Abstract
Epigenetic proteins are intently pursued targets in ligand discovery. To date, successful efforts have been limited to chromatin modifying enzymes, or so-called epigenetic “writers” and “erasers”. Potent inhibitors of histone binding modules have not yet been described. Here we report a cell-permeable small molecule (JQ1) which binds competitively to acetyl-lysine recognition motifs, or bromodomains. High potency and specificity toward a subset of human bromodomains is explained by co-crystal structures with BRD4, revealing excellent shape complementarity with the acetyl-lysine binding cavity. Recurrent translocation of BRD4 is observed in a genetically-defined, incurable subtype of human squamous carcinoma. Competitive binding by JQ1 displaces the BRD4 fusion oncoprotein from chromatin, prompting squamous differentiation and specific anti-proliferative effects in BRD4-dependent cell lines and patient-derived xenograft models. These data establish proof of concept for targeting protein-protein interactions of epigenetic “readers” and provide a versatile chemical scaffold for the development of chemical probes more broadly throughout the bromodomain family.
Gene regulation is fundamentally governed by reversible, non-covalent assembly of macromolecules1. Signal transduction to RNA polymerase requires higher-ordered protein complexes, spatially regulated by assembly factors capable of interpreting the post-translational modification states of chromatin2. Readers of epigenetic marks are structurally diverse proteins each possessing one or more evolutionarily conserved effector modules, which recognize covalent modifications of histone proteins or DNA. The ε-N-acetylation of lysine residues (Kac) on histone tails is associated with an open chromatin architecture and transcriptional activation3. Context-specific molecular recognition of acetyl-lysine is principally mediated by bromodomains.
Bromodomain-containing proteins are of substantial biological interest, as components of transcription factor complexes and determinants of epigenetic memory4. There are 41 diverse human proteins containing a total of 57 bromodomains. Despite large sequence variations, all bromodomain modules share a conserved fold comprising a left-handed bundle of four alpha helices (αZ, αA, αB, αC), linked by diverse loop regions (ZA and BC loops) that contribute to substrate specificity. Co-crystal structures with peptidic substrates showed that the acetyl-lysine is recognized by a central hydrophobic cavity and is anchored by a hydrogen bond with an asparagine residue present in most bromodomains5. The bromodomain and extra-terminal (BET) family (BRD2, BRD3, BRD4 and BRDT) shares a common domain architecture comprising two N-terminal bromodomains which exhibit high levels of sequence conservation, and a more divergent C-terminal recruitment domain (Supplementary Fig. 1)6.
Recent research has established a compelling rationale for targeting BRD4 in cancer. BRD4 remains bound to transcriptional start sites of genes expressed during the M/G1 transition, influencing mitotic progression4. BRD4 is also a critical mediator of transcriptional elongation, functioning to recruit the positive transcription elongation factor complex (P-TEFb)7,8. Cyclin dependent kinase-9, a core component of P-TEFb9–11, is a validated target in chronic lymphocytic leukemia12, and has recently been linked to c-Myc dependent transcription13. Thus, BRD4 recruits P-TEFb to mitotic chromosomes resulting in increased expression of growth promoting genes14.
Importantly, BRD4 has recently been identified as a component of a recurrent t(15;19) chromosomal translocation in an aggressive form of human squamous carcinoma15,16. Such translocations express the tandem N-terminal bromodomains of BRD4 as an in-frame chimera with the NUT (nuclear protein in testis) protein, genetically defining the so-called NUT midline carcinoma (NMC). Functional studies in patient-derived NMC cell lines have validated the essential role of the BRD4-NUT oncoprotein in maintaining the characteristic proliferation advantage and differentiation block of this uniformly fatal malignancy17. Notably, RNA silencing of BRD4-NUT arrests proliferation and prompts terminal squamous differentiation. These observations underscore the broad utility and immediate therapeutic potential of a direct-acting inhibitor of human bromodomain proteins.
A selective and potent inhibitor for the BET sub-family of bromodomains
A major collaborative focus of our research groups concerns the development of chemical probes18,19 and the optimization of therapeutic leads for the translation of small-molecule modulators of epigenetic targets as cancer therapeutics. Motivated by the above rationale, we have developed biochemical platforms for the identification of new inhibitors of bromodomain isoforms using high-throughput screening, as well as the annotation of putative ligands emerging from collaborative and published research. In the course of these studies, we learned of a remarkable observation by Mitsubishi Pharmaceuticals that simple thienodiazepines possessed binding activity for BRD420. Prior research from this group suggests these compounds emerged from anti-inflammatory phenotypic studies, such as inhibition of CD28 co-stimulation as a means of treating autoimmune diseases21,22. A rich literature has established the synthetic accessibility and favorable pharmacologic properties of this privileged class of drug-like small molecules23. Indeed, the core scaffold described appears in FDA-approved substances such as alprazolam and triazolam.
Inferring structure-activity-relationships also derived from molecular modeling of candidate ligands within the binding pocket of the apo crystal structure of the first bromodomain of BRD4 (hereafter referred to as BRD4(1); PDB ID 2OSS), we designed a prototype ligand, JQ1 (Fig. 1a). JQ1 is a novel thieno-triazolo-1,4-diazepine, possessing an appended, bulky t-butyl ester functional group at C6 in order to (1) allow for additional pendant group diversity, as needed, and (2) to mitigate binding to the central benzodiazepine receptor as predicted by published SAR23. We first established a high-yielding, seven step synthetic route to access racemic JQ1 (hereafter referred to as JQ1) and derivatives (Scheme S1, Supplemental Methods). We have also identified a route to synthesize each enantiomer, (+)-JQ1 and (−)-JQ1 (Scheme S2, Supplemental Methods).
