Abstract
Tetracyclines have been important agents in combating infectious disease since their discovery in the mid-twentieth century. Following widespread use, tetracycline resistance mechanisms emerged and continue to create a need for new derivatives that are active against resistant bacterial strains. Semisynthesis has led to second and third generation tetracycline derivatives with enhanced antibiotic activity and pharmacological properties. Recent advancement in understanding of the tetracycline biosynthetic pathway may open the door to broaden the range of tetracycline derivatives and afford analogs that are difficult to access by synthetic chemistry.
Introduction
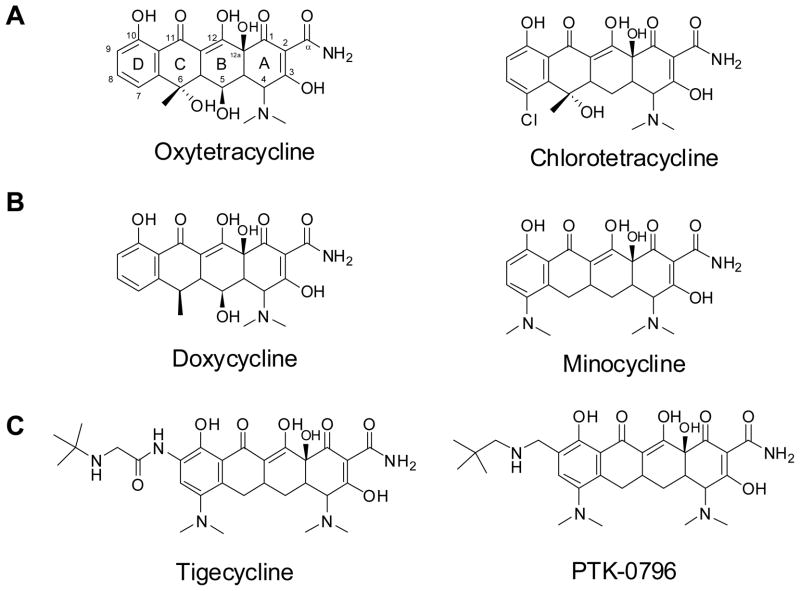
Tetracyclines are members of the polyketide family of natural products and include a number of important pharmaceutical compounds. This group is characterized by their tetracyclic ring structure, the richly substituted A ring, and a heavily oxidized periphery that includes a C11, C12 and C11a keto-enol configuration. The latter feature allows tetracycline to chelate divalent cations and bind to the 30S ribosome subunit (Brodersen et al., 2000; Chopra and Roberts, 2001). Several well-known tetracyclines, including both naturally produced and semisynthetically derived, are shown in Figure 1. Chlorotetracycline was the first of these compounds to be discovered. It was first reported by Benjamin Duggar in 1948 as an isolate from Streptomyces aureofaciens and was named aureomycin due to its yellow hue (Duggar, 1948). The discovery of oxytetracycline, produced by Streptomyces rimosus, followed shortly thereafter by A.C. Findlay in 1950 (Finlay et al., 1950). The importance of tetracyclines was immediately recognized due to their effectiveness against both gram-negative and gram-positive pathogens, and low toxicity in animals. Tetracyclines were the first major class of therapeutics to earn this distinction of “broad-spectrum antibiotic” (Chopra et al., 1992). This contributed to their widespread production and use in both human and animal medicine in the decades following their discovery. As a result, this subsequently led to emergence of resistance mechanisms and decreased effectiveness of tetracyclines as front line antibiotics.(Levy et al., 1999; Levy et al., 1989). Currently there are 40 known tetracycline resistance genes(Roberts, 2005).
Figure 1.
A) Natural product tetracyclines; B) Second-generation semisynthetic tetracycline antibiotics; C) Third generation tetracycline antibiotics based on minocycline.
The severity of bacterial resistance created an urgent need to develop new tetracycline derivatives capable of evading these resistance mechanisms. To achieve this end, there was a large push in the period around 1970–1980 to create semisynthetic variants of naturally occurring therapeutics. This period was termed by some as the “golden age of antibiotic medicinal chemistry”, where naturally occurring scaffolds were modified to achieve compounds with improved pharmacological properties (Wright, 2007). These semisynthetic variants represent the second generation tetracyclines. The most clinically valuable of these are minocycline and doxycycline, which are produced from 7-chloro-6-demethyltetracycline and oxytetracycline, respectively(Martell and Boothe, 1967). These analogs are more lipophilic than their parent compounds, which resulted in more efficient uptake into the cell. This feature was key in overcoming efflux resistance mechanism by lowering the net efflux from the cell (McMurry et al., 1982). In contrast, tetracycline and minocycline were both ineffective when exposed to resistance determinants of the ribosomal protection class (Clermont et al., 1997).
After a long gap in new tetracycline development, the glycylcyclines or so called third generation tetracycline analogs were developed recently. The sole successful member, tigecycline, was approved by the FDA in 2005 (Agwuh and MacGowan, 2006; Sum and Petersen, 1999; Sum et al., 2006). These compounds have modifications at position C-9 on the minocycline scaffold, which lead to evasion of both efflux and ribosomal protection mechanisms of resistance (Bergeron et al., 1996), making it a viable choice against tetracycline resistant infections. Another third generation derivative currently in Phase II clinical trials is PTK-0796 from Paratek Pharmaceuticals (Butler, 2008). This compound also contains a C-9 substitution and is a broad-spectrum antibiotic, including activities against multi-drug resistant MRSA and vancomycin resistant enterococci. The development of these compounds demonstrates that the tetracycline scaffold remains a valuable starting point for further drug discovery.
