Abstract
Using stabilized forms of β1 adrenergic and A2A adenosine G-protein-coupled receptors, we applied Biacore to monitor receptor activity and characterize binding constants of small-molecule antagonists spanning >20,000 fold in affinity. We also illustrate an improved method for tethering His-tagged receptors on NTA chips to yield stable, high-capacity, high-activity surfaces, as well as a novel approach to regenerate receptor-binding sites. Based on our success with this approach, we expect that the combination of stabilized receptors with biosensor technology will become a common method for characterizing members of this receptor family.
Introduction
Since 1990 Biacore biosensors have been used to study protein interactions in real time without labeling [1]. And the past five years has seen a significant surge in the application of the technology for small-molecule analysis [2,3]. In contrast, the application of biosensors to study membrane-associated systems such as G-protein-coupled receptors (GPCRs1) is still in its infancy [4–9].
The challenges of studying membrane-associated receptors with optical biosensors are two fold. First, most GPCRs are expressed at low levels and are unstable when extracted from the cell membrane. This makes it tricky to immobilize these receptors onto the sensor surface while maintaining high levels of activity. Second, the ligands for most GPCRs have low molecular weights (e.g., histamine (111 Da) and serotonin (176 Da)). This places an added burden on surface plasmon resonance (SPR) biosensor technology, which is mass based.
We envisioned that the approach of engineering stabilized GPCRs [10] for structural analysis [11,12] could provide excellent reagents for biosensor analysis. To validate this method, we used two receptors that contain point mutations which improve their thermostability and conformational homogeneity; a turkey β1 adrenergic receptor (β1AR*, with mutations R68S, Y227A, A282L, F237A, and F338M) [13,14] and a human A2A adenosine receptor (A2AR*, with mutations A54L, T88A, K122A, and V239A) [12,15].
We illustrate how Biacore technology allowed us to establish the benefits and limitations of different capturing methods and confirm the activity and stability of the immobilized receptors. In addition, we provide examples of the high-quality kinetic and affinity data available from Biacore analysis of GPCRs. Our success in combining stabilized receptors with the biosensor technology demonstrates the potential of this approach and should encourage the development of additional reagents for these challenging receptor systems.
Materials and Methods
Reagents and instrumentation
Studies were performed at 25°C using Biacore 2000 and S51 optical biosensors equipped with NTA (carboxymethylated dextran pre-immobilized with nitrilotriacetic acid) sensor chips (preconditioned with three one-minute pulses of 350 mM EDTA in running buffer and charged for 3 min with 500 uM Ni2+ in running buffer) and equilibrated with running buffer (20 mM Tris-HCl, 350 mM NaCl, 0.1% n-dodecyl-β-D-maltopyranoside, pH 7.8 supplemented with 1% or 5% DMSO). Compounds were purchased from Sigma (sigmaaldrich.com) and Tocris (tocris.com), detergent from Anatrace, sulfo-N-hydroxysuccinimide (sulfo-NHS) from BioRad (biorad.com), 1-ethyl-3-(3-dimethylaminpropyl)-carbodiimde hydrochloride (EDC) from GE Healthcare Bio-Science AB (biacore.com), and general laboratory reagents from Sigma and Fisher Scientific (fishersci.com).
Receptor expression, solubilization, and purification
Receptors were expressed in Trichoplusia ni (Tni) cells using the FastBac expression system (Invitrogen). Tni cells were grown in suspension in flasks EXCell 405 medium supplemented with 5% FBS and 1% chemically defined lipids (Invitrogen). Cells were infected with recombinant virus when cultures had reached a density 6 × 106 cells/mL, virus was added at a multiplicity of infection of 1. An equal volume of fresh medium was added immediately afterwards. Cells were harvested by centrifugation 72 h post infection.