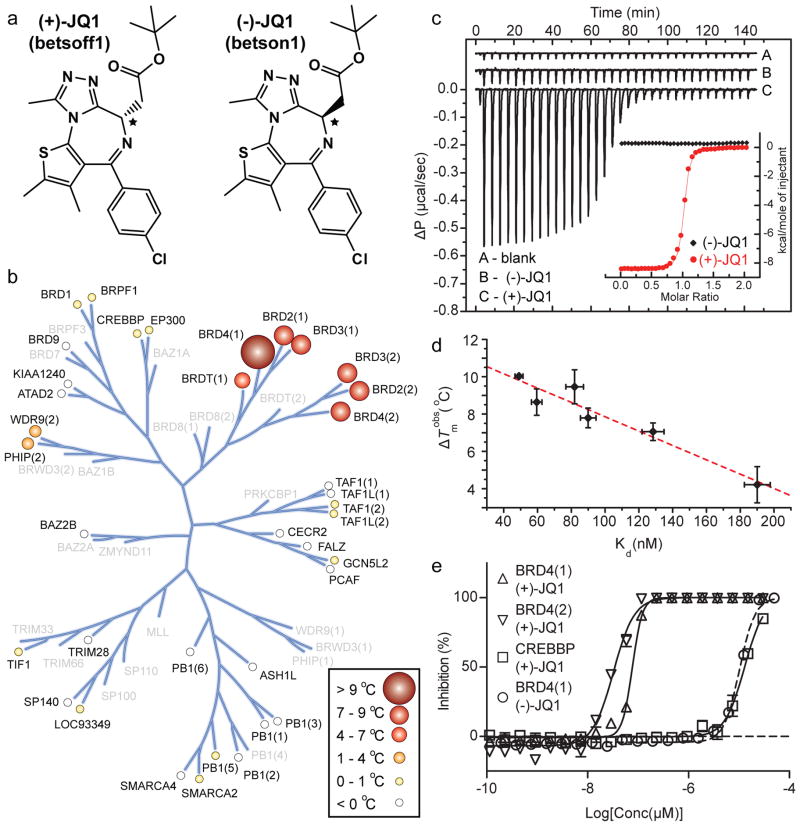
Figure 1. Structure and selectivity of JQ1.
a, Structure of the two JQ1 stereo-isomers. The stereocentre at C6 is indicated by an asterix (*). b, Assessment of inhibitor selectivity using Differential Scanning Fluorimetry (DSF). Shown are averaged temperature shifts (ΔTmobs) in °C upon binding of 10 μM (+)-JQ1. The temperature shifts are represented by spheres as indicated in the inset. Screened proteins are labelled and proteins not included in the selectivity panel are shown in grey. (−)-JQ1 did not reveal any significant temperature shifts to any of the screened bromodomains. c, Isothermal Titration Calorimetry (ITC). Raw injection heats are shown for a blank titration of (BRD4(1)) into buffer (A), and reverse titrations using the inactive isomer (−)-JQ1 (B) and the active isomer (+)-JQ1 (C). The inset shows normalized binding enthalpies corrected for the heat of dilution as a function of binding site saturation (symbols as indicated in the inset). Solid lines represent a nonlinear least squares fit using a single-site binding model. d, Thermal shifts (ΔTmobs) show good correlation to dissociation constants (Kd) determined by ITC for the BET bromodomains. The dotted red line represents a least squares fit with an R of 96 %. e, Competitive displacement of a histone peptide from human bromodomains is exhibited by JQ1 using a bead-based proximity assay. Alpha screen titrations monitoring the displacement of a tetra-acetylated histone H4 peptide by JQ1 isomers using the bromodomains BRD4(1), BRD4(2) or of an acetylated H3 peptide using CREBBP.
To establish a biochemical platform for comprehensive selectivity screening, all human bromodomains were subcloned into bacterial expression vectors. Testing of an average of 15 expression constructs per bromodomain target resulted in the identification of 37 expression systems that yielded soluble protein suitable for specificity screening and covered all bromodomain subfamilies (Supplementary Table 1). Since the specific substrates of most bromodomains are unknown, a general binding assay based on differential scanning fluorimetry (DSF) was implemented24. Binding of (+)-JQ1 significantly increased the thermal stability of all bromodomains of the BET family (Fig. 1b, Supplementary Table 2) with ΔTmobs values between 4.2 °C (BRDT(1)) and 10.1 °C (BRD4(1)). No significant stability shifts were detected for bromodomains outside the BET family, suggesting that this ligand is highly selective. In contrast, the stereoisomer (−)-JQ1 showed no significant interaction with any bromodomain present in our panel.
Within a family of proteins a linear correlation between DSF ΔTmobs values and binding constants has been observed, with temperature shifts larger than 6 °C corresponding to compounds with nM dissociation constants25,26. Since the sensitivity of this assay may vary between different protein families, isothermal titration calorimetry (ITC) was used to precisely determine binding constants. Enantiomerically pure (+)-JQ1 bound with a Kd of about 50 nM and 90 nM to the first and second bromodomains of BRD4, respectively (Fig. 1c, Supplementary Table 3). Comparable binding to both domains of BRD3 was observed, whereas the first bromodomains of BRDT and BRD2 revealed about 3-fold weaker binding. Affinities determined by ITC and ΔTmobs values showed very good correlation (Fig. 1d). Importantly, (+)-JQ1 showed no detectable binding to bromodomains that exhibited minimal temperature shifts, such as WDR9(2) and CREBBP.
To assess whether (+)-JQ1 binding was competitive with acetyl-lysine, we adapted a luminescence proximity homogeneous assay (ALPHA-screen)27 to the BET bromodomains. Binding of a tetra-acetylated Histone H4 peptide to BRD4 was strongly inhibited by (+)-JQ1, with IC50 values of 77 nM and 33 nM for the first and second bromodomain, respectively (Fig. 1e). The IC50 for the (−)-JQ1 stereoisomer against BRD4(1) and for (+)-JQ1 against CREBBP were both estimated to be above 10,000 nM (Fig. 1e). Thus, (+)-JQ1 represents a potent, highly specific and Kac competitive inhibitor for the BET family of bromodomains.
(+)-JQ1 binds to the acetyl-lysine binding site of BET-family bromodomains
In order to establish the binding mode of JQ1 we determined co-crystal structures using racemic material and purified, recombinant BRD4(1) and BRD2(2) (for data collection and refinement statistics see Supplementary Table 4). The determined high resolution structures revealed only the (+)-JQ1 enantiomer bound directly into the Kac binding site (Fig. 2, Fig. 3a, b). Similar to interactions observed in acetyl-lysine complexes28, the triazole ring formed a hydrogen bond with the evolutionarily conserved asparagine (Asn140 in BRD4(1) and Asn429 in BRD2(2); Fig. 2a). The inhibitor showed an extraordinary shape complementarity with the Kac binding site, occupying the entire binding pocket (Fig. 2c, d). In both complexes, ligand binding was stabilized by hydrophobic interactions with conserved BET residues in the ZA- and BC-loop regions (Fig. 3a, b). Structural and sequence comparison showed high conservation of the BET Kac binding pocket, but revealed a number of differences in loop regions lining the binding cavity that could be explored for future development of isoform specific inhibitors (Fig. 3a, b, c).
