling between granules and polysomes or different rate-limiting steps in a common mRNP remodeling pathway (see the Discussion section).
The importance of stress granule assembly factors varies according to the stress
Previous work suggested that stress granules are dependent on, and form in conjunction with, pre-existing P-bodies during glucose deprivation (Buchan et al., 2008). By contrast, during extreme heat-shock, yeast stress granules are more independent of P-bodies (Grousl et al., 2009), and stress granules in mammalian cells can form independently of P-bodies (Mollet et al., 2008; Ohn et al., 2008; Anderson and Kedersha, 2009). Moreover, work in mammalian cells has suggested that the assembly rules for stress granules can vary under different stress conditions (for a review, see Buchan and Parker, 2009). Given these issues, we examined how the assembly of stress granules during NaN3 treatment was affected by mutations in components of P-bodies or stress granules previously shown to affect stress granule formation during glucose deprivation. In these experiments, we compared the formation of stress granules and P-bodies during NaN3 treatment in mutant and wild-type strains transformed with a single-copy plasmid expressing a marker of stress granules (Pab1–GFP) and a marker of P-bodies (Edc3–mCh) under the control of their own promoter, thereby minimizing any affects of overexpression. These experiments led to the following observations.
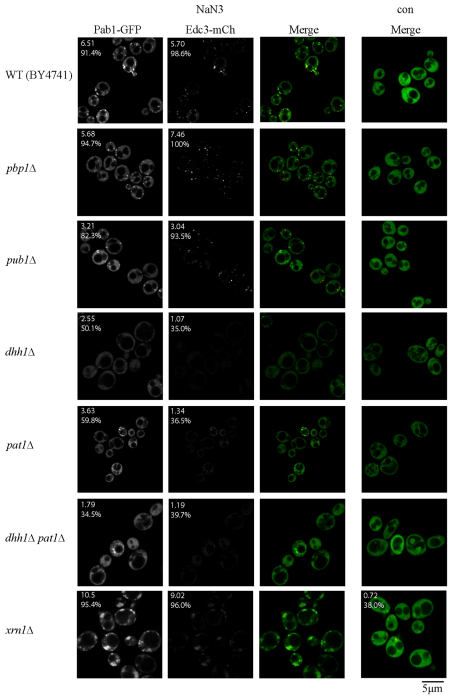
First, we observed that mutations in two stress granule components, Pbp1 and Pub1, had no or reduced effects on stress granule assembly in NaN3 treatment compared with glucose deprivation. Specifically, a pbp1Δ strain, which inhibits glucose-deprived stress granules but not P-bodies, exhibited no inhibitory effects on NaN3 stress granules or P-bodies (Fig. 5; supplementary material Tables S1, S2). Similarly, a pub1Δ strain, which is also known to show inhibited stress granule formation during glucose deprivation, only exhibited a subtle decrease in both stress granules and P-bodies, somewhat mirroring the trends observed during glucose deprivation, but with a far weaker effect overall (Fig. 5; supplementary material Tables S1, S2). We interpret these observations as indicating that stress granule assembly factors can vary in importance, depending on the stress condition.
Fig. 5.
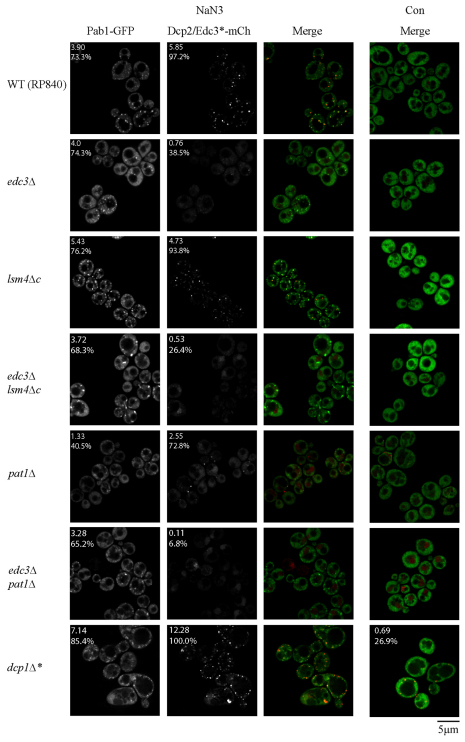
Null strain analysis of factors affecting NaN3 stress granule assembly. Exponential-phase cells, all of the BY4741 strain background, were transformed with plasmid pRP1657 and subjected to NaN3 stress [0.5% (v/v) for 30 minutes] or control conditions. Stress granules (Pab1–GFP) and P-body numbers (Edc3–mCh) were quantified in a blind manner. The number given in the top-left corner indicates the mean number of foci per cell, the percentage indicates the proportion of cells with one or more foci (see also supplementary material Tables S1, S2). con, control.