All membrane preparation and solubilization steps were carried out with ice-cold buffers with the inclusion of the protease inhibitors, 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride (0.5 mM), leupeptin (2.5 μg/ml) and pepstatin A (3.5 μg/ml). Cells were pelleted from 1.5 L culture, homogenised and resuspended in 70 ml buffer B (40 mM Tris–HCl pH 7.6, 300 mM NaCl, 5 % glycerol, 0.001% CHS, 10 μM ZM 241385 or alprenolol). Membranes were first pelleted by centrifugation at 235,000g for 1 hour, after removal of the supernatant, membranes were re-suspended in 70 ml buffer B with the addition of two tablets of Complete EDTA free protease inhibitor tablets (Roche) and subsequently solubilized by addition of 1.5% decyl-β-D-maltopyranoside (DM) for 1 hour on ice followed by centrifugation at 235,000g for 60 min to remove unsolubilized material.
All protein purification steps were carried out at 4°C. The solubilised material was applied to a 5 ml Ni-NTA superflow cartridge (Qiagen) pre-equilibrated with buffer B with the addition of 0.15% DM. The column was washed at 1 ml/min with 10 column volumes of the same buffer and then eluted with a linear gradient (15 column volumes) from 5 to 400 M imidazole in Buffer B supplemented with 0.15% DM. Protein was detected with an on-line detector to monitor A280 and column fractions were collected and analyzed by SDS PAGE gel. Fractions containing the ca. 35 kDa protein were pooled and concentrated using a YM50 Amicon ultra-filtration membrane to a final volume of 200 μl. The protein sample was applied to a 10/30 S200 size exclusion column pre-equilibrated with buffer B with the addition of 0.1% DM (or exchange detergent) and eluted at 0.5 ml/min. Protein was detected with an on-line detector to monitor A280 and column fractions were collected and analyzed by SDS PAGE gel. Fractions containing the ca. 35 kDa protein were pooled and concentrated using a YM50 Amicon ultrafiltration membrane to a final concentration of 10 mg/ml and stored at −80°C.
NTA captures of β1AR* and A2AR*
For direct NTA capture, the purified receptor (either β1AR*-His10 or A2AR*-His10) was diluted 30 – 300X in running buffer and injected at 5 uL/min to achieve capture levels of >10,000 resonance units (RU). For capture-coupling, a flow cell surface was activated for five minutes (at 10 uL/min) with 1:1 0.1 M sulfo-NHS:0.4 M EDC prior to injection of the receptor. Both the captured and captured-coupled receptor surfaces were washed with running buffer for at least one hour. Activity of the receptor surfaces were evaluated using propranolol (for β1AR*) and xanthine amine congener (XAC; for A2AR*) injected across the surfaces at 100 uL/min.
Regeneration of captured-coupled β1AR* and A2AR* surfaces
At the end of each binding cycle, the GPCR surfaces were regenerated with a weak-affinity antagonist (2X two-minute injection of 200 uM L-748,337 for β1AR* and 2X one-minute injection 100 uM PSB 1115 for A2AR*), followed by an EXTRACLEAN step and an injection of running buffer.
Kinetic characterization of β1AR* and A2AR* antagonists
High-affinity small-molecule antagonists (249 – 504 Da; KD < 1 uM) were each tested in triplicate in three-fold dilution series for binding to β1AR* or A2AR* in running buffer containing 1% DMSO. All samples were injected at a flow rate of 100 uL/min and, when necessary, the surfaces were regenerated with PSB 1115 or L-748,337.
Equilibrium analyses of lower-affinity β1AR* antagonists
Lower-affinity β1AR* analytes (KD > 1 uM) were tested in duplicate in a two-fold dilution series starting at 100 uM in running buffer containing 5% DMSO. All samples were injected at a flow rate of 90 uL/min and a DMSO calibration series was used to correct for the excluded-volume effect.