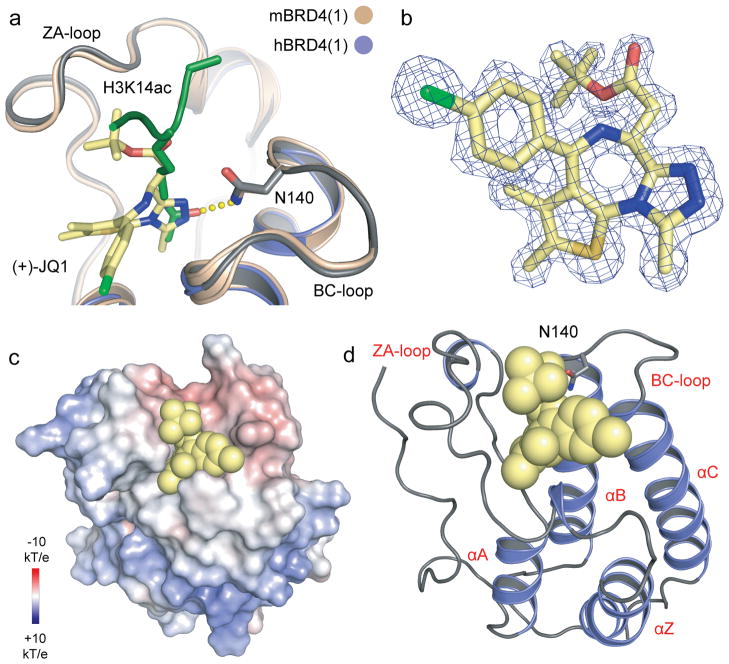
Figure 2. Characterization of BET complexes with (+)-JQ1.
a, Superimposition of the mouse BRD4(1)/H3K14ac peptide complex28 with the human BRD4(1)/(+)-JQ1 complex structure. The hydrogen bond formed to the conserved asparagine (N140) in the peptide complex is shown as yellow dots. b, 2Fo-Fc map of (+)-JQ1 in complex with BRD4(1) contoured at 2σ. c, Electrostatic surface of BRD4(1) in complex with (+)-JQ1. The ligand is shown as a CPK model demonstrating the excellent shape complimentarity with the protein acetylated lysine receptor site. d, Ribbon diagram of the complex of human BRD4(1) with (+)-JQ1 in CPK representation. The main secondary structural elements and the conserved active site asparagine side chain (N140) are labelled.
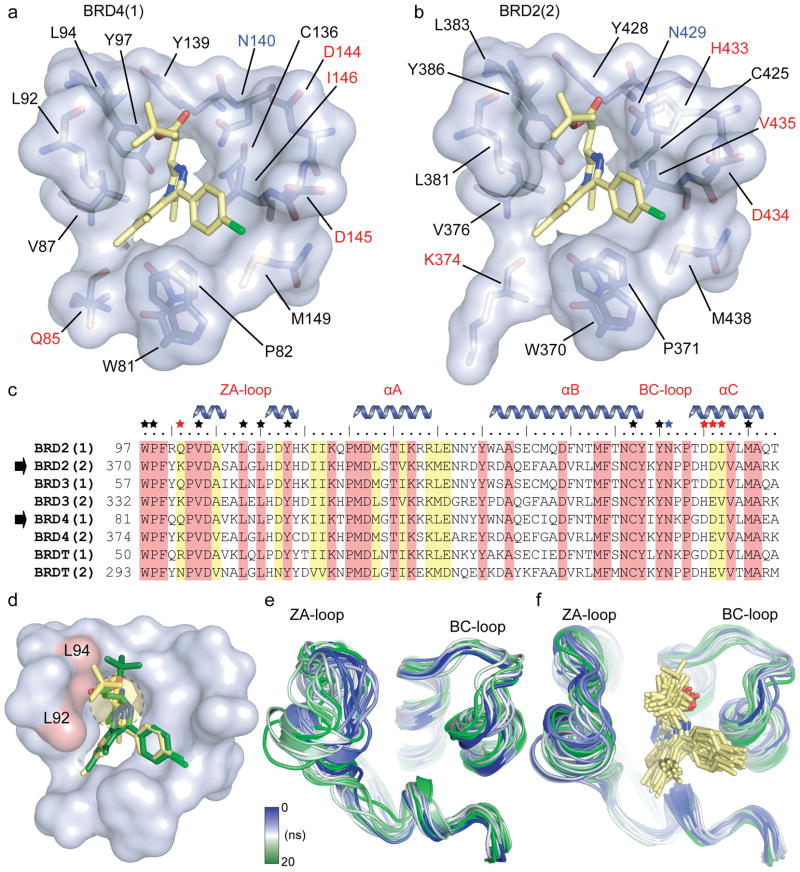
Figure 3. Binding site comparison between N- and C- terminal bromodomains in complex with (+)-JQ1.
a, The acetyl-lysine binding pocket of BRD4(1) is shown as a semitransparent surface with contact residues labelled and depicted in stick representation. Carbon atoms in (+)-JQ1 are coloured yellow to distinguish them from protein residues. b, The acetyl-lysine binding pocket of BRD2(2) is shown in identical representation and orientation as described in (a). c, Protein sequence alignment of the human BET subfamily highlighting conserved (red) and similar (yellow) residues. Major bromodomain structural elements are shown. The side-chain contacts with (+)-JQ1 are annotated with a black star. Contacts which differ between the N- and C-terminal BET bromodomains (red star) are highlighted. The family conserved asparagine is indicated by a blue star. d, Models of (+)-JQ1 (in yellow) and (−)-JQ1 (in green) docked into the BRD4(1) binding site. The steric clashes of the (−)-JQ1 stereo-isomer with Leu92 and Leu94 are highlighted in red. e, MD simulation demonstrating the flexibility of the ZA- and BC- loops of the BRD4(1) apo-structure. Shown are the backbone of BRD4(1) during a 20 ns simulation as snapshots separated by 1 ns intervals. The different structures are distinguished by colours changing from blue to green as indicated in the inset. f, MD simulation of the BRD4(1)/(+)-JQ1 complex depicted in 1 ns snapshots as described in (e).
Docking of either isomer of JQ1 to BRD4(1) resulted in excellent fit of (+)-JQ1 in a position of perfect overlap to the crystallographically determined binding mode, while (−)-JQ1 resulted in an energetically unfavourable conformation with significant distortion of the diazepine ring system due to steric clashes with residues of the ZA-loop region (Fig. 3d). To explore the dynamic features of BET bromodomains, we carried out 20 ns molecular dynamics simulations (MD) of BRD4(1) in the absence and presence of (+)-JQ1. The simulations revealed little displacement of the protein helixes, but the loop regions surrounding the acetyl-lysine binding site exhibited significant conformational flexibility. Furthermore, these loops were much more flexible in the absence (Fig. 3e) than in the presence (Fig. 3f) of the inhibitor, suggesting that (+)-JQ1 stabilized the Kac binding cavity. In all cases, MD simulation energies converged (Supplementary Fig. 2).
