Abstract
In all eukaryotic cells, 26S proteasome plays an essential role in the process of ATP-dependent protein degradation. In this review, we focus on structure characterization of the 26S proteasome. Although the progress towards a high-resolution structure of the 26S proteasome has been slow, the recently solved structures of various proteasomal subcomplexes have greatly enhanced our understanding of this large machinery. In addition to having an ATP-dependent proteolytic function, the 26S proteasome is also involved in many non-proteolytic cellular activities, which are often mediated by subunits in its 19S regulatory complex. Thus, we include a detailed discussion of the structures of 19S subunits, including proteasomal ATPases, ubiquitin receptors, deubiquitinating enzymes and subunits that contain PCI domain.
Keywords: 26S proteasome, structure
1. Introduction
1.1 Protein degradation by the ubiquitin-proteasome pathway
Intracellular protein degradation by the ubiquitin-proteasome pathway is a process that has been studied in depth. Briefly, proteins targeted for degradation by the 26S proteasome are first “labeled” by covalent linkage of a polyubiquitin chain, via a three-step cascade mechanism utilizing ubiquitin-activating enzyme E1, ubiquitin-conjugating enzyme E2, and ubiquitin-protein ligase E3 [1–4]. These polyubiquitinated substrates are recognized by the 26S proteasome and rapidly broken down into short peptides [3, 5–7]. In eukaryotic cells, this ubiquitin-mediated protein degradation pathway plays a primary role in the degradation of most short-lived proteins that are critical in the regulation of many cellular processes such as cell cycle [8–10], transcriptional regulation [11, 12], apoptosis [13], as well as in the degradation of abnormal proteins such as misfolded and damaged proteins [14]. Another role of this pathway is to break down foreign proteins to generate antigenic peptides to be presented by major histocompatibility complex (MHC) class I molecules to the T-cells of the immune system [15–19].
1.2 The 26S proteasome and its components
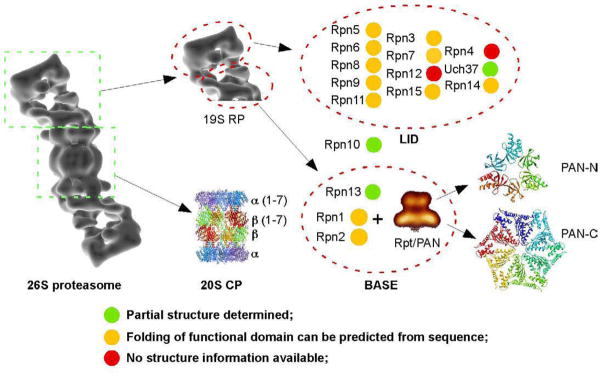
The proteolytic machinery of this ubiquitin-mediated protein degradation pathway is the 26S proteasome. It is a large and dynamic protein complex that is highly conserved in eukaryotes, in both structure and function. It consists of more than 30 different protein subunits, and has a molecular weight of over 2MDa. Figure 1 shows the major components of the 26S proteasome divided into functional groups. The middle part is a barrel-shaped 20S protease core particle (CP) that is formed by axial stacking of four heptameric rings: two identical inner β-rings, each formed by seven different β-subunits β1–7, and two identical outer α-rings each formed also by seven different α-subunits, α1–7. Capping at each end of the 20S CP is a 19S regulatory particle (RP) that regulates the proteolytic function of the protease core. The 19S RP can be further divided into base and lid subcomplexes. The base has six different ATPase subunits, Rpt1-6, and a few non-ATPase subunits, such as Rpn1, Rpn2 and Rpn13. The lid contains more than ten other Rpn subunits. The association between the base and the lid is mostly mediated by the Rpn10 subunit [20, 21]. Because the linkage between the base and lid is relative weak, under certain biochemical condition, the lid can be separated from the base to form lidless proteasomes.
Figure 1.
Schematic representation of the 26S proteasome. The components of 26S proteasome are divided into functional groups and subunits. The availability of structure information at different levels is also shown in different colors.
There are already many comprehensive reviews in the recent literature about various components or functions of the ubiquitin-proteasome system [22–27]. The main focus of this review is only on the structure characterization of the 26S proteasome. While the atomic structure of the 20S CP, as a complete protein complex, is very well characterized, high-resolution structures of the 19S RP are mostly limited to the individual subunits, or even domains. Therefore, our discussion of the 20S CP is as the complete complex, but that of the 19S RP is mainly on the individual functional subunits.
1.3 Structure of the 26S proteasome
Even with the tremendous progress made in the technology of all structure determination methods, it still has not been possible to crystallize the entire 26S proteasome for structure determination by X-ray crystallography [22]. In contrast, significant progress has been made toward determining the atomic structure of various isolated components of this complex. At relative lower resolution level, single particle cryo-electron microscopy (cryoEM) has been used to obtain three-dimensional (3D) envelopes of the 26S proteasome. The highest resolution published so far is about 20 Å [28] (with another 3D reconstruction of the human 26S proteasome at near 10 Å resolution from Edward Morris laboratory to be published soon). Such resolution is sufficient to distinguish domains, but falls short of resolving secondary structural features. In Figure 1, which summarizes the results obtained so far through these different approaches, the structures of the 26S proteasome and its components are shown as either ribbon diagrams, if their atomic structure have already been determined by X-ray crystallography, or as low-resolution density maps, if their 3D structure has been reconstructed by single particle EM. Otherwise, they are shown as colored blobs, with a color code indicating whether the functional domain of the subunits can be predicted by sequence homology to a known structure or folding, or whether their partial structures have been determined by either X-ray crystallography or NMR spectroscopy.
Functionally, the 20S CP performs the simple task of a protease, namely to degrade substrates that have entered its proteolytic chamber. The base of the 19S RP regulates the function of the 20S CP by stimulating its protease activities, recognizing substrates, unfolding substrates in an ATP-dependent manner, and translocating substrates to the 20S CP. The lid performs more complex and diverse functions during the process of protein degradation, involving ubiquitin receptors and deubiquitinating enzymes. Degradation of ubiquitinated proteins requires the lid, but the precise functions of many lid subunits are less clear than those of the subunits of the base or the core particle [21]. Many of the lid components are also involved in non-proteolytic activities. Recently, more 19S subunits have been identified, mostly associated with the lid [29, 30]. Corresponding to the functions of each subcomplex, the 20S CP is structurally the most stable complex, while the lid is the most dynamic and least stable one. Thus, it is understandable that there is more structural information about the 20S CP than about the lid of 19S RP.
2 The 20S CP
2.1 The 20S protease core
The 20S CP can be found either isolated or associated with the 19S RP [31]. Unlike the entire 26S proteasome, the 20S CP performs a relative simple protease function and is biochemically very stable. It is thus relative easy to purify a large amount of 20S CP to high purity and concentration for crystallization. Atomic structures of archaeal [32, 33], mycobacterial [34], yeast [35] and mammalian [36] 20S CP have been determined by X-ray crystallography. These structures share the same architecture: a hollow barrel-shaped structure with C2 symmetry composed of four stacked rings: two inner β rings and two outer α rings. The eukaryotic α and β rings are each composed of seven distinct homologous subunits, which form a pseudo 7-fold symmetrical structure of α1–7β1–7β1–7α1–7, with proteolytic active sites located at the N-termini of three subunits, β1, β2 and β5, of each β-ring. In contrast, the archaeal α and β rings are each composed of seven identical subunits, thus the archaeal 20S CP has true D7-symmetry. In jawed vertebrates, there is another form of proteasome, called immunoproteasome, in which three β-subunits of the normal 20S, β1, β2, β5 are replaced by three IFN-γ induced β-subunits, β1i, β2i, β5i. Immunoproteasome is responsible for breaking down foreign proteins into short antigenic peptides, which are ligands of MHC class I molecules [37]. The structure of the immunoproteasome should be very similar as the mammalian constitutive 20S proteasome [36].
The four rings of the 20S CP form three continuous chambers with proteolytic active sites sequestered in the middle chamber. There is a narrow channel in the center of the outer α-rings that only allows the passage of unfolded polypeptides to access the inner chamber. The N-terminal tails of the α-subunits form a gate, whose closed or open states prevent or allow the passages of substrates through the channel [38, 39]. The gate of free 20S CP without 19S RP is normally closed under the physiological conditions [35, 38]. Such architecture of the 20S CP prevents unregulated protein degradation [40]. Opening of the gate is regulated by the 19S RP, although it is also possible to disturb this closed gate biochemically in vitro. In the 26S proteasome, the ATPases in the base of the 19S RP regulate the opening of this gate and stimulate protease activity of the 20S CP [40]. In addition to the proteasomal ATPases, two other non-ATPase activators, 11S (PA28/REG/PA26) [41] and PA200 [42], also form complex with the 20S CP and stimulate its peptidase activity.
2.2 Assembly process of the 20S proteasome
Because of the relative simplicity of the 20S CP in both structure and function, the assembly process of this complex is also structurally well characterized [43, 44]. For the archaeal 20S CP, the assembly process is relative simple and does not require any chaperon protein. Co-expression of the α- and β- subunits in Escherichia coli produces intact and active 20S CP. Expression of only the α-subunit in E. coli without the β-subunit produces complete single heptameric α-rings that are almost identical to the ones in the 20S CP. On the other hand, expression of β-subunit in E. coli without α-subunit produces only monomeric β-subunits. This suggested that the α-ring is assembled first, and is used as a template for β-subunits to assemble upon the completed α-ring to form one half of the 20S CP [45]. The β-subunit is encoded as a precursor with a propeptide of eight residues that covers its N-terminal Thr residue. This propeptide is cleaved during the assembly of the 20S CP from the half proteasome [6].
The assembly processing of the eukaryotic 20S CP is more complicated. The α-ring is still assembled first and acts as a template for β-ring assembly. However, the α-ring has seven distinct α-subunits, each occupying a specific position in the ring. Such specific arrangement of α-subunits requires a mechanism to correctly place each α-subunit in a specific position in the α-ring. Two different dimeric protein complexes were identified as chaperones of this process in human cells: PAC1/PAC2 and PAC3/PAC4, where PAC stands for proteasome assembly chaperone. With the aid of the crystal structures of these dimeric chaperones, isolated as well as in complex with α-subunits [43, 44], the assembly mechanism of the α-ring from its constituents has been clearly revealed. A recent review by Murata et al describes in detail the assembly process of the 20S CP as well as that of the entire 26S proteasome [25].
3. 19S RP – the ATPase subunits
The 19S RP plays comparatively more complex roles in the ubiquitin-proteasome pathway than the 20S CP. Consequently its structure is more complicated and dynamic. It is also less stable biochemically and contains many more different subunits than the 20S CP. Although the intact endogenous 26S proteasome can be purified from animal tissues or yeast, so far neither the entire 26S proteasome nor the 19S RP have been crystallized. The low-resolution shape of the 19S RP is available only from single particle cryoEM data of the 26S proteasome. Currently, the resolution is less than 10 Å [28, 46]. Based on these 3D reconstructions, and even just from two-dimensional (2D) averages of images of the 26S particles, it is possible to infer that the shape of the 19S RP has a wide opening on one side [20, 21, 47], a feature often referred to as a “Chinese dragon head motif” [48].
The 19S RP is often divided into base and lid subcomplexes. It was first reported that the 19S RP can dissociate into base and lid subcomplexes in a Rpn10 knockout yeast strain [20]. The base isolated from this yeast mutant can still bind to the CP and activate the degradation of peptides or non-ubiquitinated proteins, whereas the lid is required for ubiquitin-dependent degradation. The six ATPase subunits, which are essential for the ATP-dependent protein degradation, are all located in the base subcomplex. A subsequent study also isolated the base and the lid separately from wild-type yeast and showed that the Rpn10 subunit resides in the lid subcomplex [49].
3.1 Components of the base subcomplex
The base subcomplex can be further divided into 6 ATPase subunits, Rpt1-Rpt6 [50, 51], and two non-ATPase subunits Rpn1, Rpn2. The Rpn10 and Rpn13 are also characterized as part of the base, although they are more likely linkers between the lid and the base. The six Rpt-subunits belong to the AAA+ family of ATPases, where AAA+ stands for ATPase Associated diverse cellular Activities. As expected from members of this family, the six ATPases assemble into a hexameric ring-shaped structure [28, 46, 52]. Together with other non-ATPases subunits, the base subcomplex recognizes ubiquitinated protein substrates, unfolds these globular substrates in an ATP dependent manner, opens the gate of the 20S CP, and translocates unfolded substrates to the 20S CP for degradation. However, the structure and the specific arrangement of the six Rpt-subunits in the hexameric ring-shaped base subcomplex are still not fully understood. Among all the functions performed by the ATPases, only the mechanism of gate opening has been extensively studied from other model systems [41, 53–56]. Our understanding of the mechanisms by which the ATPases in the base subcomplex unfold and translocate substrates comes only from speculative comparisons with other ATP-dependent protease systems in bacteria, which have also been described in detail [57–59].