Second, we observed that strains defective in P-body assembly [edc3Δ lsm4Δc and edc3Δ pat1Δ (Decker et al., 2007)] did not significantly inhibit NaN3 stress granule formation (Fig. 6; supplementary material Tables S1, S2). This is in contrast with glucose deprivation, where edc3Δ lsm4Δc and edc3Δ pat1Δ mutant strains are strongly inhibited for stress granule formation (Buchan et al., 2008). By contrast, P-bodies remained strongly inhibited in these strains during NaN3 stress (Fig. 6; supplementary material Tables S1, S2). This suggests that formation of large P-bodies is not as important for stress granule formation during NaN3 stress compared with glucose deprivation. Moreover, these results also imply that P-body assembly mechanisms might be more conserved across different stress conditions than that of stress granules.
Fig. 6.
Null strain analysis of factors affecting NaN3 stress granule assembly. Exponential-phase cells of yRP840 background were transformed with pRP1660, except for the dcp1Δ strain, which was transformed with pRP1657. NaN3 stress and quantification were conducted as in Fig. 5. Con, control.
Third, we observed that strains with larger P-bodies showed an increase in stress granule formation during NaN3 treatment, similar to what has been shown previously in glucose deprivation (Buchan et al., 2008). Specifically, dcp1Δ and xrn1Δ strains, which are known to have increased P-body numbers and volume owing to blocking of mRNA decay (Sheth and Parker, 2003), showed an increase in stress granule levels during NaN3 treatment (Figs 5, 6; supplementary material Tables S1, S2). Moreover, these stress granules were often colocalized with, docked to or enveloped around P-bodies, similar to prior observations during glucose deprivation stress (Buchan et al., 2008). One possibility is that mRNAs that are exiting P-bodies to return to translation accumulate in a stress granule state around P-bodies when subsequent steps in translation are inhibited. Alternatively, mRNAs that fail to undergo decay or assemble correctly into P-body mRNPs, might be diverted directly into stress granules (see the Discussion section).
A fourth observation was that dhh1Δ, pat1Δ, and dhh1Δ pat1Δ strains all exhibited decreases in stress granule and P-body numbers, with additive effects observed on stress granule numbers in the double-deletion strain (Fig. 5). In principle, this could reflect a failure to repress translation. However, polysome analysis with the dhh1Δ pat1Δ strain indicated no obvious deficiencies in global translation repression in response to NaN3 treatment (supplementary material Fig. S5), thus suggesting the existence of alternative possible roles in mRNP remodeling or granule assembly. Note that the inability to form Pab1 foci was not due to reduced expression of Pab1, as verified by western blot analysis (supplementary material Fig. S6).
Finally, in order to determine whether assembly of NaN3 stress granules was dependent on phosphorylation of eIF2α (SUI2), as is sometimes the case during various mammalian stress responses (Yamasaki and Anderson, 2008; Mazroui et al., 2006), we examined the formation of NaN3-induced stress granules in a strain lacking endogenous SUI2, with growth sustained by expression of plasmid-borne copies of either wild-type SUI2, phospho-dead (S51A) or phosphomimetic versions (S51D) (Dever et al., 1992). Our results showed that, irrespective of the SUI2 allele, NaN3 stress induced a similar number of stress granules and P-bodies (supplementary material Fig. S2B, Table S5) (note, stress granules are not induced in the absence of stress; data not shown). This demonstrates that NaN3 stress granules are assembled in a manner independent of eIF2α phosphorylation.
In summary, stress granules induced by exposure to NaN3 exhibit rules of assembly different from those applying to glucose deprivation stress granules in that they are insensitive to eIF2α phosphorylation, loss of Pbp1, Pub1 or the ability to form visible P-bodies. However, similar trends between both stresses were observed in mutants that block mRNA decay, namely dcp1Δ and xrn1Δ strains, which displayed increased levels of both P-bodies and stress granules. In addition, the translational repressors Dhh1 and Pat1 seemed to stimulate formation of both stress granules and P-bodies during NaN3 stresses.
eIF4E and eIF3 show stress specific effects on stress granule formation
Previous investigators have argued that stress granule formation in mammalian cells is dependent on eIF4E and eIF3 (Ohn et al., 2008; Mokas et al., 2009). Given this, we examined the dependence of stress granule formation in budding yeast on these factors by using temperature-sensitive alleles of eIF4E and Prt1, a core subunit of eIF3. After a shift to the restrictive temperature, with or without the addition of NaN3, we observed the following effects on stress granules and P-body formation.
In the absence of NaN3, we observed that shifting wild-type cells to either 37 or 39°C, the restrictive temperatures for the Prt1 and eIF4E alleles, respectively, led to a modest and reproducible increase in the number and size of stress granules and P-bodies (supplementary material Figs S7, S8, Table S6). This indicated that even a mild heat-shock could induce some stress granules and P-bodies, although to a lesser extent than seen with a more extreme heat-shock (Grousl et al., 2009). We also observed that strains that were temperature-sensitive for eIF4E or Prt1 showed an increase in P-bodies during the shift, as reported previously (Brengues et al., 2005). Strikingly, we observed that the temperature-sensitive eIF4E and prt1 strains showed a decrease in stress granule formation compared with that of the wild-type strains. This suggests that stress granules formed under mild heat-shock require the activity of eIF4E and eIF3 for their formation. The eIF4E and eIF3 proteins might play a direct role in targeting mRNAs to stress granules, either from P-bodies or polysomes. Alternatively, the loss of stress granules might be because the pool of nontranslating mRNAs is reduced as these conditional alleles of eIF4E and eIF3 lead to rapid mRNA degradation at the restrictive temperature (Schwartz and Parker, 1999; Schwartz and Parker, 2000).