JQ1 displaces BRD4 from nuclear chromatin in cells
To establish whether JQ1 binds bromodomains competitively with chromatin in a cellular environment, we performed fluorescence recovery after photobleaching (FRAP) experiments. Prior research has demonstrated the utility of FRAP in assessing the pace of lateral redistribution of human bromodomains17,29. Human osteosarcoma cells (U2OS) transfected with a GFP-BRD4 exhibit a time-dependent recovery of fluorescence intensity (Fig. 4a, b). In the presence of JQ1 (500 nM), observed recovery is immediate, suggesting displaced and freely diffusing nuclear BRD4 (Fig. 4a, b). Cellular FRAP studies confirmed that effects on the mobile fraction of BRD4 are limited to the biochemically active (+)-JQ1 stereoisomer (Supplemental Fig. 3).
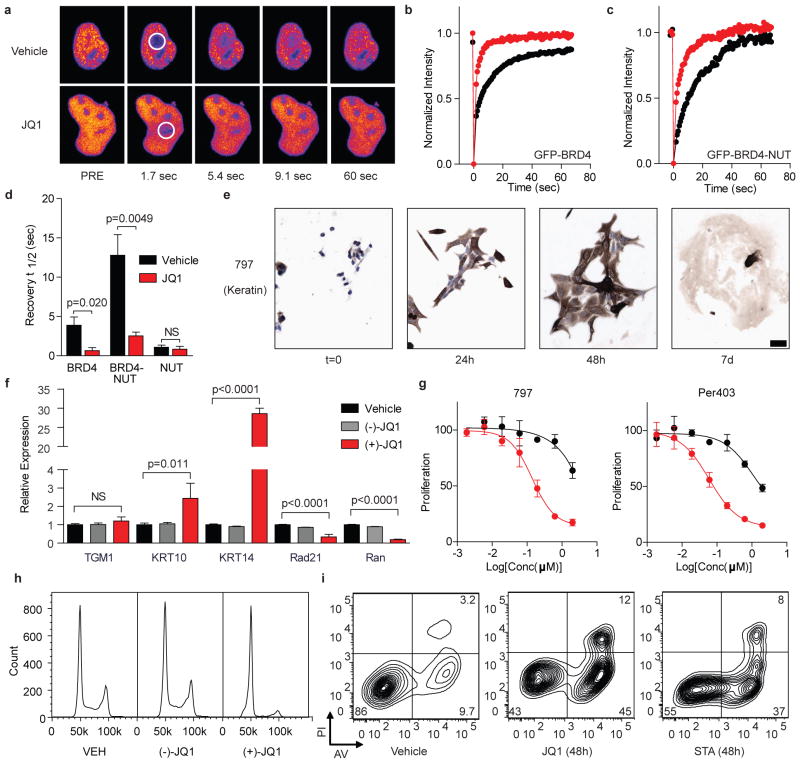
Figure 4. JQ1 binds BRD4 competitively with chromatin resulting in differentiation and growth arrest of NMC cells.
a, Fluorescence recovery after photobleaching (FRAP) of GFP-BRD4 demonstrates enhanced recovery in the presence of JQ1. Nuclei are false-colored in proportion to fluorescence intensity. White circles indicate target regions of photobleaching. b–c, JQ1 accelerates fluorescence recovery in FRAP experiments performed with transfected (b) GFP-BRD4 and (c) GFP-BRD4-NUT. d, Quantitative comparison of time to half-maximal fluorescence recovery for FRAP studies (b–c, Supplementary Fig. 3a). Data represent the mean ± s.d. (n = 5), and are annotated with p-values as obtained from a two-tailed t-test. e, Differentiation of NMC cells by JQ1 (500 nM) is prompt and characterized by a marked increase in cytokeratin expression (AE1/AE3; 10x, scale bar is 50 μm). f, Comparative gene expression studies of (+)-JQ1 (red; 250 nM, 48 h) versus (−)-JQ1 (gray; 250 nM, 48 h) and vehicle (black) confirm squamous differentiation. Data represent the mean ± s.d. (n = 3), and are annotated with p-values as obtained from a two-tailed t-test. g, Growth effects of BRD4 inhibition on BRD4-NUT dependent cell lines. Cells were incubated with (+)-JQ1 (red circles) or (−)-JQ1 (black circles) and monitored for proliferation after 72 hours. (+)-JQ1 uniquely attenuates proliferation by NMC cell lines. Data is presented as mean ± s.d. (n = 3). Curve fit was calculated by logistic regression. h, Flow cytometry for DNA content in NMC 797 cells. (+)-JQ1 (250 nM, 48 h) induces a G1 arrest compared to (−)-JQ1 (250 nM) and vehicle control. i, Flow cytometric analysis of NMC 797 squamous carcinoma cells treated with vehicle, JQ1 or staurosporine (STA), as indicated. PI, propidium iodide. AV, annexin-V.
Having demonstrated potent, selective binding to BRD4 in homogeneous and cell-based assays, we became interested to explore effects of JQ1 on disease-relevant phenotypes. Prior studies have established that the pathogenic BRD4-NUT fusion protein arising from t(15;19) translocation in NMC binds avidly to discrete foci of acetylated chromatin, conferring a proliferative advantage and differentiation block17. Using FRAP, we assessed the ability of JQ1 to target directly the BRD4-NUT oncoprotein. Compared to a vehicle control, JQ1 (500 nM) markedly accelerated time to half fluorescence recovery in photobleached regions of cells transfected with GFP-BRD4-NUT (Fig. 4c, d). Importantly, no effect was observed on redistribution of GFP-NUT (Supplementary Fig. 3). These data are consistent with competitive binding of JQ1 to BRD4 in cultured cells.