Even less is known about the tertiary or quaternary structures of Rpn-subunits in the base. As with many other Rpn-subunits in the lid, none of the four Rpn-subunits, including Rpn10, have been localized definitely to the base ring. At the level of the tertiary structure, homology models have been proposed for Rpn1 and Rpn2 [60]. Recent studies using AFM and negative-stain EM produced low-resolution information of Rpn1 and Rpn2 consistent with a prediction that they have an overall toriodal shape [61, 62]. As for the Rpn10 and Rpn13, only domain structures were determined (discussed below).
3.2 The archaeal 20S-PAN as a simpler model system
The complexity of the 26S proteasome, as well as the ubiquitin requirement, makes the ubiquitin-proteasome a difficult system to study or to manipulate in vitro. In contrast, archaea does not have ubiquitin and has a simpler proteasome system, in which the 20S CP is formed by homo-heptameric α- and β-rings. The archaeal 20S CP also has a closed gate, although not as tight as in the eukaryotic 20S CP [63]. As with the eukaryotic ATPases in the base subcomplex, an ATPases homologue named PAN (Proteasomal Activating Nucleotidase) stimulates the protease activity of the archaeal 20S CP [64]. The PAN shares 41%–45% identity with eukaryotic Rpt subunits. Six identical PAN monomers form a homo-hexamer [65] that binds to the archaeal 20S CP in a way that resembles the interaction between the base subcomplex and the eukaryotic 20S CP. The location of the PAN on the archaeal 20S CP is comparable to that of the 19S ATPase ring in the 26S proteasome [63]. Thus, the archaeal 20S-PAN serves as a simpler in vitro model for the study of many mechanistic questions, particularly related to those proteasomal ATPases of the 26S proteasome.
Although somewhat artificial, the most commonly used archaeal model system is actually a hybrid system, composed of the 20S CP from Thermoplasma acidophilum and the PAN from Mathanococcus jannaschii [64]. Since its identification, a wealth of knowledge, covering many functional and structural aspects of the proteasomal ATPases, has been built upon this model system. When mixed with T. acidophilum 20S CP and ATP, PAN from M. jannaschii stimulates the degradation of unstructured as well as globular protein substrates carrying a C-terminal ssrA recognition tag [64, 66]. Careful dissection of this process ascribed ATP binding to several events, namely binding to 20S, translocation of substrates, and gate opening of 20S [63]. The substrate unfolding is the only part that requires ATP hydrolysis by exerting mechanical force during domain movements [56, 67].
However, structurally, the PAN does not form a stable complex with the 20S CP. Rather, it waggles on the top of the α-ring of the 20S CP [63]. The mismatch between the 7-fold symmetry of the 20S CP and the 6-fold symmetry of the PAN is thought to be the cause of such unstable structure. Thus, despite its simplicity, the 20S-PAN complexes still resisted to be crystallized for structure determination.
3.3 ATPase induced gate opening in the 20S CP
To understand the mechanism of gate opening by the proteasomal ATPases, or more generally, by proteasomal activators, much efforts has been on determining the structures of non-ATPases proteasomal activators, such as the PA26 from T. brucei, a homolog of mammalian 11S proteasomal activator, and Blm10 from yeast, a homolog of mammalian PA200 activator. As a result, our understanding of the mechanisms by which proteasomal activators regulate the gate opening in the 20S CP has be improved considerably [41, 53,42] 54,. PA26 forms a homo-heptamer. It not only matches the 7-fold symmetry of the archaeal 20S CP, but also fits into the pseudo 7-fold symmetrical eukaryotic 20S CP, both resulting in opening of the 20S gate formed by N-termini of α-subunits. Two structural elements of the PA26 play different roles but operate synergistically. The C-terminus inserts itself into a pocket in the 20S CP between neighboring α-subunits, providing binding energy for the 20S-PA26 complex formation, while an activation loop of the PA26 makes contact with a reverse-turn loop of the 20S CP near the N-terminus of the α-subunit, stabilizing a four-residue cluster and locking the N-terminus in the open conformation [41, 53, 54]. The mammalian 11S, which resembles the overall structure of PA26, may bind to 20S and induce the gate opening similarly as the PA26 by using activation loop and C-terminus [54].
PAN, or eukaryotic ATPases, has neither 7-fold symmetry nor the activation loop. Thus, it works slightly different from the PA26, but probably more closely resembling the eukaryotic Rpt-subunits. The PAN C-termini bind to the same pocket in the 20S CP α-ring as the PA26 does, but such binding can open the 20S gate by itself without the activation loop [56]. This unique role is strictly dependent on the sequence of the last three C-terminal residues, dubbed as HbYX motif (Hydrophobic residue, tYrosine, and an arbitrary terminal residue, X). This motif is conserved in the archaeal PAN, as well as in the three Rpt-subunits: Rpt2, Rpt3 and Rpt5, of the eukaryotic 19S base [56]. The gate opening of the 20S CP induced by the C-terminal peptide of PAN has been visualized by single particle cryoEM at a resolution of ~6Å [55]. Clear densities of the PAN C-terminal peptide were located in the pockets between neighboring 20S α-subunits, the same pockets where the C-termini of PA26 bind. Such binding leads to a radial movement of the α-subunits, resulting in the opening of the 20S gate.
Two recent studies revealed how the C-terminal HbYX motif of PAN ATPase binds to the pocket in the 20S α-ring in atomic details. These two independent studies took a somewhat similar approach of generating a hybrid proteasomal activator using PA26 as scaffolding but substitute its C-terminus with that from PAN ATPase [68, 69]. The very similar interaction was further observed in the atomic structure of Blm10, which also contains an HbYX motif in its C-terminus, in complex with yeast 20S CP [42].
3.4 Structure of the proteasomal ATPases
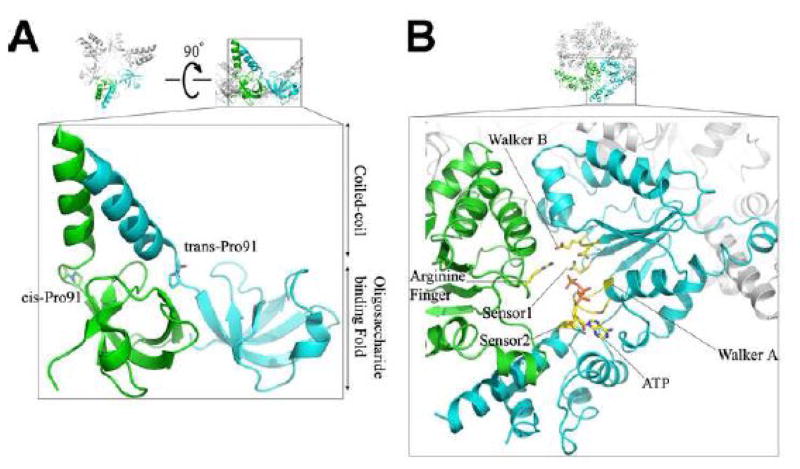
Attempts to crystallize the full-length PAN have so far yielded no success. However, the structures of individual functional domains of the PAN were determined recently [33, 70, 71]. The N-terminal substrate-binding domain of the PAN consists of a coiled-coil element, followed by the oligosaccharide-binding (OB) domain. It forms a rigid hexamer but with only a 3-fold symmetry, a trimer of dimers. The difference between two neighboring monomers in the dimer lies in a proline residue at the end of coiled-coil domain, which assumes a cis-conformation in one monomer but a trans-conformation in the other monomer [33, 70]. This proline residue appears to be critical to the formation of the coiled-coil domain between two neighboring monomers (Fig. 2A). Such conformation resembles the intra-subunit coiled-coil structure in the HslU structure [72, 73]. In both cases these coiled-coils come together to form a funnel-like shape that presumably holds protein substrate and guides the unfolded segment into the central pore. The ATPase domain of the PAN, however, forms a loose hexamer, which dissociates into monomers during purification. The monomeric crystal structure with bound ADP shows a similar architecture to that of other AAA+ ATPases [33]. A complete structure of the PAN or Rpt-subunits has been suggested by computational docking of the CC-OB ring and the ATPase domain into the ring shaped base subcomplex of the 26S proteasome [33, 74].
Figure 2.
Atomic structures of domains from archaeal PAN ATPase. (A) The CC-OB domain of the PAN assembles into a hexameric ring that has a 3-fold symmetry. The cis- and trans-proline residues in the neighboring CC-OB domain are shown in the enlarged view that shows how the coiled-coil is formed. (B) A model of a hexameric ring formed by PAN’s ATPase domain. The ATP binding site of this model is shown in the enlarged view.
Until now, we still know very little about how proteasomal ATPases unfold globular substrates and how they translocate the unfolded substrates into the 20S CP for degradation. The structure of the monomeric PAN ATPase domain provides a basis for the comparison of the proteasomal ATPases with other members of the AAA+ superfamily of proteins, which often form a hexameric ring shaped structure [75, 76]. AAA+ proteins have a structurally conserved domain [77] that consists of an N-terminal α/β Rossman fold and a C-terminal α-helical subdomain. Between these two subdomains resides the ATP binding pocket. The two subdomains work as a clamp that opens to different angles, depending on the state of the bound nucleotide (Fig. 2B). In the ATPase ring the clamp-shaped monomers fit into the concave side of their neighbors. A narrow central pore approximately 12Å in diameter is, presumably, the channel through which substrates are threaded. AAA+ ATPases have several conserved structural motifs, including the Walk-A and Walk-B ATP binding motifs. Less conserved are the so-called Second Region of Homology (SRH), including Sensor 1 on the Rossman fold, and an arginine finger on the same Rossman fold of the adjacent subunit [78, 79] (Fig. 2B).
3.5 Assembly of the Rpt-subunits into a hexameric ring
The understanding of the structure and function of PAN laid the foundations for structure study of the ATPase hexamer in the 19S RP and its interactions with the 20S CP. Rpt1-Rpt6 are six homologous, but distinct ATPases, each consisting of ~430 amino acids. From the N-terminal end, each Rpt subunit can also be divided into coiled-coil element, OB domain, ATPase domain and C-terminus. An atomic model of the ATPases was proposed by fitting domain structures generated from homology modeling into the low-resolution density maps of the base subunits segmented out from the 3D density map of the 26S proteasome [28, 74]. In the 3D reconstruction, the symmetry axes of the hexameric base and of the 20S CP are not coincident, but lie 3nm apart and are tilted 11º with respect to each other, in agreement with the previously proposed wobbling model [80].
The 19S ATPase hexamer is more complex than its PAN homolog. The six Rpt-subunits have been found to be non-redundant [51], and such non-redundancy is conserved across species [50]. Moreover, different roles of these six Rpt-subunits have been observed [51, 81]. Given these facts, a fixed arrangement of these 6 Rpt subunits seems very likely. Considerable efforts have been spent on determining the specific arrangement of Rpt subunits in the hexamer ring of the base subcomplex. Among those efforts, chemical cross-linking was used to determine the quaternary structure of the human 26S proteasome, produced a complete model of the ATPase hexamer, as well as their interactions with 20S α-subunits [52]. This model has served as a reference for many later works [81]. But as it relied solely on cross-linking, the results could be biased by possible artifacts. Other techniques such as yeast two-hybrid experiment [82], pull-down assay, filter binding, mass spectra, and co-purification have also been employed to address this question [83].