Data processing and analysis
All biosensor data processing and analysis was performed using Scrubber2 (BioLogic Software Pty Ltd.; biologic.com.au). All responses were double referenced [16]. For kinetic analyses, data were globally fit to a 1:1 interaction model including a term for mass transport to obtain binding parameters. For equilibrium analyses, the responses at equilibrium were plotted against analyte concentration and fit to a simple 1:1 binding isotherm.
Results
Capturing approaches for β1AR* and A2AR*
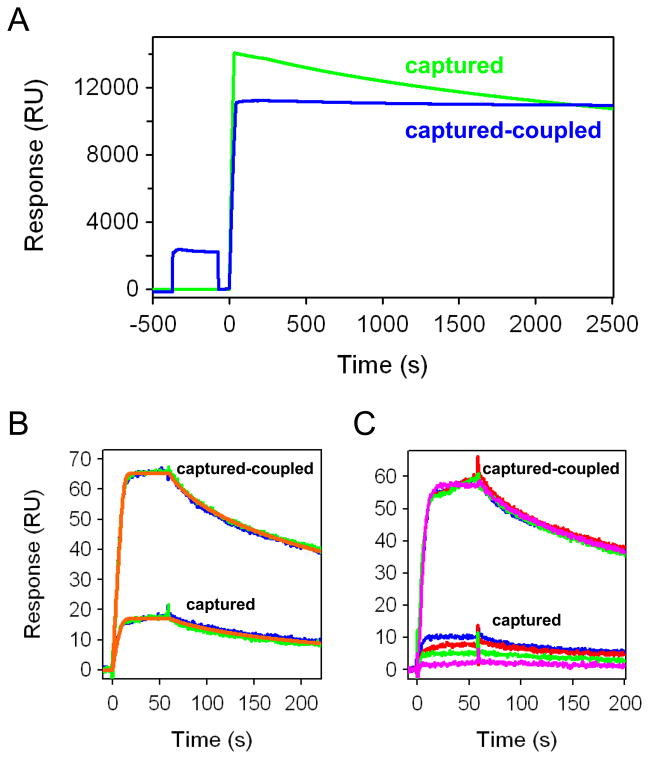
Using β1AR* as an example, Figure 1 shows two approaches for tethering His-tagged GPCRs to NTA sensor surfaces. Simply injecting β1AR*-His10 over the Ni2+-charged NTA surface produced the green response shown in Figure 1A. While the receptor was captured to a high density (>10,000 RU), it gradually dissociated from the NTA surface. This dissociation can complicate the analyte binding responses and lead to a loss of surface activity. Unfortunately, this level of dissociation from NTA surfaces is fairly typical for His-tagged proteins.
Figure 1.
NTA capture methods of His-tagged GPCRs. (A) Sensorgrams for direct capture (green) and capture-couple (blue) of His-tagged β1AR*. (B) Blue and green traces represent duplicate responses for 500 nM propranolol binding to directly captured and captured-coupled β1AR*. The first propranolol injection over both surfaces is shown in blue, the second in green. The red lines depict the fit of a 1:1 interaction model to each data set. (C) Replicate responses for 500 nM propranolol tested over 28 hours (sample order was blue, red, green, then pink) for binding to the captured-coupled and captured β1AR* surfaces.
In order to produce stable receptor surfaces, we employed an alternative approach which we call capture-coupling. This method is a hybrid of capture and amine-coupling chemistry. In capture-coupling, the nickel-charged NTA surface is activated with an EDC/sulfo-NHS mixture (from −375 to −75 sec in Fig. 1A) prior to injection of the receptor, as illustrated by the blue response in Figure 1A. The His tag serves to preconcentrate the receptor onto the surface for subsequent covalent crosslinking via the activated carboxyl groups. This method is milder than the standard preconcentration step for amine coupling, which involves dropping the pH below the isoelectric point of the protein and placing the ligand in a low-salt buffer. Using capture-coupling, the β1AR* receptor surface displayed significantly less post-immobilization drift (Fig. 1A blue sensorgram).