JQ1 induces squamous differentiation and growth arrest in BRD4-dependent carcinoma
Direct inhibition of gene products expressed from recurrent, oncogenic translocations is a validated therapeutic approach in cancer30,31. We thus endeavoured to establish the consequences of competitive inhibition of BRD4-NUT in NMC. A characteristic feature of NMC is the appearance of discrete nuclear speckles of the BRD4-NUT oncoprotein by NUT-directed immunohistochemistry (IHC)32. Treatment of the patient-derived 797 NMC cell line for 48 hours with JQ1 (500 nM) effaces nuclear foci, producing diffuse nuclear NUT staining by IHC (Supplementary Fig. 3e). In a dose- and time-dependent manner, JQ1 provokes a differentiation phenotype in NMC cell lines, featuring cell spreading and flattening, open chromatin and striking spindle morphology (Fig. 4e; Supplementary Fig. 4). Differentiation is prompt (< 24 hours) and characterized by dramatic changes in cell shape accompanied by markedly augmented expression of cytokeratin, a hallmark of squamous differentiation (Fig. 4e). After seven days in culture with sub-micromolar exposures to JQ1, terminal differentiation is observed. In this manner, JQ1 phenocopies the morphologic changes and increased keratin expression observed with BRD4-NUT silencing by RNA interference (Supplementary Fig. 5)17. Corroborating these studies, expression analysis of three canonical squamous tissue genes by RT-PCR identified marked (30-fold) induction of Keratin-14 by (+)-JQ1 in NMC 797 cells (Fig. 4f). The modest induction of keratin-10 without affecting epidermal transglutaminase (TGM1) may indicate differentiation toward thoracic squamous epithelium, consistent with the mediastinal primary tumor from which NMC 797 cells derive33. Induction of differentiation with strong (3+) keratin staining is progressive over 72 h, as determined by quantitative IHC analysis (Supplementary Fig. 6). Supporting an on-target mechanism-of-action, the (−)-JQ1 enantiomer is comparatively inactive in NMC, and a non-BRD4-dependent squamous carcinoma cell line (TE10) fails to exhibit differentiation effects of active JQ1 (Supplementary Fig. 4c).
In BRD4-dependent NMC cells, differentiation is expectedly accompanied by growth arrest, evidenced by reduced Ki67 staining (Supplementary Fig. 7), sustained inhibition of proliferation (Fig. 4g; Supplementary Fig. 8) and G1 cell cycle arrest (Fig. 4h). To further understand the observed G1 arrest and to confirm an effect of JQ1 on known BRD4-dependent genes, we performed quantitative RT-PCR for Rad21 and Ran4. (+)-JQ1 potently decreased expression of both BRD4 target genes, whereas (−)-JQ1 had no effect (Fig. 4f). Early and late apoptosis were assessed with annexin-V and propidium iodide staining to ascertain whether the antiproliferative effect and irreversible differentiation was accompanied by cell death. Indeed, JQ1 induces immediate and progressive apoptosis in BRD4-dependent human carcinoma cells, without triggering significant growth arrest or cell death in cell lines lacking the BRD-NUT fusion (Supplementary Fig. 8, 9).
Pharmacodynamic and anti-tumor efficacy of JQ1 in xenograft models of NMC
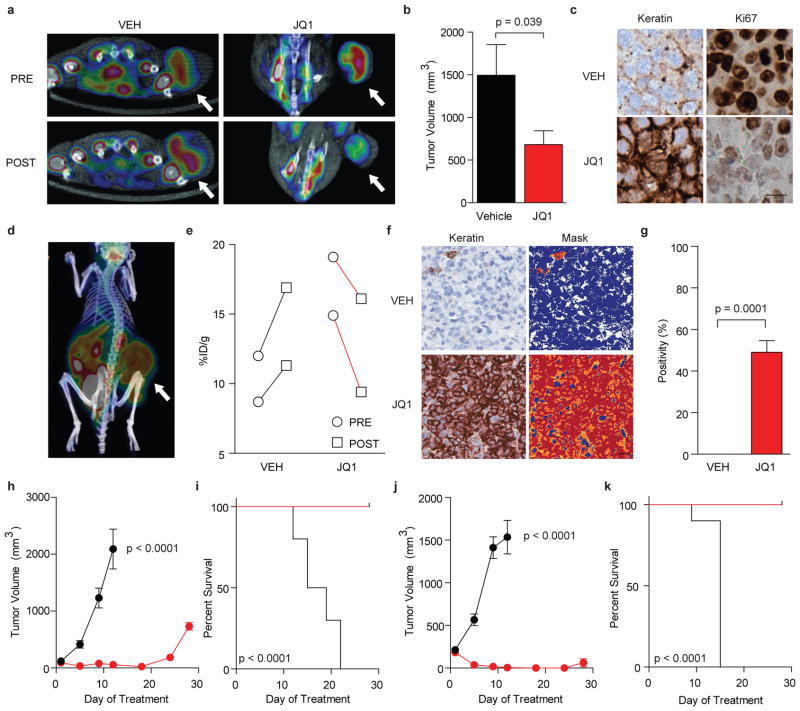
To determine whether JQ1 could attenuate the growth of BRD4-dependent carcinoma as a single agent in vivo, we developed three mouse xenograft models of NMC in mice. First, short-term treatment studies were performed in NMC 797 xenografts with positron-emission tomography (PET) imaging of 18F-fluorodeoxyglucose (FDG) uptake as a primary endpoint to explore whether activity of JQ1 could be demonstrated by non-invasive imaging. Matched cohorts of mice with established tumors were randomized to treatment with JQ1 (50 mg kg−1) or vehicle, administered by daily intraperitoneal injection. Prior to randomization, and after four days of therapy, mice were evaluated by FDG-PET imaging. A marked reduction in FDG uptake was observed with JQ1 treatment, whereas vehicle-treated animals demonstrated progressive disease (Fig. 5a). Tumor-volume measurements confirmed a reduction in tumor growth with JQ1 treatment (Fig. 5b and Supplementary Fig. 10). JQ1 was well tolerated at this dose and schedule without overt signs of toxicity or weight loss (Supplementary Fig. 10b).