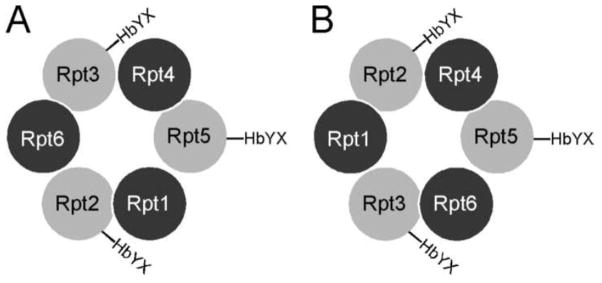
With six Rpt subunits in the base ATPase ring, there are a total of 120 (=5!) possible arrangements. Recent breakthroughs in identifying chaperones that assist the assembly of the base subcomplex have greatly enhanced our understanding. Four different laboratories have independently identified same four chaperone proteins that facilitate the formation of the Rpt intermediate subcomplexes and escort them into their destinations in the hexamer ring [84–88] (also see a review [89]). Specifically, Rpt1 and Rpt2 associated with Hsm3, Rpt4 and Rpt5 with Nas2; and Rpt3 and Rpt6 were shown to be associated with Nas6 and Rpn14 respectively. Recent data further suggested the formation of Rpt3-Rpt6 subcomplex [90]. These findings reduced the number of possible arrangements of Rpt-subunits in the hexameric ring to 16, i.e. (2 possible arrangements of 3 subcomplexes) × (2 possible directions of Rpt1-Rpt2) × (2 possible directions of Rpt4-Rpt5) × (2 possible directions of Rpt3-Rpt6). Interestingly, only one Rpt-subunit in each of these three subcomplexes, i.e. Rpt2, Rpt3, and Rpt5, contains the critical proline residue for the coiled-coil formation, as well as the C-terminal HbYX motif required for induction of the 20S gate opening. Based on the structure of archaeal PAN ATPases, we can speculate that the proline residues in these three Rpt-subunits must assume the cis-conformation, because non-proline residues in corresponding positions of the neighboring Rpt-subunits are most likely in the trans-conformation. Furthermore, these three Rpt-subunits must be separated from each other in the hexamer ring and each of them located on the left side of the subcomplex, so that it can form a similar coiled-coil structure with its partner Rpt-subunit on the right side (Fig. 2A). Considering all these assumptions, there are only two possible arrangements of Rpt-subunits in the ring: Rpt5-Rpt4-Rpt2-Rpt1-Rpt3-Rpt6 or Rpt5-Rpt4-Rpt3-Rpt6-Rpt2-Rpt1 (Fig. 3, counter-clockwise and look down from the top of the hexameric ring). A recent model, generated by combining 3D density map from cryoEM data of the 26S proteasome, homology modeling and proteomic-derived protein-protein interactions, exhibits one of the two possible orders listed above [74]. Certainly, the definitive answer to this question can only be provided by means of high-resolution structure information from the 26S proteasome or the base subcomplex of the 19S RP.
Figure 3.
Two possible arrangements of Rpt-subunits in the base subcomplex. They are viewed from the top of the hexameric ring of ATPases sits on top of the 20S CP. Rpt2, Rpt3 and Rpt5 have the conserved C-terminal HbYX motif that is required to induce gate opening in the 20S CP. They also have the cis-proline that is required to form the coiled-coil interaction with its adjacent subunit on the right side.
After submitting this review, a recent study determined the order of the Rpt subunits elegantly [91]. In this study, cysteine residues were systematically introduced to specific locations of the Rpt subunits so that the correctly located cysteine mutations can lead to the disulfide bond formation between the neighboring subunits. The final correct order of Rpt subunits determined by this approach is Rpt5-Rpt4-Rpt3-Rpt6-Rpt2-Rpt1 (Fig 3A).
4. 19S RP – non-ATPase subunits
Most of the subunits in the 19S RP are non-ATPases. They perform very diverse functions that are critical in the process of degradation of ubiquitinated substrates. Many of them are also involved in other non-proteolytic cellular processes. Ubiquitin can be linked together through isopeptide bond between its carboxyl group and the ε-amino group of a lysine residue of either another ubiquitin protein (to form polyubiquitin chain) or a target protein (ubiquitination) [2]. Among the seven lysine residues of the ubiquitin - K6, K11, K27, K29, K33, K48, and K63, polyubiquitination through the K48 and K63 are most abundant. They form well-characterized polyubiquitin chains that act as signals for either degradation by the proteasome, DNA damage tolerance, protein trafficking or ribosomal translation, etc. [92, 93]. Although there are many other atypical polyubiquitin chains in vivo [92], their functions and structures have yet to be elucidated [94].
During the process of protein degradation by the proteasome, polyubiquitinated protein substrates are recognized by the proteasomal ubiquitin receptors. Before the substrates are completely unfolded by proteasomal ATPases and degraded by the 20S CP, proteasomal deubiquitinating enzymes remove the polyubiquitin chain from the labeled substrates in a highly regulated manner. The exact locations of many of the ubiquitin receptors and deubiquitinating enzymes are not clear, but they may be closely associated with each other to perform a series of highly coordinated events, i.e. substrate recognition, removal of ubiquitin chain and unfolding of globular substrates.
4.1. Ubiquitin receptor – Rpn10/S5a and Rpn13/Adrm1
Although ubiquitin can act as a signaling molecule in a number of non-proteolytic cellular processes [95], its major role is to facilitate delivery of targeted substrates to the 26S proteasome. Polyubiquitin chains are recognized by the ubiquitin-binding domain (UBD), which is a structural motif found in a wide variety of cellular proteins. UBD has more than 20 subfamilies and very diverse structural features, such as α-helical structure, zinc fingers (ZnF), Plekstrin homology (PH) folds, or an ubiquitin-conjugating-like domain (UBC) [23]. For protein degradation by the 26S proteasome, shuttle factors such as Rad23/hHR23, Dsk2/PLIC or Ddi1, capture the ubiquitinated target protein with their UBD domain and transfer it to the ubiquitin receptors, Rpn10/S5a and Rpn13/Adrm1, in the 19S RP [24].
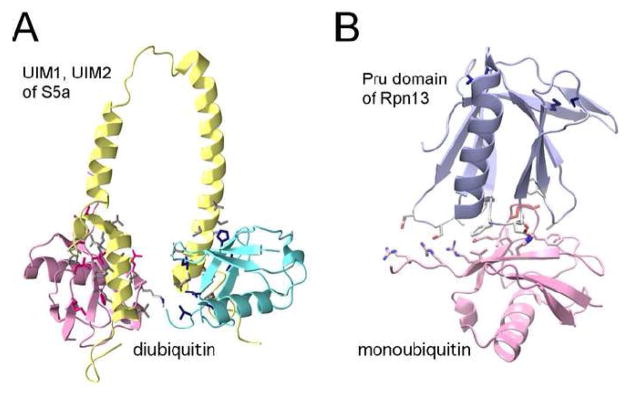
S5a was the first identified ubiquitin receptor in the 19S RP [96, 97]. It has two polyubiquitin binding site, PUbS1 and PUbS2, also called ubiquitin interacting motifs, UIM1 and UIM2. Rpn10, the Saccharomyces cerevisiae homologue of the mammalian S5a, contains UIM1 only. S5a can bind K48-linked as well as K63-linked polyubiquitin chains and has higher affinity for polyubiquitinated protein than for mono- or di-ubiquitinated proteins [96]. The S5a has three α-helices connected by flexible linkers, with first helix as UIM1 and second and third α-helices as UIM2. Such architecture allows them to adopt diverse conformations for the cooperative recognition of different ubiquitin linkages and long polyubiquitin chains [98] (Fig. 4A). S5a is a monomer in solution, two UIMs do not interact with each other but with UIM2 have higher affinity for polyubiquitin than UIM1 [98]. It was recently proposed that the two UIMs each bind simultaneously to an ubiquitin molecule of a diubiquitin chain, while the UIM2 binds predominantly to the proximal ubiquitin the UIM1 binds predominately to the distal ubiquitin (Fig. 4A) [99]. S5a has a von Willebrand factor A (vWFA) at the N-terminal region of UIM1 domain, which is also present in the Rpn10 of S. cerevisiae. Rpn10 is known as a linker between Rpn12 of lid and Rpn1 of base [20, 100]. As the structure of the complex of vWFA1 with glycoprotein Iba suggests, it is plausible that the vWFA domain of S5a is sandwiched by the concave regions of two neighboring repeating units of Rpn12 (N-term HAM domain of PCI domain, lid part, discussed below) and Rpn1 (HEAT/LRR like domain, base part) [101].
Figure 4.
Structures of ubiquitin binding domains. (A) The NMR structure of S5a UIM domain and diubiquitin (pdb code: 2KDE). Pink is the proximal ubiquitin interacting with the UIM2, and cyan is the distal ubiquitin that interacts with the UIM1. Magenta, blue and gray side chains on the interaction surface belong to proximal ubiquitin, distal ubiquitin and UIM, respectively. (B) Complex structure of Rpn13 with monoubiquitin (pdb code: 2Z59). Red side chain is the ubiquitin K48 residue. Blue side chains on the opposite side of ubiquitin represent the residues interacting with the Rpn2.
Rpn13 was identified as another novel ubiquitin receptor, from a yeast two-hybrid screening using a modified ubiquitin as bait [102, 103]. This protein was first reported as a surface glycoprotein GP110 whose expression is induced by IFN-γ [104] and involved in cell adhesion [105]. Recent work showed that Rpn13/Adrm1 interacts with Rpn2 through its 140 residue long N-terminal domain and recruits the deubiquitinating enzyme, UCH37, to the 26S proteasome through its C-terminus KEKE motif [106–108]. Crystal and NMR structures showed that the UBD of Rpn13 have a structure similar to a pleckstrin-homology domain (PHD), so this domain has been named as pleckstrin-like receptor for ubiquitin, or Pru domain. In this domain, two β-sheets orthogonally packed against each other and a long C terminal α-helix, which is flexible in the solution NMR structure, are located in the crevice of a β-sandwich (Fig. 4B) [102]. Sequence analysis reveals that this ubiquitin recognition domain, Pru domain of Rpn13, is highly conserved in multicellular organisms from S. cerevisiae to human and mouse, although the C-terminus of Rpn13 is less conserved and the Rpn13 of S. cerevisiae contains only the N-terminal Pru domain. On one side of the Pru domain, three loops, S2–S3, S4–S5 and S6–S7, bind ubiquitin, whereas the opposite side of the Pru domain interacts with the Rpn2 (Fig. 4B). The contact surface between the Pru domain and the bound ubiquitin is approximately 1300 Å2, which contributes to a tight binding of Rpn13 to the ubiquitin molecule [103]. For the K48-linked diubiquitin, Rpn13 predominately binds to the proximal ubiquitin, while two UIMs of S5a compete for one distal ubiquitin [99]. However, it is not very clear how three UBDs, one Pru domain of Rpn13 and two UIM domain of S5a, cooperatively recognize the polyubiquitin chain in the 26S proteasome.
Aside from these two ubiquitin receptors, Rpn10/S5a and Rpn13/Adrm1, one of the Rpt subunits in the base subcomplex, Rpt5/S6a, was identified as another polyubiquitin receptor in an ATP hydrolysis dependent manner [109]. Although the recombinant GST-Rpt5 alone does not directly bind to a tetra-ubiquitin, such interaction may be cooperatively modulated by another ubiquitin receptor in the 19S RP. Because the C-terminal HbYX motif of Rpt5 is thought to be critical for the axial gate opening of 20S CP [56], it is possible that the interaction of Rpt5 with ubiquitin could convey the signal from lid to 20S CP.
4.2. Deubiquitinating enzymes (DUBs) - UCH37, Rpn11, UBP6
Before a substrate is completely unfolded and degraded, the polyubiquitin chain has to be removed from the substrate. The polyubiquitin chain should also be further broken down into monoubiquitins so that ubiquitin molecules can be recycled. The process of deubiquitination and protein unfolding is highly cooperative and regulated, and it controls very precisely the rate of ubiquitin-dependent proteasomal degradation [110]. The process of deubiquitination en bloc as well as break down of the polyubiquitin chain into monoubiquitins is carried out by a variety of deubiquitinating enzymes (DUBs), which are mostly located in the lid of the 19S RP. During the process of deubiquitination, the isopeptide bonds of polyubiquitin are specifically cleaved by DUBs. Based on their sequence similarity and enzymatic property, DUBs can be classified into five different subfamilies: ubiquitin-specific processing protease (UBP), ubiquitin carboxy-terminal hydrolase (UCH), Ovarian tumor related protease (OUT), JAM/MPN/Mov34 metalloprotease (JAMM), and Josephin domain (MJD) protease [111]. Among 95 putative DUBs of human genome, three DUBs - UCH37 (UCH2), Rpn11 and USP14 (Ubp6) are associated or interact with 19S RP for the deubiquitination of substrates [111–118]. Rpn11 and UCH37 are intrinsic subunits of the 26S proteasome (Fig. 1) and UBP6 reversibly interact with the Rpn1 of the 19S base subunit via an ubiquitin-like domain at its N-terminal [115].