To establish that capture-coupling did not compromise the receptor’s activity, propranolol (259 Da) was injected across the directly NTA-captured and capture-coupled β1AR* surfaces (Fig. 1B). The antagonist bound to both surfaces, demonstrating in each case the receptor was active. The responses fit well to a 1:1 interaction model that yielded similar binding constants (ka = 9.4(6) × 105 M−1s−1, kd = 7.8(4) × 10−3 s−1, KD = 8.3(7) nM for the captured surface and ka = 1.57(4) × 106 M−1s−1, kd = 8.5(2) × 10−3 s−1, KD = 5.4(2) nM for the captured-coupled surface), indicating that capture-coupling did not alter the receptor’s binding activity.
When comparing the response levels achieved for propranolol from the two coupling methods, it is clear that the captured-coupled surface produced a significantly higher binding response than the directly captured surface (in this example, the reponse is >3 times larger for the captured-coupled surface) (Fig. 1B). The lower response for the captured receptor surface likely stems from the decay of the receptor from the NTA ligand. Importantly, the Rmax determined for the captured-coupled β1AR* surface suggests the immobilized receptor was ~75% active.
The differences in stability and activity of the two β1AR* surfaces are even more apparent in Figure 1C. Four replicate injections of propranolol (tested at seven-hour intervals) across the captured-coupled receptor surface overlay, confirming this surface was stable over >24 hours. In contrast, over the same time period the NTA-captured-only receptor surface lost all activity. Together, the data in Figure 1 demonstrate that capture-coupling produces high-density, active, and stable surfaces for this His-tagged receptor. Similar results were obtained for His-tagged A2AR* (data not shown). Based on the success of this method, capture-coupling was used to generate the data reported throughout the rest of the study.
Regeneration of GPCR surfaces
One of the limitations introduced by capture-coupling is that the receptor is now covalently immobilized to the surface. This eliminates the possibility of stripping the receptor from the surface and recapturing it as a means of regenerating tightly bound compounds. We are therefore left with two options for regeneration: (1) washing the receptor surfaces with buffer until the bound analyte dissociates (often requiring >1-hour wash phases) or (2) using a regeneration method that removes bound analyte without affecting receptor activity. Unfortunately, we find that regeneration conditions commonly used for protein/protein interactions (e.g., dilute phosphoric acid, base, or high salt) are often ineffective at regenerating small molecule binding sites [17]. And harsher conditions would likely be detrimental to the GPCR surfaces. Therefore, to regenerate the β1AR* and A2AR* systems we developed an alternative approach we refer to as “displacement regeneration”.
In displacement regeneration, bound analytes are displaced by a weak-affinity compound injected at high concentrations. This regeneration method works by saturating the available receptor binding sites, thereby blocking rebinding of the high-affinity analytes. Because this method involves passive displacement, it is most appropriate for analytes that are limited by mass transport, which we found is the case for many of the β1AR and A2AR antagonists.
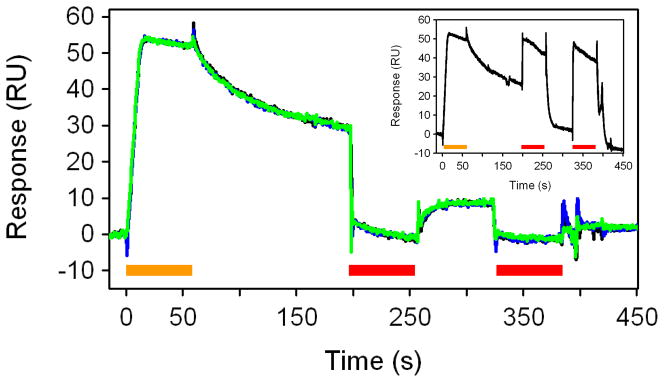
Figure 2 shows an example of the regeneration tests of A2AR*. In this work, the analyte of interest, ZM-241,385, was injected for one minute and its dissociation from the surface was monitored for two minutes. (Note that ZM-241,385 shows an apparent slow dissociation rate.) In order to displace the bound ZM-241,385, a weak A2AR antagonist, PSB 1115 (KD ~ 1 uM), was injected twice at a concentration of 100 uM during the dissociation phase. No binding response is visible for PSB 1115 in the main figure because these data were double referenced. In double referencing, buffer was injected instead of ZM-241,385 but PSB 1115 was still injected during regeneration. Once the data are fully processed this double referencing step essentially removes the binding response for PSB 1115 since it occurs in every cycle. In order to demonstrate that in fact PSB 1115 is binding during the regeneration steps, the inset in Figure 2 shows the raw data for one of the binding cycles; here the responses for PSB 1115 up to ~50 RU are apparent.