By contrast, during a heat-shift in the presence of NaN3, we observed that the temperature-sensitive translation initiation mutants formed P-bodies and stress granules to an extent similar to that of the wild-type strains (supplementary material Figs S7, S8, Table S6). These results demonstrate that the dependence of stress granule formation on eIF4E and eIF3 is stress-dependent in budding yeast. Stress induced by NaN3 might restore the ability of the eIF4E and eIF3 mutants to form stress granules by creating a more robust block to translation and/or by stabilizing the pool of mRNAs at the high temperature, as many stresses that induce P-bodies and stress granules inhibit mRNA degradation in yeast (Hilgers et al., 2006). Further work will be required to understand the stress-specific roles of translation initiation factors in stress granule formation.
Combinatorial stress addition causes stress granule composition to revert to a glucose deprivation phenotype
An unresolved issue is what the differences in composition, assembly rules and kinetics of stress granules between NaN3 and glucose deprivation stress actually reflect. One possibility is that these stress-specific forms of ‘stress granules’ actually represent fundamentally distinct mRNP granules, which assemble and disassemble through their own mRNP remodeling pathways, perhaps with specific functional differences. If so, one might anticipate that applying stresses in combination, but one before the other, would lead to the bulk of the shared repressed mRNAs between the two conditions being channeled along the stress granule assembly and disassembly pathways of whichever stress was applied first. Thus, applying NaN3 stress before glucose deprivation would result in stress granules containing eIF3 and lacking Hrp1, for example, whereas reversing this order would result in stress granules lacking eIF3 and containing Hrp1. An alternative model is that stress-specific stress granule states are related ‘steps’ on a pathway, or continuum, of mRNP remodeling events. Differences in composition, assembly and disassembly rules and kinetics, might be caused by different stresses eliciting different rate-limiting steps within a particular mRNP remodeling pathway, thus revealing compositionally distinct granules (Buchan and Parker, 2009). In this case, applying stresses in combination would always result in the phenotype of whichever stress introduces a rate-limiting step upstream of the other.
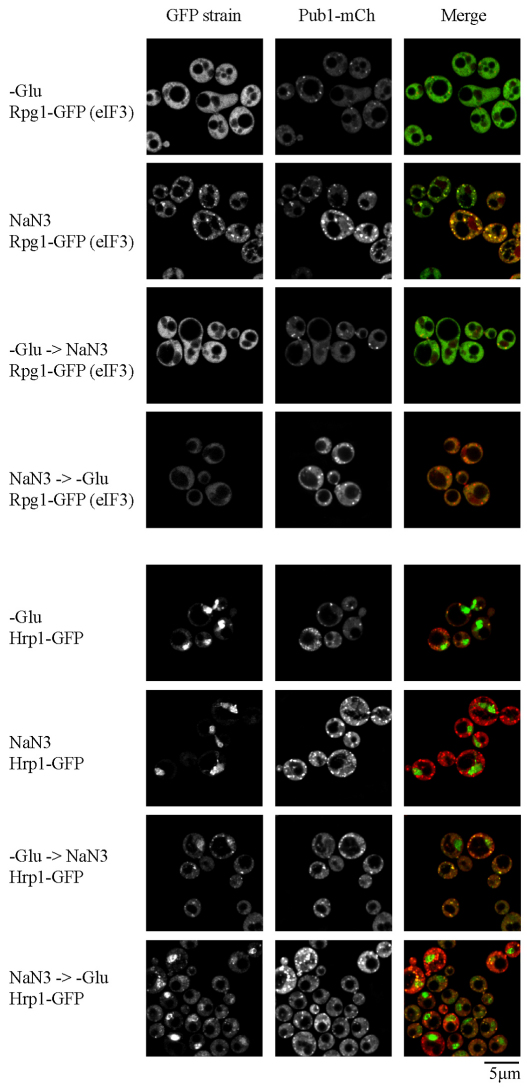
To discriminate between these two models, we applied NaN3 stress and glucose deprivation stress in combination, with one applied before the other. Importantly, irrespective of whether glucose deprivation was administered before being combined with a NaN3 stress, or vice versa, we always observed formation of Pub1 foci, indicative of stress granule assembly, which always lacked eIF3 subunits but contained Hrp1 (Fig. 7; supplementary material Fig. S9). In other words, the RNP granule phenotype observed during glucose deprivation was dominant to that observed during NaN3 stress. We interpret this observation as suggesting that both granules share an mRNP remodeling pathway, and that glucose deprivation might block translation initiation at a step earlier than that affected by NaN3 stress (see the Discussion section).
Fig. 7.