4.2.1. UCH37 (UCH-L5)/UCH2 (Human/S. pombe nomenclature)
Ubiquitin Carboxyl-terminal Hydrolase (UCH) is a cysteine protease that normally cleaves the short C-terminal extension of ubiquitin into short peptide. UCH37 (also called UCH-L5) in the 26S proteasome is a special member of UCH family that can also trim ubiquitins off from the distal end of ubiquitin chain [113, 114]. Although S. cerevisiae does not have the orthologues of UCH37, its orthologues are found in all the eukaryotes, from S. pombe to human [108, 119, 120].
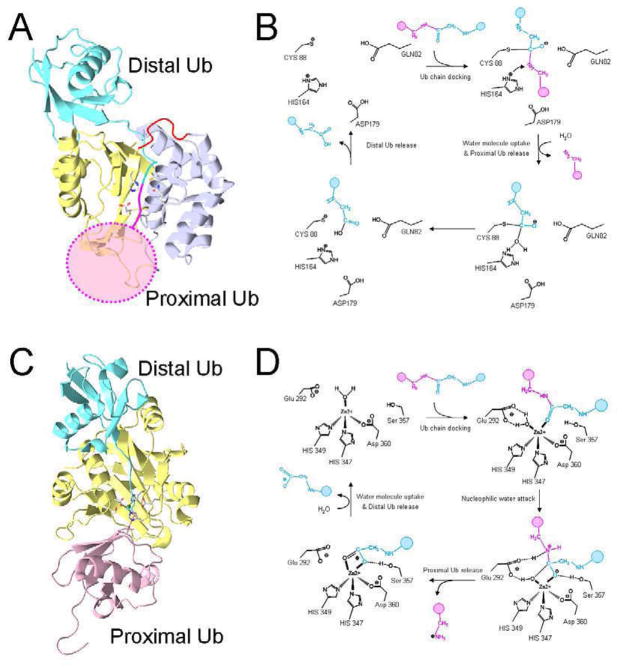
The crystal structure of the UCH37 has been recently reported [121]. As in the structures of other UCH family member [122], there is an active site in the cleft between two lobes of UCH37, which is composed of the conserved residues Gln82, Cys88, His164 and Asp179 (Fig. 5A). Being a typical cysteine protease, its scissile mechanism of breaking polyubiquitin chain can be predicted as described in Figure 5B.
Figure 5.
The catalytic mechanisms of deubiquitinating enzymes. (A) A structure model of UCH37 in complex with a diubiquitin. Two lobes of the UCH37 (pdb code: 3IHR) are colored in yellow and light blue, side chains of the catalytic residues are shown. The loop that links the two lobes is disordered, and its speculated structure is drawn in red. The distal ubiquitin is shown in cyan, which is derived by superimposing UCH37 with the complex structure of UCH-L3 and UbVME (pdb code: 1XD3). The expected position of proximal ubiquitin is drawn as a pink disk. (B) The catalytic mechanism of UCH37 predicted from similar cysteine protease: Three residues Cys88, His164 and Asp179 form a catalytic triad in which the His164 activates the thiol group of Cys88 to a nucleophile. While the main chain amide of Cys88 and Gln82 form an oxyanion hole, the nucleophilic attack to the carbonyl carbon of the ubiquitin cleaves off the proximal ubiquitin. (C) Crystal structure of the AMSH-LP and K64-ubiquitin complex (pdb code: 2ZNV). In this atomic structure Glu292 was mutated to alanine and the zinc-binding site was not occupied. The side chain of the conserved catalytic residue and zinc molecule are taken from another crystal structure of apo AMSH-LP (pdb code: 2ZNR). (D) The catalytic mechanism of AMSH-LP: A water molecule is polarized by Glu292 and Zn2+ in the catalytic site. It acts as a nucleophile to attack the carbonyl carbon of Gly76 in the distal ubiquitin. The proton from the 292Glu is transferred to the nitrogen of K49 ε-amine. The tetrahedral intermediate is then generated, where zinc is penta-coordinated, while Ser357 involves the stabilization of this intermediate by the hydrogen bond with carbonyl oxygen. Finally, the isopeptide bond is cleaved and the proximal ubiquitin is released from the distal ubiquitin [130].
The structure of the complex of any UCH family of proteins with its physiological substrate is still unknown. However, the structure of YUH1 with a potent inhibitor of UCH family, Ubal, revealed the interaction between a ubiquitin and a member of the UCH family of proteins, and showed the conformational changes of UCH upon ubiquitin binding. Ubal sits on the top of Yuh1, which corresponds to the position of a distal ubiquitin in the polyubiquitin chains. The interface between Ubal and Yuh1, which consists of numerous hydrogen bonds and van der Waals interactions, is very extensive, approximately 2500 Å2. The C-terminal end of the bound ubiquitin, which could link to the K49 of the proximal ubiquitin in the polyubiquitin chain, extends along the active site cleft [123]. The structure of UCH-L3 with ubiquitin-vinylmethylester (UbVME) also shows a similar conformation [124]. Compared with the structures of other UCH family members [122–125], UCH37 has a wider proximal ubiquitin-binding site, so that the proximal ubiquitin can easily access and fit its K48 residue into the cleft of active site. The positions of a distal ubiquitin and a hypothetic proximal ubiquitin suggest that, in order to thread the isopeptide bond into the active site, the distal ubiquitin has to pass through the flexible loop (red in Fig. 5A), which is not long enough to enable such threading. Considering the fact that UCH37 can breakdown diubiquitin into monoubiquitin only in the 26S proteasome, an unknown conformational change of the UCH37 must occur, possibly assisted by some of the other 26S subunits [121].
UCH37 bind tightly to the 26S proteasome through Rpn13. The C-terminal domain of UCH37 binds to C-terminus of the Rpn13/Adrm1 through the KEKE motif, which is known for mediating protein-protein interactions [106, 126], while the N-terminal domain of Rpn13 is associated with Rpn2 of the base subcomplex [108]. Biochemical data from fission yeast and mammalian cells suggested that the UCH37 also binds to the S5a, a mammalian homologue of Rpn10 [107, 116]. Analysis of EM images of negatively stained 26S proteasome of Drosophila melanogaster, containing gold-labeled Ubal specific for UCH37, revealed that UCH37 is located at the interface between base and lid complex [119]. Considering that Rpn2 is part of the base subunit and Rpn10 is known as a linker between lid and base [20], Rpn10, Rpn13, Uch37 may cluster around the linker between the base and the lid, and they may also regulate the rate of protein degradation.
4.2.2. Rpn11
Rpn11 belongs to the JAMM domain family. Unlike other two DUBs - UCH37 and UBP6, which are cystein proteases, Rpn11 is a metalloprotease that contains a conserved metal binding site, JAMM/MPN+ motif (EXnHS/THX7SXXD) [127, 128]. The loss of viability through knockdown of the Rpn11 gene, or mutation of its active site [117, 118, 129] suggests that this is a crucial deubiquitinating enzyme of the 26S proteasome.
The atomic structure of the Rpn11 has not been determined yet. But the crystal structure of another JAMM family DUB, AMSH-LP (Associated Molecule with SH3 domain of STAM), with a K63-linked diubiquitin was determined [130]. The mechanism of isopeptide bond cleavage of AMSH-LP is similar to the mechanism of peptide bond cleavage of other metalloproteases, such as thermolysin and angiotensin-converting enzyme (ACE) [131, 132]. It has a water molecule, His347, His349 and Asp360 residue to coordinate Zn2+ molecule in the catalytic site (Fig. 5C). The process of isopeptide bond cleavage is described in Figure 5D.
Sequence alignment of AMSH-LP and Rpn11 shows that all the residues near the active site and in the distal ubiquitin-binding site are well conserved between the two. However, the residues that are crucial for the proximal ubiquitin binding are very different. This suggests that the isopeptide bond scissile mechanism of Rpn11 is very similar to that of the AMSH-LP, but the mechanism of recognizing and binding K48-ubiquitin chain, which adopts a compact and globular conformation at neutral pH [133], to the Rpn11 could be different from that of binding K63-ubiquitin, which has an extended and linear conformation, to AMSH-LP [134]. For better understanding of deubiquitination by the Rpn11 in the 26S proteasome, the atomic structure of Rpn11 with K48 ubiquitin chain is necessary.
4.2.3. USP14/UBP6 (Human/yeast nomenclature)
USP14/UBP6, ubiquitin-specific processing protease, are also cystein proteases. In contrast to members of the UCH family which are usually small (~20–30kDa) and preferentially cleave small peptides or amino acids from the C-terminus of ubiquitin, larger UBP (60~300kDa) family members specifically cleave the ubiquitin off the substrates from the distal end of K48-linked polyubiquitin chain [110, 135]. UBP6 has an N-terminal ubiquitin-like (UBL) domain and C-terminal catalytic domain. Through this UBL domain, UBP6 reversibly associate with the Rpn1 subunit of 19S base subcomplex. The deubiquitination activity of UBP6 is dramatically enhanced upon association with the 26S proteasome [115]. The crystal structure of the core domain of human USP14, USP14 with Ubal and S. cerevisiae UBP6 (pdb code: 1VJV) show that the core domain of USP14 looks like an extended right hand with three domains: Fingers, Palm and Thumb [135]. USP14 and UBP6 have very similar structures with a backbone Cα r.m.s deviation of only 1.2Å. In contrast to the deformed active site conformation of free HAUSP, which is another member of UBP, the conserved catalytic residues, Ans109, Cys114, His435, Asp451 of USP14, are well positioned in the active site before ubiquitin binding [135]. The catalytic groove between palm and thumb domain, which is blocked by two surface loops, BL1 and BL2 in the ubiquitin-free form, is opened in the ubiquitin-bound structure. The conformational changes of BL1, BL2 induced by the interaction with some other 26S subunits upon UBP6 binding, may relieve the catalytic groove and enhance the deubiquitination activity. Unlike two other DUB - UCH37 and Rpn11, UBP6 is not an intrinsic subunit of the 26S proteasome. Therefore, it could have some other functions not directly related to protein degradation, for example, remodeling ubiquitin chain for the delay of the protein degradation [136].
4.2.4. Cooperation of DUB in the 26S proteasome
Tetra ubiquitin is the minimal signal for the delivery of the ubiquitinated target protein to the 26S proteasome for the degradation [137]. After binding of ubiquitinated protein to the 26S proteasome, the rate of substrate degradation is tightly regulated and this rate is closely related to the rate of deubiquitination and to the length of polyubiquitin chains. Ubiquitin chains can be remodeled at the 26S proteasome by ubiquitin ligase, Hul5, and deubiquitination enzyme, UBP6. Both enzymes are reversibly associated with the 26S proteasome. But they have opposite functions: one is to add ubiquitin and the other is to remove it [136, 138]. If all the ubiquitin molecules are released before the start of the unfolding or translocation of substrates, the affinity of substrate to the 26S proteasome will be reduced and the substrates will easily dissociate from 26S proteasome. If any ubiquitin is still attached to the substrate during unfolding and translocation process, substrate cannot pass the narrow axial channel of 20S CP for the degradation. Therefore, communications between the unfolding/translocation and the final deubiquitination from the substrate are crucial for the degradation of the substrates. Furthermore, the cleavage of the proximal ubiquitin chain by Rpn11 depends on the ATP hydrolysis of the Rpt-subunits [24, 139] and can release the polyubiquitin chain en bloc. This suggests that the Rpn11 might carry out the final removal of the polyubiquitin chain from the substrate right after the initiation of ATP hydrolysis by the ATPase of the base subcomplex.
4.3. PCI domain proteins –Rpn3, Rpn5, Rpn6, Rpn7, Rpn9, Rpn12
PCI domain was identified in 1998 by using a generalized profile method for searching the sequence motif. It commonly exists in the lid subunits of 19S RP, COP9 signalosome and eukaryotic translation initiation factor-3 (eIF3) [140, 141]. Among subunits in the lid of the 19S RP, subunits Rpn3, Rpn5, Rpn6, Rpn7, Rpn9 and Rpn12 each have a PCI domain [27]. The PCI domain serves as a scaffold for the protein-protein interaction in proteasome, signalosome and eIF3 [100]. The biochemical and genetic study of the Rpn subunits with PCI domain mutant suggested that these Rpn subunits play a crucial role in regulating proteasome assembly and/or maintaining the structural integrity [142–146].