In addition to the well-documented antibacterial properties of tetracyclines, some tetracycline derivatives have also been investigated for anticancer properties. Antimicrobial compounds such as oxytetracycline and doxycycline were found to also have an inhibitory effect toward human matrix metalloproteinases(Gu et al., 2001; Lokeshwar et al., 2001). These enzymes are involved in the degradation of the extracellular matrix and have implications in inflammatory disorders and cancer. Doxycycline hyclate is being used as a treatment for periodontis under the brand name Periostat® as it inhibits the enzymes that break down gum tissue (Golub et al., 1998). A family of chemically modified tetracyclines (CMTs) has been developed that lack the C-4 dimethylamino group from tetracycline and doxycycline, and thus lack antibiotic activity, but have improved activity against matrix metalloproteins.
Therefore, the tetracycline family has yielded a large number of biosynthetically and chemically modified variants, several of which have found uses as valuable pharmaceutical compounds. A large majority of these variants are semisynthetic derivatives. Total synthesis has been achieved for oxytetracycline in 1979 (Muxfeldt et al., 1979). More recently the Myers group has developed a facile method for synthesizing tetracycline and related pentacycline compounds (Charest et al., 2005). However, biosynthetic/fermentative approaches remain the most cost effective methods for large-scale production of tetracyclines (Khosla and Tang, 2005). Additionally, with increased knowledge of tetracycline gene clusters and biosynthetic pathways, engineered biosynthesis has emerged as a new tool to generate analogs that may be inaccessible using synthetic chemistry alone. Investigation of other clinically relevant type II polyketide biosynthetic pathways has led to a number of novel bioactive compounds (Lombó et al., 2006; Madduri et al., 1998; Méndez and Salas, 2001). The clinical importance of tetracyclines and their range of biological activity provide powerful motivation to utilize metabolic engineering towards the goal of developing superior tetracycline analogs.
Tetracycline biosynthesis
Rational engineering of secondary metabolites begins with a thorough understanding of the biosynthetic pathway responsible for their production. Since the initial discovery of oxytetracycline and chlorotetracycline, researchers have manipulated the tetracycline producers using the tools available to elucidate their biosynthetic origins. Much of the early knowledge about the biosynthetic pathway was investigated by McCormick et al in the 1960s through a series of blocked mutants and substrate feeding experiments (McCormick and Jensen, 1965; McCormick et al., 1968; McCormick et al., 1963a; McCormick et al., 1962; McCormick et al., 1963b). They identified several key intermediates, including 6-methylpretetramid and anhydrotetracycline (ATC), and were able to deduce valuable information about the sequence of events leading to the production of tetracycline even though the genes themselves were not known. For example, substrate feeding experiments using mutants blocked in early tetracycline biosynthesis showed that feeding of pretetramid and 6-methylpretetramid could restore tetracycline biosynthesis, however, feeding of C4-dimethylamino-pretetramid to these substrates did not restore tetracycline biosynthesis. This indicated that the dimethylaminonaphthacenes were not intermediates in the pathway and several tailoring steps stood between the pretetramid precursor and the addition of the dimethylamino group. (McCormick et al., 1963a). In another experiment with labeled substrate feeding they showed that 5a, 6-ATC can be converted to tetracycline, demonstrating that this bioconversion takes place at a late stage in the synthesis. The different substrate 5a, 6-anhydro-12a-deoxytetracycline however, was converted to 12a-deoxytetracycline, demonstrating that the 12a-hydroxylation is a key modification step that occurs prior to the C-4 amidation(McCormick et al., 1962). The next fundamental insight into tetracycline biosynthesis was accomplished in a feeding experiment using 13C-labeled acetate. Thomas and Williams verified the polyketide origin of the tetracycline backbone and supported the idea of a malonamyl-CoA as the origin of the C-2 amide (Thomas and Williams, 1983a; Thomas and Williams, 1983b).
The first heterologous expression of genes required for tetracycline biosynthesis was carried out by Binnie et al. This work demonstrated that all of the genes required for oxytetracycline synthesis were located within a S. rimosus chromosomal DNA fragment flanked by otrA and otrB (Binnie et al., 1989). This was in contrast to previous gene mapping experiments that suggested oxytetracycline biosynthetic genes reside on two separate regions on the S. rimosus genome (Rhodes et al., 1981). The Butler group also located the gene otcC (or oxyS) responsible for ATC oxygenase activity using a reverse genetics approach, and made the first attempt to rationally engineer improvements in oxytetracycline production through manipulation of gene copy number(Binnie et al., 1989).
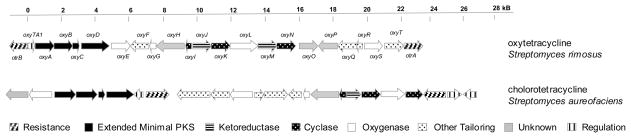
Each of these efforts made great strides in understanding the tetracycline biosynthetic pathway. The next big leap in understanding was enabled by sequencing of the gene clusters associated with chlorotetracycline (Ryan, 1999) and oxytetracycline biosynthesis (Hunter, 2002; Kim et al., 1994; Zhang et al., 2006a). The respective gene clusters are shown here for comparison in Figure 2. The annotation for the oxytetracycline cluster reflects that of Zhang et al. as published in 2006 (Zhang et al., 2006a). As the chlorotetracycline gene cluster has not been fully annotated, genes are identified here by their role in the biosynthetic pathway. As expected, there is a large degree of similarity between the two clusters, and all enzymes shown so far to be essential to oxytetracycline biosynthesis have homologs in the chlorotetracycline gene cluster. Several additional genes are included in the chlorotetracycline cluster, including those encoding additional regulatory proteins and the C-7 halogenase. We will focus on the oxytetracycline biosynthetic pathway in this review.
Figure 2.
Organization of biosynthetic gene cluster responsibly for the synthesis of oxytetracycline (top) and chlorotetracycline (bottom).