Combinatorial stress analysis reveals the dominance of glucose deprivation stress over NaN3 stress with regard to stress granule composition. Exponential-phase Rpg1–GFP or Hrp1–GFP cells, transformed with plasmid pRP1661, were subjected to each stress condition individually or in combination (see the Materials and Methods section). –Glu, glucose deprivation.
P-body assembly is not required for assembly of stress granules in response to combinatorial stress addition
Given the contrasting results of P-body dependence for stress granule assembly when glucose deprivation (Buchan et al., 2008) and NaN3 stress (the present study) were observed in isolation, we examined the effect of combination stress addition on stress granule assembly in an edc3Δ lsm4Δc mutant strain, which is defective in aggregation of P-body mRNPs (Decker et al., 2007). Consistent with previous results (Buchan et al., 2008), we observed that edc3Δ lsm4Δc strains were defective in stress granule assembly during glucose deprivation (supplementary material Fig. S10, Table S7). Strikingly, we observed that the combination of NaN3 along with glucose deprivation (independent of the order of the stress) restored stress granule formation in the edc3Δ lsm4Δc strain to levels that were approximately equal to those induced by NaN3 alone (supplementary material Fig. S10, Table S7). We also observed that P-body assembly elevated slightly during addition of combination stress in the edc3Δ lsm4Δc strain but remained significantly inhibited relative to stress granule formation in wild-type strains. Thus, the addition of NaN3 during glucose deprivation creates stress conditions that bypass the need for aggregated P-bodies for the assembly of stress granules, perhaps because of a more extreme block to mRNPs exiting stress granules (see the Discussion section).
Discussion
NaN3, stress granules and mitochondrial dysfunction
Our experiments indicate that treatment of yeast cells with NaN3 leads to a stress response wherein global translation is repressed concordant with the induction of stress granules and P-bodies. Given the known effects of NaN3 on the mitochondrial respiratory chain, the simplest interpretation of these observations is that disruption of the mitochondrial function leads to a stress response. Consistent with this interpretation, we also observed that a second inhibitor of mitochondrial function, clotrimazole, triggered the accumulation of stress granules (supplementary material Fig. S2A). Such a stress response could be caused by defects in the respiratory chain triggering a specific signal transduction pathway leading to translation inhibition or could be due to secondary effects of NaN3 and/or the consequence of defects in respiration, such as the generation of oxidative stress or energy depletion. Other observations are also consistent with mitochondria being connected to stress responses and the formation of stress granules. In human cells, proteins such as Fas-activated serine/threonine kinase (FAST) and prohibitin-2, both of which are apoptotic regulators normally resident within mitochondrial membranes, relocalize to stress granules during stress (Kedersha et al., 2005; Ohn et al., 2008). Indeed, FAST has been implicated as a possible scaffolding factor for stress granules and P-bodies (Kedersha et al., 2005) and, owing to physical and functional interactions with TIA-1, might serve as a sensor of mitochondrial stress that facilitates an appropriate posttranscriptional response (Li et al., 2004). In future work, it will be of interest to determine whether stress granules are induced in other conditions associated with mitochondrial dysfunction, including disease states such as diabetes mellitus and mitochondrial myopathies.
The composition of yeast stress granules is stress dependent
The present study and previous work (Buchan et al., 2008; Grousl et al., 2009; Hoyle et al., 2007; Brengues and Parker, 2007) demonstrate that the composition of yeast stress granules can vary depending on the stress. Specifically, during glucose deprivation yeast stress granules contain many factors, including eIF4E, eIF4G, Pab1, Pub1, Pbp1, Hrp1 and Ded1, but not eIF3 subunits, eIF5B and eIF1A (Fig. 3) (Buchan et al., 2008) (A. Hilliker, unpublished). By contrast, during NaN3 treatment, stress granules now contain all the factors seen in glucose deprivation, with the exception of Hrp1, but also contain eIF3 subunits (Prt1 and Rpg1), eIF5B and eIF1A (Fig. 3, and see supplementary material Fig. S3). This type of stress granule is similar to the granules occurring upon extreme heat-shock in budding yeast wherein eIF3 components and 40S subunits populate the stress granules (Grousl et al., 2009). The simplest interpretation of these observations is that individual mRNPs are stalled at different stages of translation initiation in different stress conditions and that this leads to the accumulation of different partial translation initiation complexes in stress granules, thereby giving a different apparent composition. Although speculative, on the basis of the microscopy data highlighted above, glucose deprivation stress might cause a block before 48S assembly on mRNPs, whereas NaN3 stress might block after assembly of the 48S complex.