Figure 5. JQ1 promotes differentiation, tumor regression and improved survival in murine models of NMC.
a, PET imaging of murine NMC 797 xenografts. FDG uptake in xenograft tumors is reduced by 50 mg kg −1 JQ1 treatment compared to vehicle control. b, Tumor volume is reduced in mice with established disease (NMC 797 xenografts) treated with 50 mg kg −1 daily JQ1 compared to vehicle control. A significant response to therapy is observed by two-tailed t-test at 14 days (p = 0.039). Data represent the mean ± s.d. (n = 7). c, Histopathological analysis of NMC 797 tumors excised from animals treated with JQ1 reveals induction of keratin expression (AE1/AE3, 40x) and impaired proliferation (Ki67, 40x), as compared to vehicle-treated animals (scale bar is 20 μm). d, Viability of patient-derived NMC 11060 xenografts was confirmed by PET imaging. e, Therapeutic response of primary 11060 NMC xenografts to (+)-JQ1 (50 mg kg −1 daily for four days) was demonstrated by PET imaging. f, Histopathological analysis of primary NMC 11060 tumors excised from animals treated with (+)-JQ1 reveals induction of keratin expression (AE1/AE3, 20x; scale bar is 20 μm), compared to vehicle-treated animals. Quantitative analysis of keratin induction was performed using image masking (f, right panel) and pixel positivity analysis (g). A significant response to therapy is observed by two-tailed t-test (p = 0.0001). Data represent the mean ± s.d. of three independent wide microscopic fields. Comparative images of stained excised tumors and quantitative masks are provided in Supplementary Figure 14. h–k, (+)-JQ1 (50 mg kg −1 daily for 18 days) produces a decrease in tumor volume (h, j) and promotes improved survival (i, k) in patient-derived 11060 (h, i) and Per403 (j, k) NMC xenograft models (n=10 in all groups). A significant response to therapy is observed for tumor volume by two-tailed t-test (p < 0.0001) and for overall survival by a log-rank test (p < 0.0001).
To confirm that the anti-neoplastic effect observed with JQ1 treatment was associated with target engagement, sectioned tumor tissue was examined for the BRD4-NUT oncoprotein. As presented in Supplementary Figure 11, JQ1 treatment resulted in effacement of NUT nuclear speckles, consistent with competitive binding to nuclear chromatin. Cell spreading and increased keratin expression confirmed induction of squamous differentiation (Fig. 5c). Decreased nuclear Ki67 and increased TUNEL staining in treated animals confirmed an ongoing anti-proliferative, pro-apoptotic effect (Supplementary Fig. 11). To quantify the pharmacodynamic biomarker of tumor keratin expression, we established protocols for automated IHC image acquisition and analysis. Paired samples from treated and untreated animals were prepared and analyzed using standardized protocols and commercially-available software (ImageScope; Aperio Technologies), demonstrating that JQ1 induced strong (3+) keratin expression in NMC 797 xenografts, (Supplementary Fig. 12).
In parallel with these studies, we had occasion to care for a 29 year-old patient with widely metastatic BRD4-NUT positive NMC arising from the mediastinum. With the goal of developing a more clinically-relevant disease model, we established short-term cultures (11060 cells) using discarded clinical material obtained from pleural fluid draining from a palliative chest tube. As presented in Supplementary Figure 13, in vitro studies confirmed the stereospecific, potent effect of (+)-JQ1 on cellular viability (IC50 = 4 nM), growth and cell cycle progression. Four animals engrafted with patient-derived tumor material developed measurable disease, which was strongly FDG-avid by PET imaging (Fig. 5d). Animals were randomly assigned to vehicle or (+)-JQ1 treatment. Prior to treatment and after four days of therapy, mice were evaluated by PET imaging. A marked response in FDG uptake was observed with (+)-JQ1 treatment, whereas vehicle-treated animals demonstrated progressive disease (Fig. 5e). Tumor material prepared for quantitative IHC analysis demonstrated induction of keratin expression following (+)-JQ1 treatment (Fig. 5f–g, Supplementary Fig. 14) in this minimally-passaged NMC xenograft model.
To confirm the translational potential of direct-acting BRD4 inhibition in NMC, we further adapted the patient-derived 11060 cells to expansion in vivo, and performed definitive efficacy studies. Marked tumor regression and improved overall survival were observed, following only 18 days of well-tolerated therapy with (+)-JQ1 (Fig. 5h–i). These results were recapitulated in a third NMC xenograft model, using Per403 cells (Fig 5j–k, Supplementary Fig. 15). Together, these data establish in vivo proof-of-concept for targeting BRD4 with JQ1 in NMC.
Discussion
Across the complex landscape of the cancer genome, recurrent chromosomal rearrangements comprise a compelling subset of clear, genetic targets in cancer. As evidenced by the successful development of first- and second-generation kinase inhibitors targeting BCR-ABL in CML, well-characterized probe compounds34,35, high-resolution crystallographic data36, translational research studies37, and informative murine models38, where available, provide an optimal platform for ligand discovery and target validation. Herein, we provide evidence supporting the BRD4-NUT fusion as a therapeutic target in an incurable, genetically-defined human squamous carcinoma, using a novel BRD4-directed small molecule inhibitor.
Beyond NUT-midline carcinoma, BET-family bromodomains contribute to other neoplastic and non-neoplastic diseases. BRD4 targets the P-TEFb complex to mitotic chromosomes resulting in expression of growth promoting genes such as c-Myc12,14 and the well established cancer target Aurora B39. BET family members have been recognized as essential genes for the replication of viruses40,41 and in mediating inflammatory responses42. Thus, the availability of (+)-JQ1 will prompt informative research broadly in developmental and disease biology. JQ1 possesses many desirable qualities of a chemical probe, such as high target potency in homogeneous and cellular assays, a well-characterized profile of selectivity, synthetic accessibility and herein proven utility in experimental biology18,19. We have also found JQ1 to exhibit few off-target effects on cellular receptors and excellent pharmacokinetic properties including 49% oral bioavailability (Supplementary Fig. 16, 17 and Supplementary Table 5), establishing the plausibility of developing drug-like derivatives for therapeutic application.
The discovery and optimization of small-molecule inhibitors of epigenetic targets is a major focus of current biomedical research. We sought to meet the challenge of developing potent, selective inhibitors of epigenetic readers. Here, we present a first, thoroughly characterized inhibitor of the BET-family of bromodomains. The approach outlined herein further establishes the feasibility of abrogating protein-protein interactions with small molecules, and targeting additional epigenetic readers for ligand discovery.