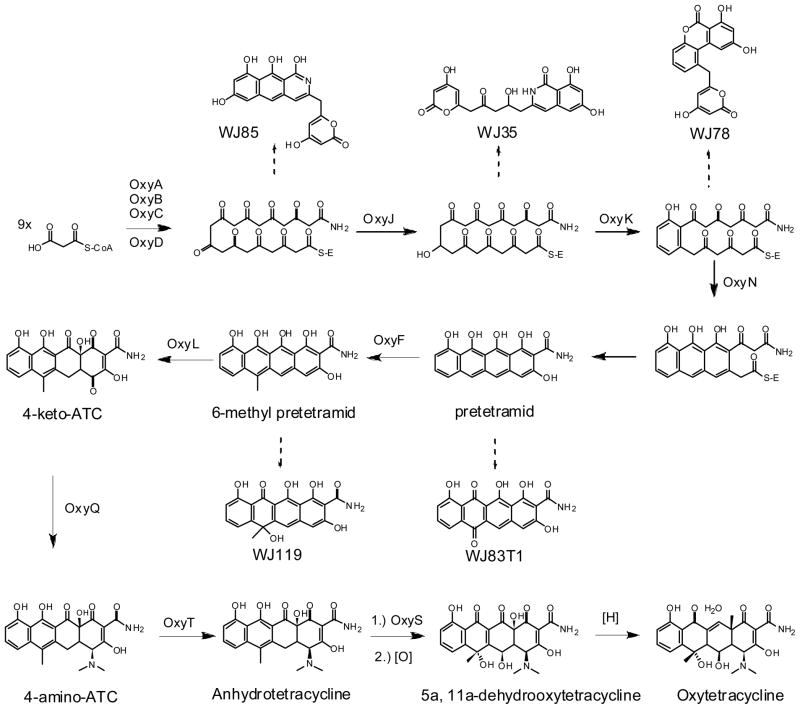
The function of several genes in the oxytetracycline pathway has been shown through genetic knockouts in S. rimosus (Peric-Concha et al., 2005; Petkovic et al., 1999) or reconstitution in heterologous Streptomyces strains. The most comprehensive study of gene function has been a systematic reconstitution of the oxytetracycline biosynthetic pathway in a heterologous host carried out by Tang and coworkers (Zhang et al., 2006a; Zhang et al., 2008; Zhang et al., 2006b; Zhang et al., 2007a; Zhang et al., 2007b). By using Streptomyces coelicolor strain CH999 (McDaniel et al., 1993), the minimal set of genes required to produce key intermediates in the pathway such as the amidated decaketide backbone, 6-methylpretetramid and ATC have been determined and are shown in Figure 3.
Figure 3.
Oxytetracycline biosynthetic pathway. Dashed arrows indicate shunt products formed at each step in the absence of the remaining enzymes.
As with all type II polyketide biosynthesis, the oxytetracycline pathway contains a minimal PKS consisting of ketosynthase (KS), chain length factor (CLF), and acyl-carrier protein (ACP)(Hertweck, 2007). In the oxytetracycline biosynthetic gene cluster, these proteins have been named OxyA, OxyB and OxyC, respectively(Zhang et al., 2006a). Together, these enzymes are responsible for the iterative condensation of malonyl-CoA to yield the poly-β-ketone backbone of tetracycline. Initiation of polyketide synthesis is proposed to involve an amide containing starter unit, mostly likely in the form of malonamyl-CoA or malonamyl-ACP. The malonamyl starter unit is reflected in the tetracycline compounds as the C-2 amide. Evidence supporting this proposal initially came from labeled substrate feeding experiments(Thomas and Williams, 1983b). Later observations of shunt products containing the amide group further supported the notion of an amide starter unit (Peric-Concha et al., 2005; Petkovic et al., 1999). Recently, Tang and coworkers showed that OxyD, which is an amidotransferase homolog, was responsible for the incorporation of this amide unit and termed the set of genes OxyA through OxyD the “extended minimal PKS”. They showed that these set of four enzymes expressed together was sufficient to produce an amidated decaketide polyketide backbone, which spontaneously cyclizes via C11-C16 regioselectivity to yield WJ85 (Zhang et al., 2006b). When OxyJ, the C-9 ketoreductase, was added to the extended minimal PKS, a C-9 reduced amidated polyketide WJ35 was biosynthesized (Zhang et al., 2006a). The isoquinolone part of WJ35 is formed through the spontaneous cyclization between carbons C13–C18. Neither WJ35 nor WJ85 exhibits the C-7 to C-12 connectivity of the D-ring of oxytetracycline, indicating the need for additional enzymes to direct the regioselectivity of the cyclization. Although these two compounds confirmed the role of OxyD, it remains unknown the exact reaction catalyzed by OxyD, as well as the exact starter unit. These insights will require in vitro biochemical investigation of OxyD properties.
Sequential cyclization of the tetracyclic scaffold is the next important step in tetracycline biosynthesis. These reactions are typically performed by dedicated cyclases (Hertweck, 2007). Surprisingly, only two cyclases, OxyK and OxyN, are required to cyclize all four rings during tetracycline biosynthesis, as shown by Zhang et al through heterologous reconstitution in CH999 (Zhang et al., 2007a). OxyK was first identified by Petkovic et al.(Petkovic et al., 1999) and confirmed by Zhang et al.(Zhang et al., 2007b) as the enzyme responsible for D-ring cyclization through C-7/C-12 connectivity. Expression of OxyABCDJKN resulted in the cyclization of all four rings to yield pretetramid, which was oxidized in vivo to yield WJ83. When OxyD was removed from the construct, the major product was the tricyclic anthracene carboxylic acid and was not cyclized in the fourth ring. This indicated an important role for the amide starter unit in directing the spontaneous cyclization of ring A. Subsequently, addition of the C-methyltransferase OxyF to OxyABCDJKN yielded the key intermediate 6-methylpretetramid, confirming the sequence of these early biosynthetic steps.