The main role of lid in 19S RP is to recognize ubiquitinated protein substrates and to release the ubiquitin molecules from the substrates. Because there is no enzymatic activity in these subunits, except Rpn11, it is difficult to infer other functions of the lid complex. With very different cellular functions, why do proteasome, COP9 signalosome and elF3 [147–149] all have this PCI motif in their subunits? Proteasome lid and COP9 signalosome have a one-to-one subunit correspondent architecture called paralogue, not only six PCI domain-subunits but also two MPN domain-subunits, Rpn8 and Rpn11. Although the studies about physical and genetic interactions between these three macrocomplexes have been started, the functional significance of interactions and the molecular mechanisms of regulation are still poorly understood [150, 151]. Recently, based on results of micro-array profiling and several yeast genetics with Sem1 mutant [152, 153], a new function of Sem1 was proposed, namely, it serves as a dynamic multitasking organizer in the three macromolecular complexes – 26S proteasome, signalosome, eIF3 [150–153].
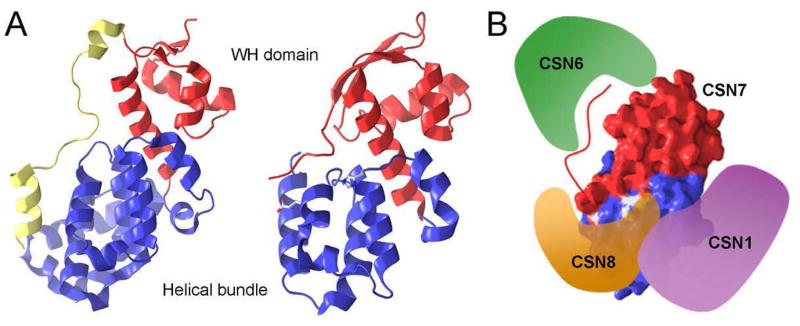
The PCI domains subunits of 19S lid subcomplex remain structurally poorly understood. The bioinformatics analysis, such as sequence alignment and secondary structure prediction, and the crystal structure of human eIF3k [154] suggested that the PCI domain consists of a conserved C-terminal part, a WH-domain (winged helix like domain) with a globular αβ structure and a less conserved N-terminal part, which is a TPR like repeat that has three anti-parallel helical hairpin structure (Fig. 6A) [155]. Based on the upstream region of the WH-domain, PCI family can be further subdivided into three subclasses, typical PCI and atypical PCI that includes Sac3 subtype and Rpn12 subtype [150]. Typical PCI have additional bihelical repeats followed by PCI. The Rpn12 subtype lacks the N-terminal helical extension. The Sac3 subtype has a conserved non-helical domain at the N-terminal of PCI domain. Recently, the crystal structure of a member of the PCI family protein has been determined. It is CSN7 in signalosome, and is the paralogue of Rpn9 (Fig. 6A) [156]. Like the eIF3k structure, the CSN7 PCI core (1~169a.a) identified by the limited proteolysis consists of two subdomains: an N-terminal helical bundle and a C-terminal WH-domain. The PCI core of CSN7 interacts with other CSN subunits, CSN1, CNS8 through its PCI domain. The C-terminal tail of CSN7, which is outside of its PCI domain, binds to the CSN6. This C-terminal tail is disordered in the crystal structure and is drawn hypothetically in the Figure 6B.
Figure 6.
Crystal structures of the PCI domains. (A) The eIF3 (left, pdb code: 1RZ4) and CSN7 (right, pdb code: 3CHM) have a similar architecture. Blue and red ribbons represent N-terminal helix bundle and C-terminal WH domain. Glu44 and His71, shown as stick in the N-terminal helical bundle of CNS7, are critical for the binding of CSN8. (B) Partial interactions between CSN subunits.
The crystal structure of CSN7, as well as biochemical interaction experiments with another signalosome subunits, provided a glimpse into the arrangement of the lid subcomplex. Results from the yeast two-hybrid screen with the lid protein from diverse organisms [82, 100, 157], mass spectra analysis and cross-linking experiments suggested an interaction map of lid complex [83]. There are two asymmetric clusters in lid subcomplex. One is a pentameric complex formed by Rpn5, Rpn6, Rpn8, Rpn9, Rpn11 in which Rpn8 is centered. The other is tetrameric complex formed by Rpn3, Rpn12, Rpn7, Rpn15 (Sem1). These two clusters are linked together by Rpn3 and Rpn5.
Negative stain EM has shown that the lid subcomplex has a non-symmetrical arrangement and central groove [158]. More detail structural organization of lid subunits have to be deduced from higher resolution structural analysis by single particle cryoEM, although the conformational and compositional heterogeneity of lid complex make this task difficult.
5. Other non-proteolytic functions of proteasome
The primary function of the 26S proteasome is for protein degradation in the ubiquitin-proteasome pathway, with the ubiquitin as the signal of substrates. However, recent emerging evidences suggest the existence of many other non-proteolytic functions of both the proteasome and ubiquitin. Such nonproteolytic functions include DNA repair, transcription initiation and transcription elongation [159].
One of such examples is that besides the proteolytic role [95], 26S proteasome also plays non-proteolytic role in regulating gene expression, particularly in pre-initiation and transcriptional elongation step [160]. It has been proposed that the 19S RP facilitates the interaction of SAGA complex (Spt-Ada-Gcn5-Acetyltransferase) with transcriptional activator on GAL1-GAL10 promoter and leads to up-regulation of gene expression level [161]. Another example is the chaperone-like activity of the 19S RP for the efficient transcriptional elongation by remodeling and stabilizing the stalled polymerase complex, RNA polymerase II and FACT elongation complex, which physically interact with proteasome [162]. Moreover, the proteasome is also involved in the transcription-coupled nucleotide excision repair, which is mediated by the repair protein Rad23 and DSS1 [163].
Ubiquitin has even more diverse non-proteolytic functions, such as membrane trafficking, protein kinase activation, DNA repair, and chromatin remodeling [95]. The relationship of nonproteolytic functions between proteasome and ubiquitin is not clear. One of the speculations is that ubiquitinated protein could recruit not only diverse UBD protein but also the 19S RP to associate with other complexes to execute yet unidentified biological functions.
6. Summary
Tremendous progress towards the structure characterization of the 26S proteasome has been made over the past twenty years. Because of its relatively simple structure and protease function, the 20S CP has been very well characterized in atomic details. In recent years, our structural understandings of the 19S RP or the 26S proteasome machinery have also made fast progress, mostly in the determinations of various subunits or associated chaperone proteins required for the assembly of the 26S proteasome. The resolution of the single particle cryoEM 3D reconstructions of the 26S proteasome has also been improved significantly. Together with the achievements from biochemical and bioinformatic studies, we have now a better understanding of the possible ways in which the subunits of proteasome might work in concert to accomplish the very complicated task of ATP-dependent protein degradation. Nevertheless, in order to fully understand the dynamic process of protein degradation by the proteasome, it is necessary to determine a high-resolution structure of the 26S proteasome. And so is the case, too, in order to understand the functions of proteasomal super-complexes formed by association with other large cellular complexes, such as ribosome, signalosome, etc. It is likely that the single particle cryoEM will continue to play an important role in achieving this goal.
Acknowledgments
Authors thank Agustin Avila-Sakar for proofreading. Work in the Cheng laboratory is supported by grants from NIH (R01 GM082893) and UCSF Program for Breakthrough Biomedical Research (Opportunity Award in Basic Science and New Technology Award).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ciechanover A, Schwartz AL. The ubiquitin-proteasome pathway: the complexity and myriad functions of proteins death. Proc Natl Acad Sci U S A. 1998;95:2727–2730. doi: 10.1073/pnas.95.6.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 3.Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 4.Glickman MH, Adir N. The proteasome and the delicate balance between destruction and rescue. PLoS Biol. 2004;2:E13. doi: 10.1371/journal.pbio.0020013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coux O, Tanaka K, Goldberg AL. Structure and functions of the 20S and 26S proteasomes. Annu Rev Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- 6.Groll M, Huber R. Substrate access and processing by the 20S proteasome core particle. Int J Biochem Cell Biol. 2003;35:606–616. doi: 10.1016/s1357-2725(02)00390-4. [DOI] [PubMed] [Google Scholar]
- 7.Voges D, Zwickl P, Baumeister W. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu Rev Biochem. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- 8.Verma R, Annan RS, Huddleston MJ, Carr SA, Reynard G, Deshaies RJ. Phosphorylation of Sic1p by G1 Cdk required for its degradation and entry into S phase. Science. 1997;278:455–460. doi: 10.1126/science.278.5337.455. [DOI] [PubMed] [Google Scholar]
- 9.Kaiser P, Moncollin V, Clarke DJ, Watson MH, Bertolaet BL, Reed SI, Bailly E. Cyclin-dependent kinase and Cks/Suc1 interact with the proteasome in yeast to control proteolysis of M-phase targets. Genes Dev. 1999;13:1190–1202. doi: 10.1101/gad.13.9.1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bech-Otschir D, Kraft R, Huang X, Henklein P, Kapelari B, Pollmann C, Dubiel W. COP9 signalosome-specific phosphorylation targets p53 to degradation by the ubiquitin system. Embo J. 2001;20:1630–1639. doi: 10.1093/emboj/20.7.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bounpheng MA, Dimas JJ, Dodds SG, Christy BA. Degradation of Id proteins by the ubiquitin-proteasome pathway. Faseb J. 1999;13:2257–2264. [PubMed] [Google Scholar]
- 12.Karin M, Ben-Neriah Y. Phosphorylation meets ubiquitination: the control of NF-[kappa]B activity. Annu Rev Immunol. 2000;18:621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- 13.Buschmann T, Fuchs SY, Lee CG, Pan ZQ, Ronai Z. SUMO-1 modification of Mdm2 prevents its self-ubiquitination and increases Mdm2 ability to ubiquitinate p53. Cell. 2000;101:753–762. doi: 10.1016/s0092-8674(00)80887-9. [DOI] [PubMed] [Google Scholar]
- 14.Hiller MM, Finger A, Schweiger M, Wolf DH. ER degradation of a misfolded luminal protein by the cytosolic ubiquitin-proteasome pathway. Science. 1996;273:1725–1728. doi: 10.1126/science.273.5282.1725. [DOI] [PubMed] [Google Scholar]
- 15.Kloetzel PM. Antigen processing by the proteasome. Nat Rev Mol Cell Biol. 2001;2:179–187. doi: 10.1038/35056572. [DOI] [PubMed] [Google Scholar]
- 16.Sijts A, Zaiss D, Kloetzel PM. The role of the ubiquitin-proteasome pathway in MHC class I antigen processing: implications for vaccine design. Curr Mol Med. 2001;1:665–676. doi: 10.2174/1566524013363230. [DOI] [PubMed] [Google Scholar]
- 17.Yewdell JW, Bennink JR. Cut and trim: generating MHC class I peptide ligands. Curr Opin Immunol. 2001;13:13–18. doi: 10.1016/s0952-7915(00)00175-8. [DOI] [PubMed] [Google Scholar]
- 18.Rock KL, York IA, Goldberg AL. Post-proteasomal antigen processing for major histocompatibility complex class I presentation. Nat Immunol. 2004;5:670–677. doi: 10.1038/ni1089. [DOI] [PubMed] [Google Scholar]
- 19.Rock KL, York IA, Saric T, Goldberg AL. Protein degradation and the generation of MHC class I-presented peptides. Adv Immunol. 2002;80:1–70. doi: 10.1016/s0065-2776(02)80012-8. [DOI] [PubMed] [Google Scholar]
- 20.Glickman MH, Rubin DM, Coux O, Wefes I, Pfeifer G, Cjeka Z, Baumeister W, Fried VA, Finley D. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell. 1998;94:615–623. doi: 10.1016/s0092-8674(00)81603-7. [DOI] [PubMed] [Google Scholar]