Several other compositional differences between the stress conditions involving initiation factors have interesting implications. For example, the presence of eIF4A and eIF4B in NaN3 stress granules, but the general absence of these proteins from glucose deprivation stress granules, was somewhat surprising given their association with other eIF4F components (Dominguez et al., 1999; Dominguez et al., 2001) and suggests a possible regulatory point in translation initiation that differs between the two stress conditions. Moreover, eIF5B, which functions in 60S subunit joining, an initiation step downstream of eIF4F and eIF3 function, accumulates in stress granules during both NaN3 stress (robustly) and glucose deprivation (weakly). Such observations could indicate a previously uncharacterized role for eIF5B in stress granule assembly and/or disassembly. Alternatively, mRNAs re-entering translation might acquire specific initiation components in a stepwise manner, which our microscopy data would suggest does not necessarily reflect the linear order in which they function during initiation that has been determined by biochemical experiments in cell-free extracts (for a review, see Sonenberg and Hinnebusch, 2009).
The kinetics and assembly rules of stress granules vary with different stresses
The rules and kinetics of stress granule assembly vary between glucose deprivation and NaN3 stress. The key differences are as follows. First, during glucose deprivation, stress granules are sensitive to loss of Pub1 or Pbp1 (Buchan et al., 2008), whereas NaN3 stress granules are only weakly sensitive to the absence of Pub1 alone (Fig. 5; supplementary material Tables S1, S2). Second, during glucose deprivation, stress granules are diminished when P-body aggregation is reduced in the edc3Δ lsm4Δc strain, whereas NaN3 stress granules are not affected (Fig. 6). Third, stress granules tend to overlap and form after and in conjunction with P-bodies during glucose deprivation, whereas, during NaN3 treatment, stress granules form earlier and are more often separate from a P-body or at most are docked and not completely overlapping (Buchan et al., 2008) (Fig. 4).
Stress granule formation also notably shares some properties in both glucose deprivation and NaN3 exposure. Specifically, some proteins are present in stress granules in both conditions (eIF4E, eIF4G, Pab1, Pbp1 and Pub1) and, in each stress condition, stress granules are reduced by mutations in Dhh1 and Pat1 (Fig. 5) and increased in dcp1Δ and xrn1Δ strains (Figs 5, 6). In addition, note that different yeast strain backgrounds (e.g. BY4741, yRP840 and yRP2537) show relative differences in the induction of stress granules and P-bodies (compare the wild-type data in supplementary material Tables S1, S2 and S5), a phenomenon also seen in different mammalian cell lines.
An unresolved issue is what the difference in stress granule assembly, composition and kinetics reveals about the dynamics of mRNP formation and aggregation into subcellular bodies. In principle, these results can be interpreted in two extreme models. In one view, each stress response leads to a specific redistribution of translating mRNAs into a nontranslating pool that accumulates into a cytoplasmic mRNP. For example, during glucose deprivation, the evidence suggests that mRNAs exit the translation pool and accumulate predominantly in P-bodies before a transition to a stress granule state (Buchan et al., 2008). By contrast, the rapid accumulation of stress granules, and their independence from P-body aggregation factors during NaN3 exposure, could be explained by translating mRNAs ceasing translation and directly accumulating in stress granules without passing through a biochemical state analogous to the P-body. However, such a model does not easily explain why stress granule formation would be reduced in dhh1Δ, pat1Δ or dhh1Δ pat1Δ strains during NaN3 exposure (Fig. 5), given that these factors have more typically been implicated in P-body assembly both in yeast and mammals (Coller and Parker, 2005; Serman et al., 2007; Scheller et al., 2007; Ohn et al., 2008). Additionally, in this ‘independent’ states model, it is not clear why the glucose deprivation phenotype is dominant over the NaN3 phenotype, even when NaN3 is applied first (Fig. 7).
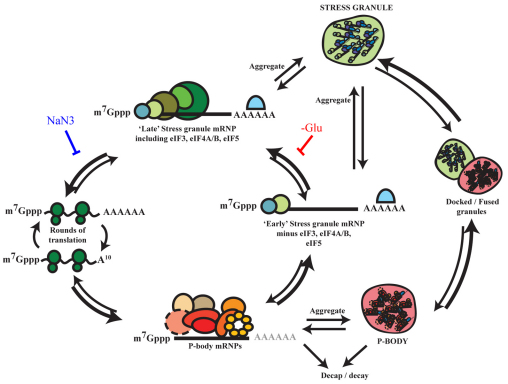
An alternative model to interpret these differences is that mRNPs broadly undergo the same cycle of rearrangement and aggregation in different stress conditions, but that different stresses affect individual steps in that cycle to different extents, thereby leading to stress-specific behavior in a consistent ‘mRNA cycle’ (Fig. 8). For example, during glucose deprivation, stress granule assembly might depend on pre-existing P-bodies owing to the rate of mRNPs exiting stress granules typically being faster than their aggregation into stress granules, and therefore only mRNPs that are first highly concentrated in P-bodies go on to aggregate as they undergo the transition to a stress granule mRNPs. By contrast, during NaN3 stress, the rate of mRNAs exiting stress granules might be dramatically slower, and therefore sufficient time might exist for these mRNPs to aggregate. This would be the case even if they were formed from an mRNP bound to P-body factors but were not present and/or previously concentrated in an aggregated P-body. Consistent with this view, NaN3 stress generally leads to the formation of larger, more intense and more numerous stress granules than those formed during glucose deprivation.
Fig. 8.
