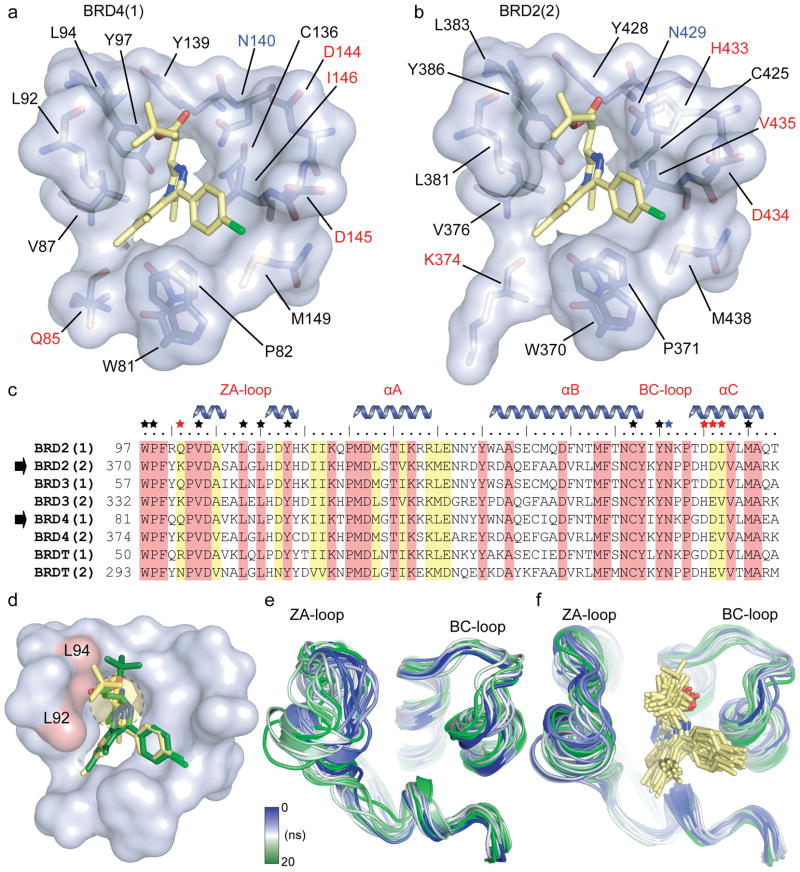
Figure 3. Binding site comparison between N- and C- terminal bromodomains in complex with (+)-JQ1.
a, The acetyl-lysine binding pocket of BRD4(1) is shown as a semitransparent surface with contact residues labelled and depicted in stick representation. Carbon atoms in (+)-JQ1 are coloured yellow to distinguish them from protein residues. b, The acetyl-lysine binding pocket of BRD2(2) is shown in identical representation and orientation as described in (a). c, Protein sequence alignment of the human BET subfamily highlighting conserved (red) and similar (yellow) residues. Major bromodomain structural elements are shown. The side-chain contacts with (+)-JQ1 are annotated with a black star. Contacts which differ between the N- and C-terminal BET bromodomains (red star) are highlighted. The family conserved asparagine is indicated by a blue star. d, Models of (+)-JQ1 (in yellow) and (−)-JQ1 (in green) docked into the BRD4(1) binding site. The steric clashes of the (−)-JQ1 stereo-isomer with Leu92 and Leu94 are highlighted in red. e, MD simulation demonstrating the flexibility of the ZA- and BC- loops of the BRD4(1) apo-structure. Shown are the backbone of BRD4(1) during a 20 ns simulation as snapshots separated by 1 ns intervals. The different structures are distinguished by colours changing from blue to green as indicated in the inset. f, MD simulation of the BRD4(1)/(+)-JQ1 complex depicted in 1 ns snapshots as described in (e).