- 21.Glickman MH, Rubin DM, Fried VA, Finley D. The regulatory particle of the Saccharomyces cerevisiae proteasome. Mol Cell Biol. 1998;18:3149–3162. doi: 10.1128/mcb.18.6.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng Y. Toward an atomic model of the 26S proteasome. Curr Opin Struct Biol. 2009;19:203–208. doi: 10.1016/j.sbi.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dikic I, Wakatsuki S, Walters KJ. Ubiquitin-binding domains - from structures to functions. Nat Rev Mol Cell Biol. 2009;10:659–671. doi: 10.1038/nrm2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murata S, Yashiroda H, Tanaka K. Molecular mechanisms of proteasome assembly. Nat Rev Mol Cell Biol. 2009;10:104–115. doi: 10.1038/nrm2630. [DOI] [PubMed] [Google Scholar]
- 26.Schrader EK, Harstad KG, Matouschek A. Targeting proteins for degradation. Nat Chem Biol. 2009;5:815–822. doi: 10.1038/nchembio.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tanaka K. The proteasome: overview of structure and functions. Proc Jpn Acad Ser B Phys Biol Sci. 2009;85:12–36. doi: 10.2183/pjab.85.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nickell S, Beck F, Scheres SH, Korinek A, Forster F, Lasker K, Mihalache O, Sun N, Nagy I, Sali A, Plitzko JM, Carazo JM, Mann M, Baumeister W. Insights into the molecular architecture of the 26S proteasome. Proc Natl Acad Sci U S A. 2009;106:11943–11947. doi: 10.1073/pnas.0905081106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takeuchi J, Chen H, Hoyt MA, Coffino P. Structural elements of the ubiquitin-independent proteasome degron of ornithine decarboxylase. Biochem J. 2008;410:401–407. doi: 10.1042/BJ20071239. [DOI] [PubMed] [Google Scholar]
- 30.Besche H, Haas W, Gygi S, Goldberg A. Isolation of mammalian 26S proteasomes and p97/VCP complexes using the ubiquitin-like domain from HHR23B reveals novel proteasome-associated proteins. Biochemistry. 2009 doi: 10.1021/bi802198q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hough R, Pratt G, Rechsteiner M. Purification of two high molecular weight proteases from rabbit reticulocyte lysate. J Biol Chem. 1987;262:8303–8313. [PubMed] [Google Scholar]
- 32.Lowe J, Stock D, Jap B, Zwickl P, Baumeister W, Huber R. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 A resolution. Science. 1995;268:533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- 33.Zhang F, Hu M, Tian G, Zhang P, Finley D, Jeffrey PD, Shi Y. Structural insights into the regulatory particle of the proteasome from Methanocaldococcus jannaschii. Mol Cell. 2009;34:473–484. doi: 10.1016/j.molcel.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu G, Lin G, Wang M, Dick L, Xu RM, Nathan C, Li H. Structure of the Mycobacterium tuberculosis proteasome and mechanism of inhibition by a peptidyl boronate. Mol Microbiol. 2006;59:1417–1428. doi: 10.1111/j.1365-2958.2005.05036.x. [DOI] [PubMed] [Google Scholar]
- 35.Groll M, Ditzel L, Lowe J, Stock D, Bochtler M, Bartunik HD, Huber R. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 36.Unno M, Mizushima T, Morimoto Y, Tomisugi Y, Tanaka K, Yasuoka N, Tsukihara T. The structure of the mammalian 20S proteasome at 2.75 A resolution. Structure. 2002;10:609–618. doi: 10.1016/s0969-2126(02)00748-7. [DOI] [PubMed] [Google Scholar]
- 37.Rock KL, Goldberg AL. Degradation of cell proteins and the generation of MHC class I-presented peptides. Annu Rev Immunol. 1999;17:739–779. doi: 10.1146/annurev.immunol.17.1.739. [DOI] [PubMed] [Google Scholar]
- 38.Groll M, Bajorek M, Kohler A, Moroder L, Rubin DM, Huber R, Glickman MH, Finley D. A gated channel into the proteasome core particle. Nat Struct Biol. 2000;7:1062–1067. doi: 10.1038/80992. [DOI] [PubMed] [Google Scholar]
- 39.Groll M, Brandstetter H, Bartunik H, Bourenkow G, Huber R. Investigations on the maturation and regulation of archaebacterial proteasomes. J Mol Biol. 2003;327:75–83. doi: 10.1016/s0022-2836(03)00080-9. [DOI] [PubMed] [Google Scholar]
- 40.Kohler A, Cascio P, Leggett DS, Woo KM, Goldberg AL, Finley D. The axial channel of the proteasome core particle is gated by the Rpt2 ATPase and controls both substrate entry and product release. Mol Cell. 2001;7:1143–1152. doi: 10.1016/s1097-2765(01)00274-x. [DOI] [PubMed] [Google Scholar]
- 41.Whitby FG, Masters EI, Kramer L, Knowlton JR, Yao Y, Wang CC, Hill CP. Structural basis for the activation of 20S proteasomes by 11S regulators. Nature. 2000;408:115–120. doi: 10.1038/35040607. [DOI] [PubMed] [Google Scholar]
- 42.Sadre-Bazzaz K, Whitby FG, Robinson H, Formosa T, Hill CP. Structure of a Blm10 complex reveals common mechanisms for proteasome binding and gate opening. Mol Cell. 2010;37:728–735. doi: 10.1016/j.molcel.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hirano Y, Hayashi H, Iemura S, Hendil KB, Niwa S, Kishimoto T, Kasahara M, Natsume T, Tanaka K, Murata S. Cooperation of multiple chaperones required for the assembly of mammalian 20S proteasomes. Mol Cell. 2006;24:977–984. doi: 10.1016/j.molcel.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 44.Yashiroda H, Mizushima T, Okamoto K, Kameyama T, Hayashi H, Kishimoto T, Niwa S, Kasahara M, Kurimoto E, Sakata E, Takagi K, Suzuki A, Hirano Y, Murata S, Kato K, Yamane T, Tanaka K. Crystal structure of a chaperone complex that contributes to the assembly of yeast 20S proteasomes. Nat Struct Mol Biol. 2008;15:228–236. doi: 10.1038/nsmb.1386. [DOI] [PubMed] [Google Scholar]
- 45.Zwickl P, Kleinz J, Baumeister W. Critical elements in proteasome assembly. Nat Struct Biol. 1994;1:765–770. doi: 10.1038/nsb1194-765. [DOI] [PubMed] [Google Scholar]
- 46.da Fonseca PC, Morris EP. Structure of the human 26S proteasome: subunit radial displacements open the gate into the proteolytic core. J Biol Chem. 2008;283:23305–23314. doi: 10.1074/jbc.M802716200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walz J, Erdmann A, Kania M, Typke D, Koster AJ, Baumeister W. 26S proteasome structure revealed by three-dimensional electron microscopy. J Struct Biol. 1998;121:19–29. doi: 10.1006/jsbi.1998.3958. [DOI] [PubMed] [Google Scholar]
- 48.Mochida K, Tres LL, Kierszenbaum AL. Structural features of the 26S proteasome complex isolated from rat testis and sperm tail. Mol Reprod Dev. 2000;57:176–184. doi: 10.1002/1098-2795(200010)57:2<176::AID-MRD9>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 49.Saeki Y, Toh-e A, Yokosawa H. Rapid isolation and characterization of the yeast proteasome regulatory complex. Biochem Biophys Res Commun. 2000;273:509–515. doi: 10.1006/bbrc.2000.2980. [DOI] [PubMed] [Google Scholar]
- 50.Fu H, Doelling JH, Rubin DM, Vierstra RD. Structural and functional analysis of the six regulatory particle triple-A ATPase subunits from the Arabidopsis 26S proteasome. Plant J. 1999;18:529–539. doi: 10.1046/j.1365-313x.1999.00479.x. [DOI] [PubMed] [Google Scholar]
- 51.Rubin DM, Glickman MH, Larsen CN, Dhruvakumar S, Finley D. Active site mutants in the six regulatory particle ATPases reveal multiple roles for ATP in the proteasome. EMBO J. 1998;17:4909–4919. doi: 10.1093/emboj/17.17.4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hartmann-Petersen R, Tanaka K, Hendil KB. Quaternary structure of the ATPase complex of human 26S proteasomes determined by chemical cross-linking. Arch Biochem Biophys. 2001;386:89–94. doi: 10.1006/abbi.2000.2178. [DOI] [PubMed] [Google Scholar]
- 53.Forster A, Masters EI, Whitby FG, Robinson H, Hill CP. The 1.9 A structure of a proteasome-11S activator complex and implications for proteasome-PAN/PA700 interactions. Mol Cell. 2005;18:589–599. doi: 10.1016/j.molcel.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 54.Forster A, Whitby FG, Hill CP. The pore of activated 20S proteasomes has an ordered 7-fold symmetric conformation. Embo J. 2003;22:4356–4364. doi: 10.1093/emboj/cdg436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rabl J, Smith DM, Yu Y, Chang SC, Goldberg AL, Cheng Y. Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases. Mol Cell. 2008;30:360–368. doi: 10.1016/j.molcel.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smith DM, Chang SC, Park S, Finley D, Cheng Y, Goldberg AL. Docking of the proteasomal ATPases’ carboxyl termini in the 20S proteasome’s alpha ring opens the gate for substrate entry. Mol Cell. 2007;27:731–744. doi: 10.1016/j.molcel.2007.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Baker TA, Sauer RT. ATP-dependent proteases of bacteria: recognition logic and operating principles. Trends Biochem Sci. 2006;31:647–653. doi: 10.1016/j.tibs.2006.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sauer RT, Bolon DN, Burton BM, Burton RE, Flynn JM, Grant RA, Hersch GL, Joshi SA, Kenniston JA, Levchenko I, Neher SB, Oakes ES, Siddiqui SM, Wah DA, Baker TA. Sculpting the proteome with AAA(+) proteases and disassembly machines. Cell. 2004;119:9–18. doi: 10.1016/j.cell.2004.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zolkiewski M. A camel passes through the eye of a needle: protein unfolding activity of Clp ATPases. Mol Microbiol. 2006;61:1094–1100. doi: 10.1111/j.1365-2958.2006.05309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kajava AV. What curves alpha-solenoids? Evidence for an alpha-helical toroid structure of Rpn1 and Rpn2 proteins of the 26 S proteasome. J Biol Chem. 2002;277:49791–49798. doi: 10.1074/jbc.M204982200. [DOI] [PubMed] [Google Scholar]
- 61.Effantin G, Rosenzweig R, Glickman MH, Steven AC. Electron microscopic evidence in support of alpha-solenoid models of proteasomal subunits Rpn1 and Rpn2. J Mol Biol. 2009;386:1204–1211. doi: 10.1016/j.jmb.2009.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rosenzweig R, Osmulski PA, Gaczynska M, Glickman MH. The central unit within the 19S regulatory particle of the proteasome. Nat Struct Mol Biol. 2008;15:573–580. doi: 10.1038/nsmb.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smith DM, Kafri G, Cheng Y, Ng D, Walz T, Goldberg AL. ATP binding to PAN or the 26S ATPases causes association with the 20S proteasome, gate opening, and translocation of unfolded proteins. Mol Cell. 2005;20:687–698. doi: 10.1016/j.molcel.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 64.Benaroudj N, Goldberg AL. PAN, the proteasome-activating nucleotidase from archaebacteria, is a protein-unfolding molecular chaperone. Nat Cell Biol. 2000;2:833–839. doi: 10.1038/35041081. [DOI] [PubMed] [Google Scholar]
- 65.Zwickl P, Ng D, Woo KM, Klenk HP, Goldberg AL. An archaebacterial ATPase, homologous to ATPases in the eukaryotic 26 S proteasome, activates protein breakdown by 20 S proteasomes. J Biol Chem. 1999;274:26008–26014. doi: 10.1074/jbc.274.37.26008. [DOI] [PubMed] [Google Scholar]
- 66.Benaroudj N, Zwickl P, Seemuller E, Baumeister W, Goldberg AL. ATP hydrolysis by the proteasome regulatory complex PAN serves multiple functions in protein degradation. Mol Cell. 2003;11:69–78. doi: 10.1016/s1097-2765(02)00775-x. [DOI] [PubMed] [Google Scholar]
- 67.Horwitz AA, Navon A, Groll M, Smith DM, Reis C, Goldberg AL. ATP-induced structural transitions in PAN, the proteasome-regulatory ATPase complex in Archaea. J Biol Chem. 2007;282:22921–22929. doi: 10.1074/jbc.M702846200. [DOI] [PubMed] [Google Scholar]
- 68.Stadtmueller BM, Ferrell K, Whitby FG, Heroux A, Robinson H, Myszka DG, Hill CP. Structural models for interactions between the 20S proteasome and its PAN/19S activators. J Biol Chem. 2010;285:13–17. doi: 10.1074/jbc.C109.070425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu Y, Smith DM, Kim HM, Rodriguez V, Goldberg AL, Cheng Y. Interactions of PAN’s C-termini with archaeal 20S proteasome and implications for the eukaryotic proteasome-ATPase interactions. Embo J. 2010;29:692–702. doi: 10.1038/emboj.2009.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Djuranovic S, Hartmann MD, Habeck M, Ursinus A, Zwickl P, Martin J, Lupas AN, Zeth K. Structure and activity of the N-terminal substrate recognition domains in proteasomal ATPases. Mol Cell. 2009;34:580–590. doi: 10.1016/j.molcel.2009.04.030. [DOI] [PubMed] [Google Scholar]