Figure 2.
Regeneration of captured-coupled GPCR surfaces. Main panel: Overlay of three binding cycles for A2AR*: injection of 300 nM ZM-241,385 (highlighted by the orange bar) followed by buffer wash for two minutes and two one-min injections of 100 uM PSB 1115 (highlighted by the red bars). Inset: Raw responses for these binding cycles.
By the end of the second PSB 1115 injection, all ZM-241,385 appears to be displaced from the receptor surface (Fig. 2, main panel). The ZM-241,385 binding and PBS 1115 regeneration cycles were repeated three times to demonstrate this regeneration condition is sufficient to remove bound ZM-241,385 from A2AR* without reducing receptor activity (note the excellent overlay of the black, green, and blue sensorgrams in the main panel of Fig. 2). A similar regeneration approach was successfully developed for β1AR* using 200 uM L-748,337 as the displacement analyte (data not shown).
Kinetic analyses of β1AR* and A2AR* antagonists
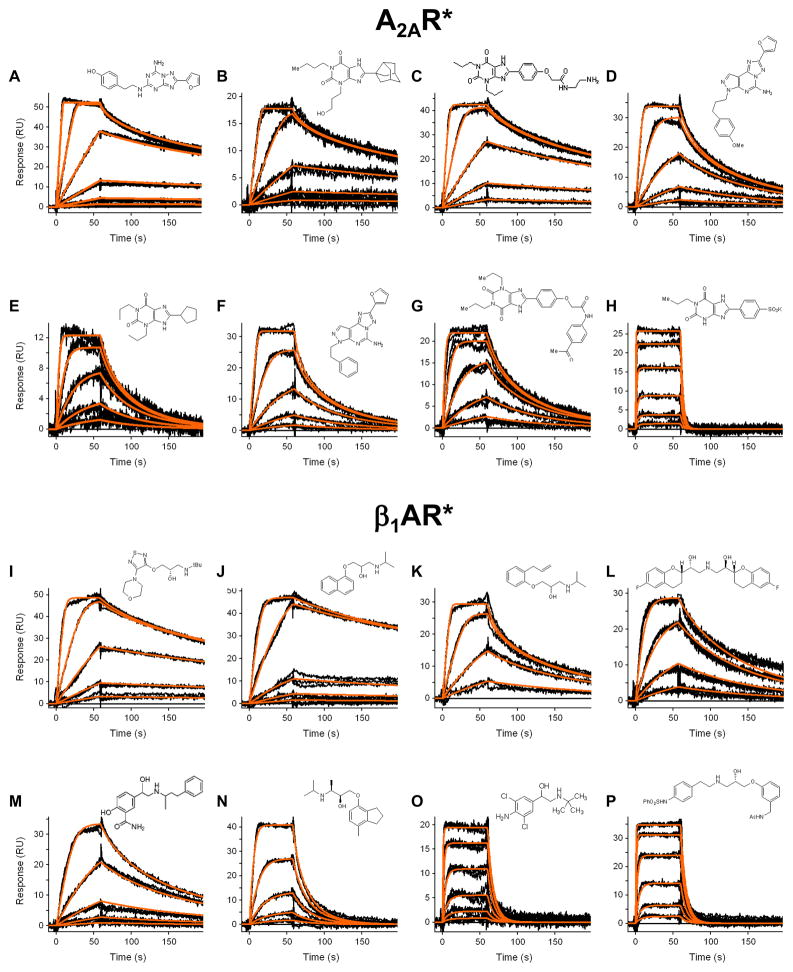
Having optimized immobilization and regeneration conditions for β1AR* and A2AR*, we next applied the Biacore assay to characterize the binding kinetics of eight antagonists (which ranged in size from 249 to 504 Da) to each receptor. Figure 3 presents the response data for a concentration series of each compound. Triplicate injections of each concentration overlay very well, demonstrating the observed binding responses were reproducible. In addition, each data set could be globally fit to a 1:1 interaction model. A summary of the binding constants is provided in Table 1.