Theoretical models of mRNA remodeling during glucose deprivation and NaN3 stress. Glucose deprivation (–Glu) and NaN3 stress granules could in principle form through distinct assembly pathways, which are dependent and independent, respectively, of visible P-body aggregation. Alternatively, different rate-limiting steps in a cycle of broadly conserved mRNP remodeling events could lead to the formation of subtly different, but related, granules (e.g. ‘early’ and ‘late’ stress granules) during glucose deprivation and NaN3 stress, as indicated by the proposed points at which glucose deprivation and NaN3 stress could block the mRNA cycle on the basis of our observations. Note, poly(A) tails are shown in light gray within P-bodies to highlight the fact that P-body mRNPs are likely to be a mixture of adenylated and deadenylated species, the latter of which are more likely to undergo decay.
This mRNA cycle model provides a possible explanation for several observations. First, it provides a rationale for the increased level of stress granules seen in dcp1Δ and xrn1Δ strains during NaN3 exposure simply because mRNA can exit P-bodies directly to stress granules even during NaN3 stress. Second, it provides an explanation for why the glucose deprivation phenotype is dominant over the NaN3 phenotype – because the glucose deprivation block is upstream of the primary block to the cycle caused by NaN3 stress. Third, it could explain why stress granules formed during NaN3 stress are affected by the P-body components Dhh1 and Pat1 as mRNPs would still cycle through a P-body mRNP before forming a stress granule even during NaN3 stress. Fourth, it could explain why stress granules form independently of P-body aggregation during a combination of NaN3 treatment and glucose deprivation, wherein the rate of mRNPs exiting stress granules would be sufficiently reduced by the combination stress such that individual ‘stress granule’ mRNPs would have time to aggregate even independently of being in a pre-aggregated P-body. In order to determine rigorously the flux of mRNAs between different biochemical and aggregated states, studies following the passage of individual mRNAs through different mRNP and granule states will be required.
Finally, note that the differential localization of the nuclear factor Hrp1 suggests that processes such as transcription, mRNA export and shuttling of nuclear proteins might be altered under different stress conditions. In theory, newly exported mRNPs could feed directly into non-translating pools such as stress granules or P-bodies and thus could contribute to the different behaviors in assembly and kinetics observed between the granules (Buchan and Parker, 2009; Scarcelli et al., 2008; Cuenca-Bono et al., 2010). Notably, Hrp1 accumulated in stress granules to a higher degree during glucose deprivation plus NaN3 combination stress experiments compared with glucose deprivation alone (although no foci were observed with NaN3 alone; Fig. 7). The significance of this is unknown, but it could indicate a synergistic effect of the two stresses, wherein increased export of Hrp1 mRNPs, sufficient to allow their observation in stress granules, depends exclusively on glucose deprivation stress, but that remodeling of Hrp1 mRNPs and subsequent re-import of Hrp1 back into the nucleus can also be impaired by NaN3 stress, thus enhancing accumulation of Hrp1 in stress granules. On a related note, a temperature-sensitive allele of DBP5, which localized in P-body-distinct foci termed RNA export granules (REGs), notably contained Pab1 in some genetic backgrounds, which could then merge with P-bodies at higher stress temperatures (Scarcelli et al., 2008). Therefore it seems quite feasible that these REGs are related to, or are in fact, stress granules. Further work addressing the integration of exported nuclear mRNPs into cytoplasmic mRNP remodeling pathways will be of considerable interest.
Materials and Methods
Yeast strains and growth conditions
Yeast strains used in the present study are listed in supplementary material Table S8. All growth conditions in SC media, and transformations, were as described previously (Buchan et al., 2008).
Plasmids
Plasmids used in the present study are listed in supplementary material Table S8. Construction of plasmids pRP1574-75 (Edc3–mCh), pRP1657, pRP1659 (Pab1–GFP and Edc3–mCh), pRP1660 (Pab1–GFP and Dcp2–mCh) and pRP1661-62 (Pub1–mCh) was performed as described previously (Buchan et al., 2008).
Microscopy and image quantification
Glucose deprivation stress was conducted as described previously (Buchan et al., 2008). NaN3 stress was administered by addition of NaN3 to exponentially growing cultures (D600 of 0.3–0.5) at a final concentration of 0.5% (v/v), from a 10% (v/v) stock in water. After 30 minutes, cultures were concentrated without any wash steps and examined immediately, except during time-lapse experiments, when NaN3 was added immediately before spotting onto slides. For combination stress experiments, each stress was sequentially performed as described above, while ensuring that during the second stress, the first stress was maintained (e.g. in NaN3 treatment plus glucose deprivation experiments, cells were washed and resuspended in glucose-deprived medium additionally containing 0.5% NaN3).
All experiments were conducted on a Deltavision RT system, with image capture and processing, including blind quantification scoring, as described previously (Buchan et al., 2008), with one exception: all experimental images were captured as Z-stacks of eight images [Buchan et al. (Buchan et al., 2008) typically used single-plane data], which were collapsed and used for image quantification. However, images presented in figures are single-plane images so as to visualize better the colocalization between P-bodies and stress granules. A minimum of three replicate experiments for each strain was conducted, with >100 cells for each strain counted.