- 71.Zhang F, Wu Z, Zhang P, Tian G, Finley D, Shi Y. Mechanism of substrate unfolding and translocation by the regulatory particle of the proteasome from Methanocaldococcus jannaschii. Mol Cell. 2009;34:485–496. doi: 10.1016/j.molcel.2009.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang J, Song JJ, Franklin MC, Kamtekar S, Im YJ, Rho SH, Seong IS, Lee CS, Chung CH, Eom SH. Crystal structures of the HslVU peptidase-ATPase complex reveal an ATP-dependent proteolysis mechanism. Structure. 2001;9:177–184. doi: 10.1016/s0969-2126(01)00570-6. [DOI] [PubMed] [Google Scholar]
- 73.Wang J, Song JJ, Seong IS, Franklin MC, Kamtekar S, Eom SH, Chung CH. Nucleotide-dependent conformational changes in a protease-associated ATPase HsIU. Structure. 2001;9:1107–1116. doi: 10.1016/s0969-2126(01)00670-0. [DOI] [PubMed] [Google Scholar]
- 74.Forster F, Lasker K, Beck F, Nickell S, Sali A, Baumeister W. An atomic model AAA-ATPase/20S core particle sub-complex of the 26S proteasome. Biochem Biophys Res Commun. 2009;388:228–233. doi: 10.1016/j.bbrc.2009.07.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ogura T, Wilkinson AJ. AAA+ superfamily ATPases: common structure--diverse function. Genes Cells. 2001;6:575–597. doi: 10.1046/j.1365-2443.2001.00447.x. [DOI] [PubMed] [Google Scholar]
- 76.Vale RD. AAA proteins. Lords of the ring. J Cell Biol. 2000;150:F13–19. doi: 10.1083/jcb.150.1.f13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Beyer A. Sequence analysis of the AAA protein family. Protein Sci. 1997;6:2043–2058. doi: 10.1002/pro.5560061001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Karata K, Inagawa T, Wilkinson AJ, Tatsuta T, Ogura T. Dissecting the role of a conserved motif (the second region of homology) in the AAA family of ATPases. Site-directed mutagenesis of the ATP-dependent protease FtsH. J Biol Chem. 1999;274:26225–26232. doi: 10.1074/jbc.274.37.26225. [DOI] [PubMed] [Google Scholar]
- 79.Neuwald AF, Aravind L, Spouge JL, Koonin EV. AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- 80.Saeki Y, Tanaka K. Unlocking the proteasome door. Mol Cell. 2007;27:865–867. doi: 10.1016/j.molcel.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 81.Gillette TG, Kumar B, Thompson D, Slaughter CA, DeMartino GN. Differential roles of the COOH termini of AAA subunits of PA700 (19 S regulator) in asymmetric assembly and activation of the 26 S proteasome. J Biol Chem. 2008;283:31813–31822. doi: 10.1074/jbc.M805935200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen C, Huang C, Chen S, Liang J, Lin W, Ke G, Zhang H, Wang B, Huang J, Han Z, Ma L, Huo K, Yang X, Yang P, He F, Tao T. Subunit-subunit interactions in the human 26S proteasome. Proteomics. 2008;8:508–520. doi: 10.1002/pmic.200700588. [DOI] [PubMed] [Google Scholar]
- 83.Sharon M, Taverner T, Ambroggio XI, Deshaies RJ, Robinson CV. Structural organization of the 19S proteasome lid: insights from MS of intact complexes. PLoS Biol. 2006;4:e267. doi: 10.1371/journal.pbio.0040267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Funakoshi M, Tomko RJ, Kobayashi H, Hochstrasser M. Multiple assembly chaperones govern biogenesis of the proteasome regulatory particle base. Cell. 2009;137:887–899. doi: 10.1016/j.cell.2009.04.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kaneko T, Hamazaki J, Iemura S, Sasaki K, Furuyama K, Natsume T, Tanaka K, Murata S. Assembly pathway of the Mammalian proteasome base subcomplex is mediated by multiple specific chaperones. Cell. 2009;137:914–925. doi: 10.1016/j.cell.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 86.Park S, Roelofs J, Kim W, Robert J, Schmidt M, Gygi SP, Finley D. Hexameric assembly of the proteasomal ATPases is templated through their C termini. Nature. 2009;459:866–870. doi: 10.1038/nature08065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Roelofs J, Park S, Haas W, Tian G, McAllister FE, Huo Y, Lee BH, Zhang F, Shi Y, Gygi SP, Finley D. Chaperone-mediated pathway of proteasome regulatory particle assembly. Nature. 2009;459:861–865. doi: 10.1038/nature08063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Saeki Y, Toh-E A, Kudo T, Kawamura H, Tanaka K. Multiple proteasome-interacting proteins assist the assembly of the yeast 19S regulatory particle. Cell. 2009;137:900–913. doi: 10.1016/j.cell.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 89.Besche HC, Peth A, Goldberg AL. Getting to first base in proteasome assembly. Cell. 2009;138:25–28. doi: 10.1016/j.cell.2009.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hendil KB, Kriegenburg F, Tanaka K, Murata S, Lauridsen AM, Johnsen AH, Hartmann-Petersen R. The 20S Proteasome as an Assembly Platform for the 19S Regulatory Complex. J Mol Biol. 2009;394:320–328. doi: 10.1016/j.jmb.2009.09.038. [DOI] [PubMed] [Google Scholar]
- 91.Tomko RJ, Jr, Funakoshi M, Schneider K, Wang J, Hochstrasser M. Heterohexameric ring arrangement of the eukaryotic proteasomal ATPases: implications for proteasome structure and assembly. Mol Cell. 2010;38:393–403. doi: 10.1016/j.molcel.2010.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- 93.Pickart CM, Fushman D. Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol. 2004;8:610–616. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 94.Ikeda F, Dikic I. Atypical ubiquitin chains: new molecular signals. ‘Protein Modifications: Beyond the Usual Suspects’ review series. EMBO Rep. 2008;9:536–542. doi: 10.1038/embor.2008.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chen ZJ, Sun LJ. Nonproteolytic functions of ubiquitin in cell signaling. Mol Cell. 2009;33:275–286. doi: 10.1016/j.molcel.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 96.Deveraux Q, Ustrell V, Pickart C, Rechsteiner M. A 26 S protease subunit that binds ubiquitin conjugates. J Biol Chem. 1994;269:7059–7061. [PubMed] [Google Scholar]
- 97.Young P, Deveraux Q, Beal RE, Pickart CM, Rechsteiner M. Characterization of two polyubiquitin binding sites in the 26 S protease subunit 5a. J Biol Chem. 1998;273:5461–5467. doi: 10.1074/jbc.273.10.5461. [DOI] [PubMed] [Google Scholar]
- 98.Wang Q, Young P, Walters KJ. Structure of S5a bound to monoubiquitin provides a model for polyubiquitin recognition. J Mol Biol. 2005;348:727–739. doi: 10.1016/j.jmb.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 99.Zhang N, Wang Q, Ehlinger A, Randles L, Lary JW, Kang Y, Haririnia A, Storaska AJ, Cole JL, Fushman D, Walters KJ. Structure of the s5a:k48-linked diubiquitin complex and its interactions with rpn13. Mol Cell. 2009;35:280–290. doi: 10.1016/j.molcel.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fu H, Reis N, Lee Y, Glickman MH, Vierstra RD. Subunit interaction maps for the regulatory particle of the 26S proteasome and the COP9 signalosome. Embo J. 2001;20:7096–7107. doi: 10.1093/emboj/20.24.7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Huizinga EG, Tsuji S, Romijn RA, Schiphorst ME, de Groot PG, Sixma JJ, Gros P. Structures of glycoprotein Ibalpha and its complex with von Willebrand factor A1 domain. Science. 2002;297:1176–1179. doi: 10.1126/science.107355. [DOI] [PubMed] [Google Scholar]
- 102.Husnjak K, Elsasser S, Zhang N, Chen X, Randles L, Shi Y, Hofmann K, Walters KJ, Finley D, Dikic I. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature. 2008;453:481–488. doi: 10.1038/nature06926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Schreiner P, Chen X, Husnjak K, Randles L, Zhang N, Elsasser S, Finley D, Dikic I, Walters KJ, Groll M. Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction. Nature. 2008;453:548–552. doi: 10.1038/nature06924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shimada S, Ogawa M, Schlom J, Greiner JW. Identification of a novel tumor-associated Mr 110,000 gene product in human gastric carcinoma cells that is immunologically related to carcinoembryonic antigen. Cancer Res. 1991;51:5694–5703. [PubMed] [Google Scholar]
- 105.Simins AB, Weighardt H, Weidner KM, Weidle UH, Holzmann B. Functional cloning of ARM-1, an adhesion-regulating molecule upregulated in metastatic tumor cells. Clin Exp Metastasis. 1999;17:641–648. doi: 10.1023/a:1006790912877. [DOI] [PubMed] [Google Scholar]
- 106.Hamazaki J, Iemura S, Natsume T, Yashiroda H, Tanaka K, Murata S. A novel proteasome interacting protein recruits the deubiquitinating enzyme UCH37 to 26S proteasomes. Embo J. 2006;25:4524–4536. doi: 10.1038/sj.emboj.7601338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Qiu XB, Ouyang SY, Li CJ, Miao S, Wang L, Goldberg AL. hRpn13/ADRM1/GP110 is a novel proteasome subunit that binds the deubiquitinating enzyme, UCH37. Embo J. 2006;25:5742–5753. doi: 10.1038/sj.emboj.7601450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yao T, Song L, Xu W, DeMartino GN, Florens L, Swanson SK, Washburn MP, Conaway RC, Conaway JW, Cohen RE. Proteasome recruitment and activation of the Uch37 deubiquitinating enzyme by Adrm1. Nat Cell Biol. 2006;8:994–1002. doi: 10.1038/ncb1460. [DOI] [PubMed] [Google Scholar]
- 109.Lam YA, Lawson TG, Velayutham M, Zweier JL, Pickart CM. A proteasomal ATPase subunit recognizes the polyubiquitin degradation signal. Nature. 2002;416:763–767. doi: 10.1038/416763a. [DOI] [PubMed] [Google Scholar]
- 110.Amerik AY, Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim Biophys Acta. 2004;1695:189–207. doi: 10.1016/j.bbamcr.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 111.Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, Bernards R. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 112.Borodovsky A, Kessler BM, Casagrande R, Overkleeft HS, Wilkinson KD, Ploegh HL. A novel active site-directed probe specific for deubiquitylating enzymes reveals proteasome association of USP14. Embo J. 2001;20:5187–5196. doi: 10.1093/emboj/20.18.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lam YA, DeMartino GN, Pickart CM, Cohen RE. Specificity of the ubiquitin isopeptidase in the PA700 regulatory complex of 26 S proteasomes. J Biol Chem. 1997;272:28438–28446. doi: 10.1074/jbc.272.45.28438. [DOI] [PubMed] [Google Scholar]
- 114.Lam YA, Xu W, DeMartino GN, Cohen RE. Editing of ubiquitin conjugates by an isopeptidase in the 26S proteasome. Nature. 1997;385:737–740. doi: 10.1038/385737a0. [DOI] [PubMed] [Google Scholar]
- 115.Leggett DS, Hanna J, Borodovsky A, Crosas B, Schmidt M, Baker RT, Walz T, Ploegh H, Finley D. Multiple associated proteins regulate proteasome structure and function. Mol Cell. 2002;10:495–507. doi: 10.1016/s1097-2765(02)00638-x. [DOI] [PubMed] [Google Scholar]
- 116.Stone M, Hartmann-Petersen R, Seeger M, Bech-Otschir D, Wallace M, Gordon C. Uch2/Uch37 is the major deubiquitinating enzyme associated with the 26S proteasome in fission yeast. J Mol Biol. 2004;344:697–706. doi: 10.1016/j.jmb.2004.09.057. [DOI] [PubMed] [Google Scholar]
- 117.Verma R, Aravind L, Oania R, McDonald WH, Yates JR, 3rd, Koonin EV, Deshaies RJ. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298:611–615. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- 118.Yao T, Cohen RE. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419:403–407. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