Recently, the late stage tailoring steps leading to the production of anhydrotetracycline (ATC) have been elucidated. Again using the heterologous host/vector pair, it was shown that starting from 6-methylpretetramid, the enzymes OxyL, OxyQ and OxyT catalyze the C-12a hydroxylation, C-4 amidation, and C-4 N,N-dimethylation steps, respectively, to afford ATC (Zhang et al., 2008). An in vitro reaction of OxyL with 6-methylpretetramid as a substrate showed that this single enzyme is responsible for the double hydroxylation (C12a and C4) of ring A, resulting in the unstable product 4-keto-ATC. Addition of OxyQ to the host producing 4-keto-ATC (OxyABCDJKNFL) led to the reductive amination of C-4 and synthesis of 4-amino-ATC, confirming the function of OxyQ. In vitro addition of OxyT to 4-amino-ATC resulted in a N, N-dimethylation modification as confirmed by the isolation of ATC. Finally, in vivo expression of the entire construct OxyABCDJKNFLQT proved this to be the minimal set of enzymes required to produce ATC in heterologous host CH999 (Zhang et al., 2008).
The final steps converting ATC to oxytetracycline require two oxygenations and a dehydration step. OxyS (OtcC) has been shown to be responsible for the oxygenation of ATC (Binnie et al., 1989). In vitro studies of the homologous gene in chlorotetracycline biosynthesis showed that purified enzyme could convert ATC to 5a,11a-dehydrotetracycline (DHTC) (Binnie et al., 1989; Vancurová et al., 1988). A second oxygenation reaction is needed in the oxytetracycline pathway to add the C-5 hydroxyl group resulting in 5a, 11a-dehydrooxytetracycline(DHOTC). The enzyme responsible for this specific tailoring step is unknown. The final reduction step converting 5a, 11a-DHOTC to oxytetracycline is still not entirely clear. Nakono et al. reported a tetracycline dehydrogenase gene from S. aureofaciens external to the CTC gene cluster was capable of performing this last step (Nakano et al., 2004). This was shown through the identification of a mutant deficient in the reduction step, and subsequent restoration of function by integrating TchA into the host genome. A homologous ORF was identified in the S. rimosus genome, but its function has not been verified. The most up to date oxytetracycline biosynthetic pathway is shown in Figure 3. A number of steps in the pathway are unresolved and further genetic and biochemical studies are needed to complete our understanding of tetracycline biosynthesis.
Manipulation of Oxytetracycline Pathway
Throughout the recent studies of the tetracycline pathways, several new compounds/shunt products have been generated and the substrate specificities of several enzymes in the pathway have been examined. The first such study was the expression of the oxytetracycline minimal PKS in heterologous host CH999. OxyABC were expressed along with the act C-9 ketoreductase and resulted in an acetate primed decaketide product previously identified as RM20b (Fu et al., 1994). This was significant as it demonstrated the relaxed starter unit specificity of oxytetracycline minimal PKS, as well as its functional interaction with a heterologous tailoring enzyme act KR (Fu et al., 1994). This prompted Zhang et al. to inquire if the amidotransferase OxyD would be able to introduce an amidated starter unit to heterologous minimal PKSs to generate amidated polyketides of different sizes. When OxyD, OxyC and act KR were coexpressed with the act KS-CLF and the tcm KS-CLF in engineered strain CH999, however, no amidated polyketides were produced (Zhang et al., 2006a).
The Hunter group constructed S. rimosus knockouts strains lacking oxyK (Petkovic et al., 1999) and oxyS (Peric-Concha et al., 2005). Surprisingly, both strains produced new polyketides with truncated carbon backbones. Although these results revealed the functions of OxyK and OxyS, they also demonstrated the inherent unpredictability of genetic manipulation of the oxytetracycline gene cluster in the native host. Several new products and shunt products (see previous section) were also formed through the systematic reconstitution of oxytetracycline pathway enzymes by the Tang group. Though many are interesting for their novel cyclization and structure features, the most useful from a metabolic engineering standpoint are the intermediates that can be used as building blocks for the generation of tetracycline analogs. These include 6-methylpretetramid, 4-amino-ATC, and ATC. 6-desmethyl-ATC was also produced through rational engineering of a heterologous construct lacking the oxyF gene (Zhang et al., 2008). While compounds lacking the C-6 methyl were previously produced by blocked-mutant studies, this is the first rational production of 6-desmethyl-ATC in a heterologous host. This also confirmed the relaxed specificity of the downstream tailoring enzymes toward this position. The ability to reliably produce intermediates and variants in the tetracycline pathway will undoubtedly lead to engineered biosynthesis of other tetracycline derived compounds.
In addition to producing building blocks for tetracycline biosynthesis, the unique elements of the oxytetracycline biosynthetic pathway have also been exploited for the production of other chemical moieties. In a 2007 paper, Zhang and coworkers reported the utilization of OxyD as a new mechanism for benzopyrone formation (Zhang et al., 2007b). This mechanism involves the displacement of the amide starter unit by the nucleophilic phenol group, which is in contrast to the nucleophilic attack of the free amide on a ketone carbonyl. Once this mechanism was identified, they set out to rationally biosynthesize an analog of HMG-CoA reductase inhibitor pannorin via an amidated backbone. This is an example of how genes unique to the tetracycline pathway can serve as enzymatic tools towards creating other classes of compounds.
Metabolic Engineering of Tetracycline Biosynthesis
As the naturally occurring tetracyclines have been of such great commercial importance over the past 60 years, there had been much effort dedicated to strain improvement and titer optimization. However, most of the strain improvement was performed in industrial settings, and little has been made available to the scientific community. Nearly all strain improvement efforts rely on mutagenesis techniques, which is a black-box approach that provided few, if any genotypic information. Studies on the optimization of the use of NTG for mutagenesis of S. rimosus were detailed by Delić et al in 1970 (Delic et al., 1970). The randomly introduced mutations can be beneficial in a number of ways, not only through increased production but also give rise to strain phenotypes that display improved fermentation properties. In one instance, genetic recombination was used to produce a strain that resulted in no foaming in the fermentation medium, as well as increased oxytetracycline production (Mindlin et al., 1961). In several studies phage resistant mutants have been found which can be of great benefit in industrial fermentation (Vesligaj et al., 1981).