Polysome analysis
Polysome analysis was conducted as described previously (Brengues et al., 2005). Exponentially growing cells were treated for 10 minutes with NaN3 at a final concentration of 0.5% (v/v), or were subjected to glucose deprivation, after which cell pellets were quickly harvested and frozen in liquid nitrogen in the presence of cycloheximide. For recovery experiments, cells were washed and resuspended in fresh medium and allowed to recover for 10 minutes, before harvesting, as above.
Western blot analysis
Pab1 was detected with a rabbit anti-Pab1 antibody, a gift of Allan Jacobson (School of Medicine, University of Massachusetts, Worcester, MA), at a dilution of 1:5000, and with a goat anti-(rabbit Ig) conjugated to horseradish peroxidase (HRP) (Pierce) at a concentration of 1:1000. As a loading control, Pgk1 was detected with a mouse monoclonal antibody (Molecular Probes) at a dilution of 1:500 and goat anti-(mouse Ig)–HRP conjugate (Pierce) at a concentration of 1:1000.
Acknowledgments
We thank Alan Hinnebusch for kindly providing us with strains and plasmids, Alan Jacobson for providing us with an anti-Pab1 antibody, and all members of the Parker laboratory for useful feedback and discussion of the manuscript data. This work was supported by the Howard Hughes Medical Institute (J.R.B., R.P.) and the National Institutes of Health (J.-H.Y. grant number R37GM45443). Deposited in PMC for release after 6 months.
Footnotes
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/124/2/228/DC1
References
- Anderson P., Kedersha N. (2009). RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat. Rev. Mol. Cell Biol. 10, 430-436 [DOI] [PubMed] [Google Scholar]
- Brengues M., Parker R. (2007). Accumulation of polyadenylated mRNA, Pab1p, eIF4E, and eIF4G with P-bodies in Saccharomyces cerevisiae. Mol. Biol. Cell 18, 2592-2602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brengues M., Teixeira D., Parker R. (2005). Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science 310, 486-489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan J. R., Parker R. (2009). Eukaryotic stress granules: the ins and outs of translation. Mol. Cell 36, 932-941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan J. R., Muhlrad D., Parker R. (2008). P bodies promote stress granule assembly in Saccharomyces cerevisiae. J. Cell Biol. 183, 441-455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller J., Parker R. (2005). General translational repression by activators of mRNA decapping. Cell 122, 875-886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuenca-Bono B., Garcia-Molinero V., Pascual-Garcia P., Garcia-Oliver E., Llopis A., Rodriguez-Navarro S. (2010). A novel link between Sus1 and the cytoplasmic mRNA decay machinery suggests a broad role in mRNA metabolism. BMC Cell Biol. 11, 19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker C. J., Teixeira D., Parker R. (2007). Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J. Cell Biol. 179, 437-449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever T. E., Feng L., Wek R. C., Cigan A. M., Donahue T. F., Hinnebusch A. G. (1992). Phosphorylation of initiation factor 2 alpha by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell 68, 585-596 [DOI] [PubMed] [Google Scholar]
- Dominguez D., Altmann M., Benz J., Baumann U., Trachsel H. (1999). Interaction of translation initiation factor eIF4G with eIF4A in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 274, 26720-26726 [DOI] [PubMed] [Google Scholar]
- Dominguez D., Kislig E., Altmann M., Trachsel H. (2001). Structural and functional similarities between the central eukaryotic initiation factor (eIF)4A-binding domain of mammalian eIF4G and the eIF4A-binding domain of yeast eIF4G. Biochem. J. 355, 223-230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan H. M., Mackler B. (1966). Electron transport systems of yeast. 3. Preparation and properties of cytochrome oxidase. J. Biol.. Chem. 241, 1694-1697 [PubMed] [Google Scholar]
- Grant C. M., MacIver F. H., Dawes I. W. (1997). Mitochondrial function is required for resistance to oxidative stress in the yeast Saccharomyces cerevisiae. FEBS Lett. 410, 219-222 [DOI] [PubMed] [Google Scholar]
- Grousl T., Ivanov P., Frydlova I., Vasicova P., Janda F., Vojtova J., Malinska K., Malcova I., Novakova L., Janoskova D., et al. (2009). Robust heat shock induces eIF2alpha-phosphorylation-independent assembly of stress granules containing eIF3 and 40S ribosomal subunits in budding yeast, Saccharomyces cerevisiae. J. Cell Sci. 122, 2078-2088 [DOI] [PubMed] [Google Scholar]