- 119.Holzl H, Kapelari B, Kellermann J, Seemuller E, Sumegi M, Udvardy A, Medalia O, Sperling J, Muller SA, Engel A, Baumeister W. The regulatory complex of Drosophila melanogaster 26S proteasomes. Subunit composition and localization of a deubiquitylating enzyme. J Cell Biol. 2000;150:119–130. doi: 10.1083/jcb.150.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Li T, Naqvi NI, Yang H, Teo TS. Identification of a 26S proteasome-associated UCH in fission yeast. Biochem Biophys Res Commun. 2000;272:270–275. doi: 10.1006/bbrc.2000.2767. [DOI] [PubMed] [Google Scholar]
- 121.Nishio K, Kim SW, Kawai K, Mizushima T, Yamane T, Hamazaki J, Murata S, Tanaka K, Morimoto Y. Crystal structure of the deubiquitinating enzyme UCH37 (human UCH-L5) catalytic domain. Biochem Biophys Res Commun. 2009;390:855–860. doi: 10.1016/j.bbrc.2009.10.062. [DOI] [PubMed] [Google Scholar]
- 122.Johnston SC, Larsen CN, Cook WJ, Wilkinson KD, Hill CP. Crystal structure of a deubiquitinating enzyme (human UCH-L3) at 1.8 A resolution. Embo J. 1997;16:3787–3796. doi: 10.1093/emboj/16.13.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Johnston SC, Riddle SM, Cohen RE, Hill CP. Structural basis for the specificity of ubiquitin C-terminal hydrolases. Embo J. 1999;18:3877–3887. doi: 10.1093/emboj/18.14.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Misaghi S, Galardy PJ, Meester WJ, Ovaa H, Ploegh HL, Gaudet R. Structure of the ubiquitin hydrolase UCH-L3 complexed with a suicide substrate. J Biol Chem. 2005;280:1512–1520. doi: 10.1074/jbc.M410770200. [DOI] [PubMed] [Google Scholar]
- 125.Das C, Hoang QQ, Kreinbring CA, Luchansky SJ, Meray RK, Ray SS, Lansbury PT, Ringe D, Petsko GA. Structural basis for conformational plasticity of the Parkinson’s disease-associated ubiquitin hydrolase UCH-L1. Proc Natl Acad Sci U S A. 2006;103:4675–4680. doi: 10.1073/pnas.0510403103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Realini C, Rogers SW, Rechsteiner M. KEKE motifs. Proposed roles in protein-protein association and presentation of peptides by MHC class I receptors. FEBS Lett. 1994;348:109–113. doi: 10.1016/0014-5793(94)00569-9. [DOI] [PubMed] [Google Scholar]
- 127.Ambroggio XI, Rees DC, Deshaies RJ. JAMM: a metalloprotease-like zinc site in the proteasome and signalosome. PLoS Biol. 2004;2:E2. doi: 10.1371/journal.pbio.0020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Maytal-Kivity V, Reis N, Hofmann K, Glickman MH. MPN+, a putative catalytic motif found in a subset of MPN domain proteins from eukaryotes and prokaryotes, is critical for Rpn11 function. BMC Biochem. 2002;3:28. doi: 10.1186/1471-2091-3-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Koulich E, Li X, DeMartino GN. Relative structural and functional roles of multiple deubiquitylating proteins associated with mammalian 26S proteasome. Mol Biol Cell. 2008;19:1072–1082. doi: 10.1091/mbc.E07-10-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sato Y, Yoshikawa A, Yamagata A, Mimura H, Yamashita M, Ookata K, Nureki O, Iwai K, Komada M, Fukai S. Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains. Nature. 2008;455:358–362. doi: 10.1038/nature07254. [DOI] [PubMed] [Google Scholar]
- 131.Kim HM, Shin DR, Yoo OJ, Lee H, Lee JO. Crystal structure of Drosophila angiotensin I-converting enzyme bound to captopril and lisinopril. FEBS Lett. 2003;538:65–70. doi: 10.1016/s0014-5793(03)00128-5. [DOI] [PubMed] [Google Scholar]
- 132.Pelmenschikov V, Blomberg MR, Siegbahn PE. A theoretical study of the mechanism for peptide hydrolysis by thermolysin. J Biol Inorg Chem. 2002;7:284–298. doi: 10.1007/s007750100295. [DOI] [PubMed] [Google Scholar]
- 133.Eddins MJ, Varadan R, Fushman D, Pickart CM, Wolberger C. Crystal structure and solution NMR studies of Lys48-linked tetraubiquitin at neutral pH. J Mol Biol. 2007;367:204–211. doi: 10.1016/j.jmb.2006.12.065. [DOI] [PubMed] [Google Scholar]
- 134.Datta AB, Hura GL, Wolberger C. The Structure and Conformation of Lys63-Linked Tetraubiquitin. J Mol Biol. 2009;392:1117–1124. doi: 10.1016/j.jmb.2009.07.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hu M, Li P, Song L, Jeffrey PD, Chenova TA, Wilkinson KD, Cohen RE, Shi Y. Structure and mechanisms of the proteasome-associated deubiquitinating enzyme USP14. Embo J. 2005;24:3747–3756. doi: 10.1038/sj.emboj.7600832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hanna J, Hathaway NA, Tone Y, Crosas B, Elsasser S, Kirkpatrick DS, Leggett DS, Gygi SP, King RW, Finley D. Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell. 2006;127:99–111. doi: 10.1016/j.cell.2006.07.038. [DOI] [PubMed] [Google Scholar]
- 137.Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. Embo J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Crosas B, Hanna J, Kirkpatrick DS, Zhang DP, Tone Y, Hathaway NA, Buecker C, Leggett DS, Schmidt M, King RW, Gygi SP, Finley D. Ubiquitin chains are remodeled at the proteasome by opposing ubiquitin ligase and deubiquitinating activities. Cell. 2006;127:1401–1413. doi: 10.1016/j.cell.2006.09.051. [DOI] [PubMed] [Google Scholar]
- 139.Eytan E, Armon T, Heller H, Beck S, Hershko A. Ubiquitin C-terminal hydrolase activity associated with the 26 S protease complex. J Biol Chem. 1993;268:4668–4674. [PubMed] [Google Scholar]
- 140.Bucher P, Karplus K, Moeri N, Hofmann K. A flexible motif search technique based on generalized profiles. Comput Chem. 1996;20:3–23. doi: 10.1016/s0097-8485(96)80003-9. [DOI] [PubMed] [Google Scholar]
- 141.Hofmann K, Bucher P. The PCI domain: a common theme in three multiprotein complexes. Trends Biochem Sci. 1998;23:204–205. doi: 10.1016/s0968-0004(98)01217-1. [DOI] [PubMed] [Google Scholar]
- 142.Bailly E, Reed SI. Functional characterization of rpn3 uncovers a distinct 19S proteasomal subunit requirement for ubiquitin-dependent proteolysis of cell cycle regulatory proteins in budding yeast. Mol Cell Biol. 1999;19:6872–6890. doi: 10.1128/mcb.19.10.6872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Isono E, Saeki Y, Yokosawa H, Toh-e A. Rpn7 Is required for the structural integrity of the 26 S proteasome of Saccharomyces cerevisiae. J Biol Chem. 2004;279:27168–27176. doi: 10.1074/jbc.M314231200. [DOI] [PubMed] [Google Scholar]
- 144.Isono E, Saito N, Kamata N, Saeki Y, Toh EA. Functional analysis of Rpn6p, a lid component of the 26 S proteasome, using temperature-sensitive rpn6 mutants of the yeast Saccharomyces cerevisiae. J Biol Chem. 2005;280:6537–6547. doi: 10.1074/jbc.M409364200. [DOI] [PubMed] [Google Scholar]
- 145.Takeuchi J, Fujimuro M, Yokosawa H, Tanaka K, Toh-e A. Rpn9 is required for efficient assembly of the yeast 26S proteasome. Mol Cell Biol. 1999;19:6575–6584. doi: 10.1128/mcb.19.10.6575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Yen HC, Espiritu C, Chang EC. Rpn5 is a conserved proteasome subunit and required for proper proteasome localization and assembly. J Biol Chem. 2003;278:30669–30676. doi: 10.1074/jbc.M302093200. [DOI] [PubMed] [Google Scholar]
- 147.Hinnebusch AG. eIF3: a versatile scaffold for translation initiation complexes. Trends Biochem Sci. 2006;31:553–562. doi: 10.1016/j.tibs.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 148.Sharon M, Mao H, Boeri Erba E, Stephens E, Zheng N, Robinson CV. Symmetrical modularity of the COP9 signalosome complex suggests its multifunctionality. Structure. 2009;17:31–40. doi: 10.1016/j.str.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 149.Zhou M, Sandercock AM, Fraser CS, Ridlova G, Stephens E, Schenauer MR, Yokoi-Fong T, Barsky D, Leary JA, Hershey JW, Doudna JA, Robinson CV. Mass spectrometry reveals modularity and a complete subunit interaction map of the eukaryotic translation factor eIF3. Proc Natl Acad Sci U S A. 2008;105:18139–18144. doi: 10.1073/pnas.0801313105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Pick E, Hofmann K, Glickman MH. PCI complexes: Beyond the proteasome, CSN, and eIF3 Troika. Mol Cell. 2009;35:260–264. doi: 10.1016/j.molcel.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 151.Pick E, Pintard L. In the land of the rising sun with the COP9 signalosome and related Zomes. Symposium on the COP9 signalosome, Proteasome and eIF3. EMBO Rep. 2009;10:343–348. doi: 10.1038/embor.2009.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Faza MB, Kemmler S, Jimeno S, Gonzalez-Aguilera C, Aguilera A, Hurt E, Panse VG. Sem1 is a functional component of the nuclear pore complex-associated messenger RNA export machinery. J Cell Biol. 2009;184:833–846. doi: 10.1083/jcb.200810059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Wilmes GM, Bergkessel M, Bandyopadhyay S, Shales M, Braberg H, Cagney G, Collins SR, Whitworth GB, Kress TL, Weissman JS, Ideker T, Guthrie C, Krogan NJ. A genetic interaction map of RNA-processing factors reveals links between Sem1/Dss1-containing complexes and mRNA export and splicing. Mol Cell. 2008;32:735–746. doi: 10.1016/j.molcel.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wei Z, Zhang P, Zhou Z, Cheng Z, Wan M, Gong W. Crystal structure of human eIF3k, the first structure of eIF3 subunits. J Biol Chem. 2004;279:34983–34990. doi: 10.1074/jbc.M405158200. [DOI] [PubMed] [Google Scholar]
- 155.Scheel H, Hofmann K. Prediction of a common structural scaffold for proteasome lid, COP9-signalosome and eIF3 complexes. BMC Bioinformatics. 2005;6:71. doi: 10.1186/1471-2105-6-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Dessau M, Halimi Y, Erez T, Chomsky-Hecht O, Chamovitz DA, Hirsch JA. The Arabidopsis COP9 signalosome subunit 7 is a model PCI domain protein with subdomains involved in COP9 signalosome assembly. Plant Cell. 2008;20:2815–2834. doi: 10.1105/tpc.107.053801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Davy A, Bello P, Thierry-Mieg N, Vaglio P, Hitti J, Doucette-Stamm L, Thierry-Mieg D, Reboul J, Boulton S, Walhout AJ, Coux O, Vidal M. A protein-protein interaction map of the Caenorhabditis elegans 26S proteasome. EMBO Rep. 2001;2:821–828. doi: 10.1093/embo-reports/kve184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kapelari B, Bech-Otschir D, Hegerl R, Schade R, Dumdey R, Dubiel W. Electron microscopy and subunit-subunit interaction studies reveal a first architecture of COP9 signalosome. J Mol Biol. 2000;300:1169–1178. doi: 10.1006/jmbi.2000.3912. [DOI] [PubMed] [Google Scholar]
- 159.Baker SP, Grant PA. The proteasome: not just degrading anymore. Cell. 2005;123:361–363. doi: 10.1016/j.cell.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 160.Collins GA, Tansey WP. The proteasome: a utility tool for transcription? Curr Opin Genet Dev. 2006;16:197–202. doi: 10.1016/j.gde.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 161.Lee D, Ezhkova E, Li B, Pattenden SG, Tansey WP, Workman JL. The proteasome regulatory particle alters the SAGA coactivator to enhance its interactions with transcriptional activators. Cell. 2005;123:423–436. doi: 10.1016/j.cell.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 162.Ferdous A, Gonzalez F, Sun L, Kodadek T, Johnston SA. The 19S regulatory particle of the proteasome is required for efficient transcription elongation by RNA polymerase II. Mol Cell. 2001;7:981–991. doi: 10.1016/s1097-2765(01)00250-7. [DOI] [PubMed] [Google Scholar]
- 163.Krogan NJ, Lam MH, Fillingham J, Keogh MC, Gebbia M, Li J, Datta N, Cagney G, Buratowski S, Emili A, Greenblatt JF. Proteasome involvement in the repair of DNA double-strand breaks. Mol Cell. 2004;16:1027–1034. doi: 10.1016/j.molcel.2004.11.033. [DOI] [PubMed] [Google Scholar]