Some of the naturally occurring variation in oxytetracycline production levels has been attributed to the genetic instability of the oxytetracycline biosynthetic cluster. Genetic instability is commonly observed among Streptomyces species and in the case of the oxytetracycline gene cluster, is likely due in part to its location on the end of the linear chromosome of S. rimosus. The gene cluster is located approximately 600 kb from one end of the chromosome (Petkovic et al., 2006). These instability variants were classified into three groups based on oxytetracycline self-resistance (Gravius et al., 1993). Class I (99%) showed unchanged resistance from the parent strain, but differed in a wide variety of morphologies including oxytetracycline production. Class II (1%) showed increased sensitivity to oxytetracycline, and class III (.1%) showed increased resistance to oxytetracycline and higher titer of oxytetracycline. Class II mutants were shown to have deletion of the oxytetracycline cluster through a fusion of two inverted copies of the chromosome both lacking the end with the antibiotic biosynthetic genes. Class III mutants, on the other hand resulted from increased copies of the oxytetracycline biosynthetic genes. Though class III mutants themselves can then revert to class I variants, this illustrates the possibility of using amplified DNA sequences to increase tetracycline production (Petkovic et al., 2006). The Butler group made the first attempt to use a gene dosing approach to increasing oxytetracycline production in S. rimosus (Butler et al., 1990). Here they constructed a low copy plasmid containing the oxytetracycline biosynthetic pathway between otrA and otrB. Transforming this plasmid into strains with lower productivity than the donor strain resulted in increased production of oxytetracycline in the low producing strains, however, production in excess of the parent strain was not achieved.
Other approaches have focused on genes external to the tetracycline biosynthetic pathway. One example is the overexpression of the alpha-amylase gene in S. rimosus. This was motivated by the observation that high alpha-amylase activity was observed during oxytetracycline production(Yang and Wang, 1999). They report an increase of amylase activity of up to two-fold and an increase of oxytetracycline production of up to 2.5 fold as a result of overexpression of this gene (Cheng et al., 2000). The underlying biochemical reasons for this increase remain unknown.
Another interesting strategy for increased antibiotic production is the ribosomal engineering technique outlined by Ochi (Ochi, 2007). In this method, the rRNA is the target for mutation rather than the pathway genes themselves. One mutation in the S12 rRNA resulted in preferential transcription of secondary metabolite regulation genes such as ActII-ORF4 and RedD in S. coelicolor. These SARP (Streptomyces Antibiotic Regulatory Protein) genes are directly involved in the up-regulation of specific biosynthetic genes cluster and are the terminal activators in the regulatory signaling cascade(Bibb, 2005). This concept was demonstrated in a recent example by Chen and coworkers. The production of another type II polyketide, fredericamycin, was increased by a factor of 5.6 in the wild-type host by overexpression of the SARP FdmR1(Chen et al., 2008). Both the oxytetracycline and chlorotetracycline pathways include SARP-like regulatory proteins. Their respective influences on tetracycline production and regulation have yet to be investigated.
Other Tetracycline Natural Products
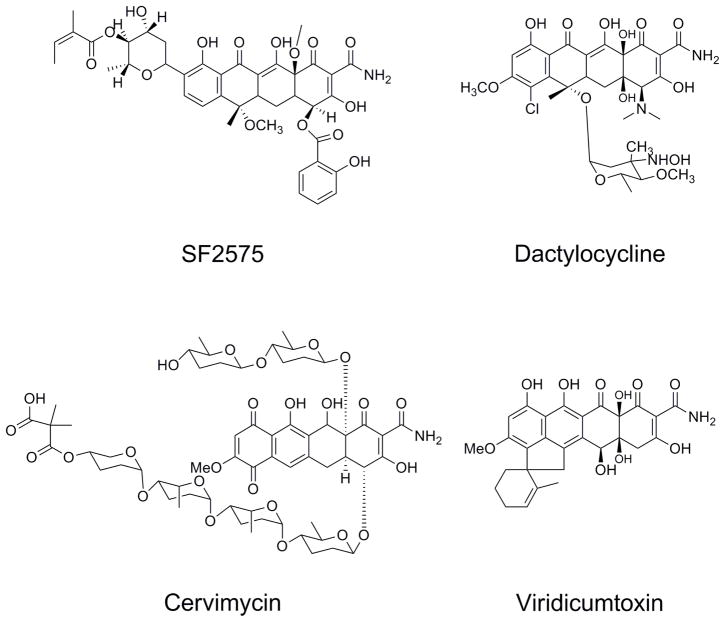
There are other tetracycline natural products, which have been more recently identified, with very interesting structural features and bioactivity (Figure 4). Though they have not yet achieved the clinical and commercial success of the classic tetracyclines discussed here, a detailed understanding of their biosynthetic pathways may add to the enzyme toolbox that one can use to construct and diversify new tetracycline molecules. SF2575, identified by Hatsu and coworkers, is an intriguing tetracycline compound containing a salicylic acid substitution at C4 and a C-deoxysugar at C-8(Hatsu et al., 1992a; Hatsu et al., 1992b). The core of the molecule is also methylated at numerous hydroxyl positions. While this compound showed antibiotic activity only against gram-positive bacteria, it was demonstrated to have potent antitumor activity against P388 leukemia cells in mice (Hatsu et al., 1992b). Around the same time, another group reported isolation of dactylocyclines, from Dactylosporangium Sp. SC14051(Tymiak et al., 1993). In addition to their novel structure and glycosylation patterns, they showed antibiotic activity against tetracycline resistant gram-positive bacteria. Cervimycin C is also an aromatic polyketide antibiotic with a tetracycline-like core, and also contains extensive decoration and modification by deoxysugars. It was reported to be very effective against vancomycin-resistant enterococci and has a different mode of action from that of traditional tetracycline antibiotics, making it an attractive antibiotic target (Herold et al., 2004). The Hertwerk group has begun biosynthetic investigation of this compound, starting with the origin of the rare dimethylmalonyl group (Herold et al., 2004). Finally, the structurally related compound viridicatumtoxin is produced by fungal species Penicillium viridicatum (Raju et al., 1982). While it is structurally similar to the tetracycline family, the polyketide backbone exhibits a distinct folding mechanism (type F) compared to the Streptomyces tetracyclines (type S)(Thomas, 2001). Further genetic studies with these gene clusters would provide a fascinating comparison to known tetracycline pathways.