- Hilgers V., Teixeira D., Parker R. (2006). Translation-independent inhibition of mRNA deadenylation during stress in Saccharomyces cerevisiae. RNA 12, 1835-1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyle N. P., Castelli L. M., Campbell S. G., Holmes L. E., Ashe M. P. (2007). Stress-dependent relocalization of translationally primed mRNPs to cytoplasmic granules that are kinetically and spatially distinct from P-bodies. J. Cell Biol. 179, 65-74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Stoecklin G., Ayodele M., Yacono P., Lykke-Andersen J., Fritzler M. J., Scheuner D., Kaufman R. J., Golan D. E., Anderson P. (2005). Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169, 871-884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotyk A., Rihova L., Ponec M. (1971). Uptake of amino acids by actidione-treated yeast cells. II. Effect of incubation conditions and metabolic inhibitors. Folia Microbiol. (Praha) 16, 445-450 [DOI] [PubMed] [Google Scholar]
- Li W., Simarro M., Kedersha N., Anderson P. (2004). FAST is a survival protein that senses mitochondrial stress and modulates TIA-1-regulated changes in protein expression. Mol. Cell. Biol. 24, 10718-10732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazroui R., Sukarieh R., Bordeleau M. E., Kaufman R. J., Northcote P., Tanaka J., Gallouzi I., Pelletier J. (2006). Inhibition of ribosome recruitment induces stress granule formation independently of eukaryotic initiation factor 2alpha phosphorylation. Mol. Biol. Cell 17, 4212-4219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokas S., Mills J. R., Garreau C., Fournier M. J., Robert F., Arya P., Kaufman R. J., Pelletier J., Mazroui R. (2009). Uncoupling stress granule assembly and translation initiation inhibition. Mol. Biol. Cell 20, 2673-2683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollet S., Cougot N., Wilczynska A., Dautry F., Kress M., Bertrand E., Weil D. (2008). Translationally repressed mRNA transiently cycles through stress granules during stress. Mol. Biol. Cell 19, 4469-4479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohn T., Kedersha N., Hickman T., Tisdale S., Anderson P. (2008). A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. Nat. Cell Biol. 10, 1224-1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R., Sheth U. (2007). P bodies and the control of mRNA translation and degradation. Mol. Cell 25, 635-646 [DOI] [PubMed] [Google Scholar]
- Penso J., Beitner R. (1998). Clotrimazole and bifonazole detach hexokinase from mitochondria of melanoma cells. Eur. J. Pharmacol. 342, 113-117 [DOI] [PubMed] [Google Scholar]
- Rikhvanov E. G., Varakina N. N., Rusaleva T. M., Rachenko E. I., Kiseleva V. A., Voinikov V. K. (2001). Effect of sodium azide on heat-shock resistance in Saccharomyces cerevisiae and Debaryomyces vanriji yeasts. Mikrobiologiia 70, 300-304 [PubMed] [Google Scholar]
- Rikhvanov E. G., Varakina N. N., Rusaleva T. M., Rachenko E. I., Voinikov V. K. (2002). Sodium azide reduces the thermotolerance of respiratively grown yeasts. Curr. Microbiol. 45, 394-399 [DOI] [PubMed] [Google Scholar]
- Scarcelli J. J., Viggiano S., Hodge C. A., Heath C. V., Amberg D. C., Cole C. N. (2008). Synthetic genetic array analysis in Saccharomyces cerevisiae provides evidence for an interaction between RAT8/DBP5 and genes encoding P-body components. Genetics 179, 1945-1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheller N., Resa-Infante P., de la Luna S., Galao R. P., Albrecht M., Kaestner L., Lipp P., Lengauer T., Meyerhans A., Diez J. (2007). Identification of PatL1, a human homolog to yeast P body component Pat1. Biochim. Biophys. Acta 1773, 1786-1792 [DOI] [PubMed] [Google Scholar]
- Schuster I. (1985). The interaction of representative members from two classes of antimycotics-the azoles and the allylamines-with cytochromes P-450 in steroidogenic tissues and liver. Xenobiotica 15, 529-546 [DOI] [PubMed] [Google Scholar]
- Schwartz D. C., Parker R. (1999). Mutations in translation initiation factors lead to increased rates of deadenylation and decapping of mRNAs in Saccharomyces cerevisiae. Mol. Cell. Biol. 19, 5247-5256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D. C., Parker R. (2000). mRNA decapping in yeast requires dissociation of the cap binding protein, eukaryotic translation initiation factor 4E. Mol. Cell. Biol. 20, 7933-7942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serman A., Le Roy F., Aigueperse C., Kress M., Dautry F., Weil D. (2007). GW body disassembly triggered by siRNAs independently of their silencing activity. Nucleic Acids Res. 35, 4715-4727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U., Parker R. (2003). Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300, 805-808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N., Hinnebusch A. G. (2009). Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136, 731-745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira D., Sheth U., Valencia-Sanchez M. A., Brengues M., Parker R. (2005). Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA 11, 371-382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson D. F., Chance B. (1967). Azide inhibition of mitochondrial electron transport. I. The aerobic steady state of succinate oxidation. Biochim. Biophys. Acta 131, 421-430 [DOI] [PubMed] [Google Scholar]
- Yamasaki S., Anderson P. (2008). Reprogramming mRNA translation during stress. Curr. Opin. Cell Biol. 20, 222-226 [DOI] [PMC free article] [PubMed] [Google Scholar]