Methods Summary
The inhibitor JQ1 was synthesized in both racemic and enantiomerically pure format using the synthetic route outlined in Scheme S1 and Scheme S2 and its structure was fully characterized. Human bromodomains were expressed in bacteria as His-tagged proteins and were purified by nickel-affinity and gel-filtration chromatography. Proteins integrity was assessed by SDS-PAGE and Electro-spray Mass Spectrometry on an Agilent 1100 Series LC/MSD TOF. All crystallizations were carried out at 4 °C using the sitting drop vapour-diffusion method. X-ray diffraction data were collected at the Swiss Light source beamline X10SA, or using a Rigaku FR-E generator. Structures were determined by molecular replacement. Isothermal titration calorimetry experiments were performed at 15 °C on a VP-ITC titration microcalorimeter (MicroCal™). Thermal melting experiments were carried out on a Mx3005p RT- PCR machine (Stratagene) using SYPRO Orange as a fluorescence probe. Dose-ranging small-molecule studies of proliferation were performed in white, 384-well plates (Corning) in DMEM media containing 10 % FBS. Compounds were delivered with a PerkinElmer JANUS pin-transfer robot and Envision multilabel plate-reader, using a commercial assay (Cell TiterGlo). Murine xenografts were established by injecting NMC cells in 30 % Matrigel (BD Biosciences) into the flank of 6 week-old female NCr nude mice (Charles River Laboratories). Tumor measurements were assessed by caliper measurements, and volume calculated using the formula Vol = 0.5 × L × W2. All mice were humanely euthanized, and tumors were fixed in 10 % formalin for histopathological examination. Quantitative immunohistochemistry was performed using the Aperio Digital Pathology Environment (Aperio Technologies, Vista, CA) at the DF/HCC Core Laboratory at the Brigham and Women’s Hospital.
Supplementary Material
Acknowledgments
We are grateful to Drs. Udo Oppermann, Susanne Müller, Stephen Sallan, Christopher Lathan, Peter Rahl, Richard Young, Kevin Lee and Katharin Shaw for thoughtful discussions and sharing unpublished information; Kachicholu Agu, Stephen Johnston and Li Li for analytical chemistry support; John Daley for flow cytometry support; Teri Bowman, Tyler Caron, Corey Marvin and Dr. Scott Rodig for IHC; and Adam Bass for sharing cell lines. The Structural Genomics Consortium is a registered charity (number 1097737) that receives funds from the Canadian Institutes for Health Research, the Canadian Foundation for Innovation, Genome Canada through the Ontario Genomics Institute, GlaxoSmithKline, Karolinska Institutet, the Knut and Alice Wallenberg Foundation, the Ontario Innovation Trust, the Ontario Ministry for Research and Innovation, Merck & Co., Inc., the Novartis Research Foundation, the Swedish Agency for Innovation Systems, the Swedish Foundation for Strategic Research and the Wellcome Trust. This research was supported by a Graduate Fellowship from the Chemistry-Biochemistry-Biology Interface Program at the University of Notre Dame, NIGMS T32-075762 (to Y.S.), the DF/HCC (to C.A.F. and J.E.B.), the National Institutes of Health, the Burroughs Wellcome Fund, and the Leukemia & Lymphoma Society (to J.E.B.).
Footnotes
Supplementary Information accompanies the paper on www.nature.com/nature.
Author Contributions P.F., J.Q., S.K. and J.E.B. designed the study, analyzed data and wrote the manuscript. P.F. and S.P. performed and analyzed biophysical studies. J.Q. and J.E.B. designed JQ1 and established the synthetic routes. Y.S. and O.W. completed docking and molecular dynamics studies. O.F. performed and analyzed DSF. M.P. and T.H. performed and analyzed alpha-screen assays. W.B.S., M.J.C. and J.E.B. performed in vitro NMC studies and IHC. E.M.M performed flow cytometry studies. E.M.M. and N.W. performed proliferation studies. T.T.H., M.J.C., C.A.F. and J.E.B. completed FRAP studies. M.M. and B.S. performed expression analysis. Y.W., A.L.C. and A.L.K. completed in vivo efficacy studies. T.K. and I.F. expressed and purified proteins. S.K. and J.E.B. supervised the research.
Author Information Atomic coordinates and structure factors for the reported crystal structures have been deposited with the Protein Data Bank under accession codes 2OSS (BRD4(1)), 3MXF (BRD4(1)/(+)-JQ1) and 3ONI (BRD2(2)/(+)-JQ1). Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
Full Methods and any associated references are available online.
References
- 1.Ptashne M. Binding reactions: epigenetic switches, signal transduction and cancer. Curr Biol. 2009;19:R234–241. doi: 10.1016/j.cub.2009.02.015. [DOI] [PubMed] [Google Scholar]
- 2.Schreiber SL, Bernstein BE. Signaling network model of chromatin. Cell. 2002;111:771–778. doi: 10.1016/s0092-8674(02)01196-0. [DOI] [PubMed] [Google Scholar]
- 3.Marushige K. Activation of chromatin by acetylation of histone side chains. Proc Natl Acad Sci U S A. 1976;73:3937–3941. doi: 10.1073/pnas.73.11.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dey A, Nishiyama A, Karpova T, McNally J, Ozato K. Brd4 marks select genes on mitotic chromatin and directs postmitotic transcription. Mol Biol Cell. 2009;20:4899–4909. doi: 10.1091/mbc.E09-05-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Owen DJ, et al. The structural basis for the recognition of acetylated histone H4 by the bromodomain of histone acetyltransferase gcn5p. Embo J. 2000;19:6141–6149. doi: 10.1093/emboj/19.22.6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zeng L, Zhou MM. Bromodomain: an acetyl-lysine binding domain. FEBS Lett. 2002;513:124–128. doi: 10.1016/s0014-5793(01)03309-9. [DOI] [PubMed] [Google Scholar]
- 7.Yang XJ. Multisite protein modification and intramolecular signaling. Oncogene. 2005;24:1653–1662. doi: 10.1038/sj.onc.1208173. [DOI] [PubMed] [Google Scholar]
- 8.Yang Z, et al. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell. 2005;19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 9.Peng J, Zhu Y, Milton JT, Price DH. Identification of multiple cyclin subunits of human P-TEFb. Genes Dev. 1998;12:755–762. doi: 10.1101/gad.12.5.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marshall NF, Price DH. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J Biol Chem. 1995;270:12335–12338. doi: 10.1074/jbc.270.21.12335. [DOI] [PubMed] [Google Scholar]
- 11.Marshall NF, Peng J, Xie Z, Price DH. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J Biol Chem. 1996;271:27176–27183. doi: 10.1074/jbc.271.43.27176. [DOI] [PubMed] [Google Scholar]