Figure 4.
Additional natural products containing the tetracycline scaffold.
Concluding Remarks
The unique structural features of tetracyclines provide exciting opportunities for synthetic chemists and biomolecular engineers alike. The recent knowledge gained from sequencing of the tetracycline producing gene clusters has afforded the biosynthetic blueprints to these pathways. At the same time, the systematic investigation and rational engineering of the oxytetracycline biosynthesis have provided the first glimpse into the combinatorial potential of these polyketide pathways. Metabolic engineering and engineered biosynthesis, combined with chemical synthesis, will play valuable roles in the discovery and production of future generations of tetracyclines.
Acknowledgments
Research in our lab on this topic has been supported by grants from NSF (CBET #0545860).
References
- Agwuh KN, MacGowan A. Pharmacokinetics and pharmacodynamics of the tetracyclines including glycylcyclines. J Antimicrob Chemother. 2006;58:256–265. doi: 10.1093/jac/dkl224. [DOI] [PubMed] [Google Scholar]
- Bergeron J, et al. Glycylcyclines bind to the high-affinity tetracycline ribosomal binding site and evade Tet(M)- and Tet(O)-mediated ribosomal protection. Antimicrob Agents Chemother. 1996;40:2226–2228. doi: 10.1128/aac.40.9.2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibb MJ. Regulation of secondary metabolism in streptomycetes. Curr Opin Microbiol. 2005;8:208–215. doi: 10.1016/j.mib.2005.02.016. [DOI] [PubMed] [Google Scholar]
- Binnie C, et al. Cloning and heterologous expression in Streptomyces lividans of Streptomyces rimosus genes involved in oxytetracycline biosynthesis. J Bacteriol. 1989;171:887–895. doi: 10.1128/jb.171.2.887-895.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen DE, et al. The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit. Cell. 2000;103:1143–1154. doi: 10.1016/s0092-8674(00)00216-6. [DOI] [PubMed] [Google Scholar]
- Butler MJ, et al. Genetic manipulation of the oxytetracycyline biosynthetic pathway genes. Dev Ind Microbiol. 1990;31:41–50. [Google Scholar]
- Butler MS. Natural products to drugs: natural product-derived compounds in clinical trials. Nat Prod Rep. 2008;25:475–516. doi: 10.1039/b514294f. [DOI] [PubMed] [Google Scholar]
- Charest MG, et al. A convergent enantioselective route to structurally diverse 6- deoxytetracycline antibiotics. Science. 2005;308:395–398. doi: 10.1126/science.1109755. [DOI] [PubMed] [Google Scholar]
- Chen Y, et al. Identification and utility of FdmR1 as a SARP activator for fredericamycin production in Streptomyces griseus ATCC49344 and heterologous hosts. J Bacteriol. 2008;190:5587–5596. doi: 10.1128/JB.00592-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng CW, et al. Cloning and expression of the α-amylase gene and oxytetracycline production in Streptomyces rimosus. World J Microbiol Biotechnol. 2000;16:225–230. [Google Scholar]
- Chopra I, et al. Tetracyclines, molecular and clinical aspects. J Antimicrob Chemother. 1992;29:245–277. doi: 10.1093/jac/29.3.245. [DOI] [PubMed] [Google Scholar]
- Chopra I, Roberts M. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol Mol Biol Rev. 2001;65:232–260. doi: 10.1128/MMBR.65.2.232-260.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clermont D, et al. New tetracycline resistance determinants coding for ribosomal protection in streptococci and nucleotide sequence of tet(T) isolated from Streptococcus pyogenes A498. Antimicrob Agents Chemother. 1997;41:112–116. doi: 10.1128/aac.41.1.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delic V, et al. Mutagenesis by N-methyl-N′-nitro-N-nitrosoguanidine (NTG) in Streptomyces coelicolor. Mutat Res Fund Mol Mech. 1970;9:167–182. doi: 10.1016/0027-5107(70)90055-2. [DOI] [PubMed] [Google Scholar]
- Duggar BM. Aureomycin; a product of the continuing search for new antibiotics. Ann NY Acad Sci. 1948;51:177–181. doi: 10.1111/j.1749-6632.1948.tb27262.x. [DOI] [PubMed] [Google Scholar]
- Finlay AC, et al. Terramycin, a new antibiotic. Science. 1950;111:85. doi: 10.1126/science.111.2874.85. [DOI] [PubMed] [Google Scholar]
- Fu H, et al. Relaxed specificity of the oxytetracycline polyketide synthase for an acetate primer in the absence of a malonamyl primer. J Am Chem Soc. 1994;116:6443–6444. [Google Scholar]
- Golub LM, et al. Tetracyclines inhibit connective tissue breakdown by multiple non-antimicrobial mechanisms. Adv Dent Res. 1998;12:12–26. doi: 10.1177/08959374980120010501. [DOI] [PubMed] [Google Scholar]
- Gravius B, et al. Genetic instability and strain degeneration in Streptomyces rimosus. Appl Environ Microbiol. 1993;59:2220–2228. doi: 10.1128/aem.59.7.2220-2228.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, et al. Inhibition of tumor cell invasiveness by chemically modified tetracyclines. Curr Med Chem. 2001;8:261–70. doi: 10.2174/0929867013373642. [DOI] [PubMed] [Google Scholar]
- Hatsu M, et al. A new tetracycline antibiotic with antitumor activity. II The structural elucidation of SF2575. J Antibiotics. 1992a;45:325–330. doi: 10.7164/antibiotics.45.325. [DOI] [PubMed] [Google Scholar]
- Hatsu M, et al. A new tetracycline antibiotic with antitumor activity. I Taxonomy and fermentation of the producing strain, isolation and characterization of SF2575. J Antibiotics. 1992b;45:320–324. doi: 10.7164/antibiotics.45.320. [DOI] [PubMed] [Google Scholar]
- Herold K, et al. Biosynthesis of cervimycin C, an aromatic polyketide antibiotic bearing an unusual dimethylmalonyl moity. Org Biomol Chem. 2004;2:2411–2414. doi: 10.1039/B409221J. [DOI] [PubMed] [Google Scholar]
- Hertweck C. Type II polyketide synthases: gaining a deeper insight into enzymatic teamwork. Nat Prod Rep. 2007;24:162–190. doi: 10.1039/b507395m. [DOI] [PubMed] [Google Scholar]
- Hunter IS. Tetracyclines. In: Fierro F, Martin JF, editors. Microbial Secondary Metabolites: Biosynthesis, Genetics and Regulation. Research Signpost; Lucknow: 2002. pp. 141–166. [Google Scholar]
- Khosla C, Tang Y. Chemistry. A new route to designer antibiotics. Science. 2005;308:367–368. doi: 10.1126/science.1111415. [DOI] [PubMed] [Google Scholar]
- Kim ES, et al. Sequences of the oxytetracycline polyketide synthase-encoding otc genes from Streptomyces rimosus. Gene. 1994;141:141–142. doi: 10.1016/0378-1119(94)90144-9. [DOI] [PubMed] [Google Scholar]
- Levy SB, et al. Nomenclature for new tetracycline resistance determinants. Antimicrob Agents Chemother. 1999;43:1523–1524. doi: 10.1128/aac.43.6.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy SB, et al. Nomenclature for tetracycline resistance determinants. Antimicrob Agents Chemother. 1989;33:1373–1374. doi: 10.1128/aac.33.8.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokeshwar BL, et al. Cytotoxic activity and inhibition of tumor cell invasion by derivatives of a chemically modified tetracycline CMT-3 (COL-3) Curr Med Chem. 2001;8:271–279. doi: 10.2174/0929867013373516. [DOI] [PubMed] [Google Scholar]
- Lombó F, et al. The aureolic acid family of antitumor compounds: structure, mode of action, biosynthesis, and novel derivatives. Appl Environ Microbiol. 2006;73:1–14. doi: 10.1007/s00253-006-0511-6. [DOI] [PubMed] [Google Scholar]
- Madduri K, et al. Production of the antitumor drug epirubicin (4′-epidoxorubicin) and its precursor by a genetically engineered strain of Streptomyces peucetius. Nat Biotech. 1998;16:69–74. doi: 10.1038/nbt0198-69. [DOI] [PubMed] [Google Scholar]
- Martell MJ, Boothe JH. The 6-deoxytetracyclines. VII alkylated aminotetracyclines possessing unique antibacterial activity. J Med Chem. 1967;10:44–46. doi: 10.1021/jm00313a009. [DOI] [PubMed] [Google Scholar]
- McCormick JRD, Jensen ER. Biosynthesis of the tetracyclines. VIII Characterization of 4-hydroxy-6-methylpretetramid. J Am Chem Soc. 1965;87:1794–1795. doi: 10.1021/ja01086a034. [DOI] [PubMed] [Google Scholar]
- McCormick JRD, et al. Biosynthesis of the tetracyclines. IX 4-Aminodedimethylaminoanhydrodemethylchlortetracycline from a mutant of Streptomyces aureofaciens. J Am Chem Soc. 1968;90:2201–2202. doi: 10.1021/ja01010a063. [DOI] [PubMed] [Google Scholar]
- McCormick JRD, et al. Biosynthesis of the tetracyclines. V Naphthacenic precursors. J Am Chem Soc. 1963a;85:1692–1694. [Google Scholar]
- McCormick JRD, et al. Biosynthesis of the tetracyclines. IV Biological rehydration of the 5a,6-anhydrotetracyclines. J Am Chem Soc. 1962;84:3023–3025. [Google Scholar]
- McCormick JRD, et al. Biosynthesis of the tetracyclines. VI Total synthesis of a naphthacenic precursor: 1,3,10,11,12-pentahydroxynaphthacene-2-carboxamide. J Am Chem Soc. 1963b;85:1694–1695. [Google Scholar]
- McDaniel R, et al. Engineered biosynthesis of novel polyketides. Science. 1993;262:1546–1550. doi: 10.1126/science.8248802. [DOI] [PubMed] [Google Scholar]
- McMurry LM, et al. Transport of lipophilic analog minocycline differs from that of tetracycline in susceptible and resistant Escherichia coli strains. Antimicrob Agents Chemother. 1982;22:791–799. doi: 10.1128/aac.22.5.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méndez C, Salas JA. Altering the glycosylation pattern of bioactive compounds. Trends Biotechnol. 2001;19:449–456. doi: 10.1016/s0167-7799(01)01765-6. [DOI] [PubMed] [Google Scholar]
- Mindlin SZ, et al. A new hybrid strain of an oxytetracycline-producing organism, Streptomyces rimosus. Appl Microbiol. 1961;9:349–353. doi: 10.1128/am.9.4.349-353.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muxfeldt H, et al. Tetracyclines. 9. Total synthesis of dl-terramycin. J Am Chem Soc. 1979;101:689–701. [Google Scholar]