- 12.Phelps MA, et al. Clinical response and pharmacokinetics from a phase 1 study of an active dosing schedule of flavopiridol in relapsed chronic lymphocytic leukemia. Blood. 2009;113:2637–2645. doi: 10.1182/blood-2008-07-168583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rahl PB, et al. c-Myc Regulates Transcriptional Pause Release. Cell. 2010;141:4323–4445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Z, He N, Zhou Q. Brd4 recruits P-TEFb to chromosomes at late mitosis to promote G1 gene expression and cell cycle progression. Mol Cell Biol. 2008;28:967–976. doi: 10.1128/MCB.01020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.French CA, Miyoshi I, Aster JC, et al. BRD4 bromodomain gene rearrangement in aggressive carcinoma with translocation t (15;19) Am J Pathol. 2001;159(6):1987–1992. doi: 10.1016/S0002-9440(10)63049-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.French CA, et al. BRD4-NUT fusion oncogene: a novel mechanism in aggressive carcinoma. Cancer Res. 2003;63:304–307. [PubMed] [Google Scholar]
- 17.French CA, et al. BRD-NUT oncoproteins: a family of closely related nuclear proteins that block epithelial differentiation and maintain the growth of carcinoma cells. Oncogene. 2008;27:2237–2242. doi: 10.1038/sj.onc.1210852. [DOI] [PubMed] [Google Scholar]
- 18.Frye SV. The art of the chemical probe. Nat Chem Biol. 6:159–161. doi: 10.1038/nchembio.296. [DOI] [PubMed] [Google Scholar]
- 19.Oprea TI, et al. A crowdsourcing evaluation of the NIH chemical probes. Nat Chem Biol. 2009;5:441–447. doi: 10.1038/nchembio0709-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyoshi S, Ooike S, Iwata K, Hikawa H, Sugaraha K. ANTITUMOR AGENT. MITSUBISHI TANABE PHARMA CORPORATION; 2009. pp. 1–37. [Google Scholar]
- 21.Adachi K, et al. MITSUBISHI TANABE PHARMA CORPORATION; 2006. pp. 1–240. [Google Scholar]
- 22.Sueoka H, Komatsu H, Kobayashi H, Ehara S. Thienotriazolodiazepine compounds and medicinal uses thereof. YOSHITOMI PHARMACEUTICAL INDUSTRIES, LTD; 1998. pp. 1–50. [Google Scholar]
- 23.VonVoigtlander PF, Straw RN. Alprazolam: Review of Pharmacological, Pharmacokinetic and Clinical Data. Drug Development Research. 1985;6:1–12. [Google Scholar]
- 24.Niesen FH, Berglund H, Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat Protoc. 2007;2:2212–2221. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 25.Fedorov O, et al. A systematic interaction map of validated kinase inhibitors with Ser/Thr kinases. Proc Natl Acad Sci U S A. 2007;104:20523–20528. doi: 10.1073/pnas.0708800104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bullock AN, et al. Structural basis of inhibitor specificity of the human protooncogene proviral insertion site in moloney murine leukemia virus (PIM-1) kinase. J Med Chem. 2005;48:7604–7614. doi: 10.1021/jm0504858. [DOI] [PubMed] [Google Scholar]
- 27.Quinn AM, et al. A homogeneous method for investigation of methylation-dependent protein-protein interactions in epigenetics. Nucleic Acids Res. 2010;38:e11. doi: 10.1093/nar/gkp899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vollmuth F, Blankenfeldt W, Geyer M. Structures of the dual bromodomains of the P-TEFb-activating protein Brd4 at atomic resolution. J Biol Chem. 2009;284:36547–36556. doi: 10.1074/jbc.M109.033712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dey A, et al. A bromodomain protein, MCAP, associates with mitotic chromosomes and affects G(2)-to-M transition. Mol Cell Biol. 2000;20:6537–6549. doi: 10.1128/mcb.20.17.6537-6549.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang ME, et al. Use of all-trans retinoic acid in the treatment of acute promyelocytic leukemia. Blood. 1988;72:567–572. [PubMed] [Google Scholar]
- 31.Druker BJ, et al. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med. 2001;344:1031–1037. doi: 10.1056/NEJM200104053441401. [DOI] [PubMed] [Google Scholar]
- 32.Haack H, et al. Diagnosis of NUT midline carcinoma using a NUT-specific monoclonal antibody. Am J Surg Pathol. 2009;33:984–991. doi: 10.1097/PAS.0b013e318198d666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Toretsky JA, et al. Translocation (11;15;19): a highly specific chromosome rearrangement associated with poorly differentiated thymic carcinoma in young patients. Am J Clin Oncol. 2003;26:300–306. doi: 10.1097/01.COC.0000020960.98562.84. [DOI] [PubMed] [Google Scholar]
- 34.Buchdunger E, et al. Selective inhibition of the platelet-derived growth factor signal transduction pathway by a protein-tyrosine kinase inhibitor of the 2-phenylaminopyrimidine class. Proc Natl Acad Sci U S A. 1995;92:2558–2562. doi: 10.1073/pnas.92.7.2558. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Buchdunger E, et al. Inhibition of the Abl protein-tyrosine kinase in vitro and in vivo by a 2-phenylaminopyrimidine derivative. Cancer Res. 1996;56:100–104. [PubMed] [Google Scholar]
- 36.Schindler T, et al. Structural mechanism for STI-571 inhibition of abelson tyrosine kinase. Science. 2000;289:1938–1942. doi: 10.1126/science.289.5486.1938. [DOI] [PubMed] [Google Scholar]
- 37.Druker BJ, et al. Effects of a selective inhibitor of the Abl tyrosine kinase on the growth of Bcr-Abl positive cells. Nat Med. 1996;2:561–566. doi: 10.1038/nm0596-561. [DOI] [PubMed] [Google Scholar]
- 38.le Coutre P, et al. In vivo eradication of human BCR/ABL-positive leukemia cells with an ABL kinase inhibitor. J Natl Cancer Inst. 1999;91:163–168. doi: 10.1093/jnci/91.2.163. [DOI] [PubMed] [Google Scholar]
- 39.You J, et al. Regulation of aurora B expression by the bromodomain protein Brd4. Mol Cell Biol. 2009;29:5094–5103. doi: 10.1128/MCB.00299-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.You J, et al. Kaposi’s sarcoma-associated herpesvirus latency-associated nuclear antigen interacts with bromodomain protein Brd4 on host mitotic chromosomes. J Virol. 2006;80:8909–8919. doi: 10.1128/JVI.00502-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abbate EA, Voitenleitner C, Botchan MR. Structure of the papillomavirus DNA-tethering complex E2:Brd4 and a peptide that ablates HPV chromosomal association. Mol Cell. 2006;24:877–889. doi: 10.1016/j.molcel.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 42.Huang B, Yang XD, Zhou MM, Ozato K, Chen LF. Brd4 coactivates transcriptional activation of NF-kappaB via specific binding to acetylated RelA. Mol Cell Biol. 2009;29:1375–1387. doi: 10.1128/MCB.01365-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cole PA. Chemical probes for histone-modifying enzymes. Nat Chem Biol. 2008;4:590–597. doi: 10.1038/nchembio.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.