- Nakano T, et al. Identification and cloning of the gene involved in the final step of chlortetracycline biosynthesis in Streptomyces aureofaciens. Biosci Biotechnol Biochem. 2004;68:1345–1352. doi: 10.1271/bbb.68.1345. [DOI] [PubMed] [Google Scholar]
- Ochi K. From microbial differentiation to ribosome engineering. Biosci Biotechnol Biochem. 2007;71:1373–1386. doi: 10.1271/bbb.70007. [DOI] [PubMed] [Google Scholar]
- Peric-Concha N, et al. Ablation of the otcC gene encoding a post-polyketide hydroxylase from the oxytetracyline biosynthetic pathway in Streptomyces rimosus results in novel polyketides with altered chain length. J Biol Chem. 2005;280:37455–37460. doi: 10.1074/jbc.M503191200. [DOI] [PubMed] [Google Scholar]
- Petkovic H, et al. Genetics of Streptomyces rimosus, the oxytetracycline producer. Microbiol Mol Biol Rev. 2006;70:704–728. doi: 10.1128/MMBR.00004-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkovic H, et al. Disruption of an aromatase/cyclase from the oxytetracycline gene cluster of Streptomyces rimosus results in production of novel polyketides with shorter chain lengths. J Biol Chem. 1999;274:32829–32834. [PubMed] [Google Scholar]
- Raju MS, et al. Microbial transformations of natural antitumor agents. 20. Glycosylation of viridicatumtoxin. J Nat Prod. 1982;45:321–327. [Google Scholar]
- Rhodes PM, et al. Biochemical and genetic characterization of Streptomyces rimosus mutants impaired in oxytetracycline biosynthesis. J Gen Microbiol. 1981;124:329–338. [Google Scholar]
- Roberts MC. Update on acquired tetracycline resistance genes. FEMS Microbiol Lett. 2005;245:195–203. doi: 10.1016/j.femsle.2005.02.034. [DOI] [PubMed] [Google Scholar]
- Ryan MJ, et al. Cloning of the biosynthetic pathway for chlortetracycline and tetracycline formation and cosmids useful therein. 5,866,410. US patent. 1999 February;
- Sum PE, Petersen P. Synthesis and structure-activity relationship of novel glycylcycline derivatives leading to the discovery of GAR-936. Bioorg Med Chem Lett. 1999;9:1459–1462. doi: 10.1016/s0960-894x(99)00216-4. [DOI] [PubMed] [Google Scholar]
- Sum PE, et al. Synthesis and antibacterial activity of 9-substituted minocycline derivatives. Bioorg Med Chem Lett. 2006;16:400–403. doi: 10.1016/j.bmcl.2005.09.078. [DOI] [PubMed] [Google Scholar]
- Thomas R. A biosynthetic classification of fungal and Streptomycete fused-ring aromatic polyketides. ChemBioChem. 2001;2:612–627. doi: 10.1002/1439-7633(20010903)2:9<612::AID-CBIC612>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Thomas R, Williams DJ. Oxytetracycline biosynthesis: mode of incorporation of [1- 13 C]- and [1,2–13C2]-acetate. J Chem Soc Chem Commun. 1983a:128–130. [Google Scholar]
- Thomas R, Williams DJ. Oxytetracycline biosynthesis: origin of the carboxamide substituent. J Chem Soc. 1983b:677–679. [Google Scholar]
- Tymiak AA, et al. Dactylocyclines: novel tetracycline glycosides active against tetracycline-resistant bacteria. J Org Chem. 1993;58:535–537. [Google Scholar]
- Vancurová I, et al. Isolation of pure anhydrotetracycline oxygenase from Streptomyces aureofaciens. Biochem J. 1988;253:263–267. doi: 10.1042/bj2530263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vesligaj M, et al. Isolation of Streptomyces rimosus mutants with reduced actinophage susceptibility. Appl Environ Microbiol. 1981;41:986–991. doi: 10.1128/aem.41.4.986-991.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright GD. The antibiotic resistome: the nexus of chemical and genetic diversity. Nat Rev Microbiol. 2007;5:175–186. doi: 10.1038/nrmicro1614. [DOI] [PubMed] [Google Scholar]
- Yang SS, Wang JY. Protease and amylase production by Streptomyces rimosus in submerged and solid substrate cultivation. Bot Bull Acad Sin. 1999;40:259–265. [Google Scholar]
- Zhang W, et al. Engineered biosynthesis of a novel amidated polyketide, using the malonamyl-specific initiation module from the oxytetracycline polyketide synthase. Appl Environ Microbiol. 2006a;72:2573–2580. doi: 10.1128/AEM.72.4.2573-2580.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, et al. Identifying the minimal enzymes required for anhydrotetracycline biosynthesis. J Am Chem Soc. 2008;130:6068–6069. doi: 10.1021/ja800951e. [DOI] [PubMed] [Google Scholar]
- Zhang W, et al. Heterologous biosynthesis of amidated polyketides with novel cyclization regioselectivity from oxytetracycline polyketide synthase. J Nat Prod. 2006b;69:1633–1636. doi: 10.1021/np060290i. [DOI] [PubMed] [Google Scholar]
- Zhang W, et al. Investigation of early tailoring reactions in the oxytetracycline biosynthetic pathway. J Biol Chem. 2007a;282:25717–25725. doi: 10.1074/jbc.M703437200. [DOI] [PubMed] [Google Scholar]
- Zhang W, et al. A new mechanism for benzopyrone formation in aromatic polyketide biosynthesis. J Am Chem Soc. 2007b;129:9304–9305. doi: 10.1021/ja0736919. [DOI] [PubMed] [Google Scholar]