Figure 3.
Kinetic analyses of A2AR* and β1AR* antagonists. Each compound was tested in triplicate in a three-fold dilution series and the responses were fit to a 1:1 interaction model. Antagonist structures are shown in the insets; compound identities and binding parameters determined from the fits are listed in Table 1.
Table 1.
Binding constants for GPCR/compound interactions determined at 25 °C.
| panela | A2AR* analytes | m.w. (Da) | ka (M−1s−1) | kd (s−1) | KD (nM) |
|---|---|---|---|---|---|
| 3A | ZM-241,385 | 337 | 1.33(6) × 107,b | 3.8(2) × 10−2,b | 2.8(2)b |
| 3B | PSB 36 | 386 | 5.1(6) × 106 | 3.6(4) × 10−2 | 7(1) |
| 3C | XAC | 428 | 1.39(2) × 106 | 1.12(1) × 10−2 | 8.1(1) |
| 3D | SCH 442416 | 389 | 2.60(8) × 106 | 5.3(2) × 10−2 | 20.2(9) |
| 3E | DPCPX | 304 | 1.2(5) × 107 | 6(3) × 10−1 | 50(30) |
| 3F | SCH 58261 | 345 | 1.00(3) × 106 | 6.6(2) × 10−2 | 66(1) |
| 3G | MRS 1706 | 504 | 3.36(7) × 105 | 3.47(8) × 10−2 | 103(3) |
| 3H | PSB 1115 | 388 | 3.9(1) × 105 | 3.18(9) × 10−1 | 810(30) |
| panel | β1AR* analytes | m.w. (Da) | ka (M−1s−1) | kd (s−1) | KD (nM) |
| 3I | timolol | 316 | 9.4(1) × 105 | 6.94(8) × 10−3 | 7.4(1) |
| 3J | propranolol | 259 | 4.5(1) × 105 | 3.28(4) × 10−3 | 7.2(2) |
| 3K | alprenolol | 249 | 1.31(4) × 106 | 2.49(7) × 10−2 | 19.0(1) |
| 3L | nebivolol | 405 | 2.76(5) × 105 | 1.50(3) × 10−2 | 31.8(1) |
| 3M | labetalol | 328 | 2.64(6) × 105 | 2.51(5) × 10−2 | 95(3) |
| 3N | ICI 118,551 | 277 | 2.40(7) × 105 | 8.5(3) × 10−2 | 350(10) |
| 3O | clenbuterol | 277 | 2.47(9) × 105 | 1.33(5) × 10−1 | 540(30) |
| 3P | L-748,337 | 498 | 2.37(6) × 105 | 1.41(4) × 10−1 | 600(20) |
| 4A | butoxamine | 267 | ---c | ---c | 5150(20) |
| 4B | serotonin | 176 | --- | --- | 14,170(50) |
| 4C | atenolol | 266 | --- | --- | 49,200(200) |
| 4D | methylergonovine | 339 | --- | --- | 49,600(300) |
| 4E | ergonovine | 325 | --- | --- | 84,100(800) |
Equilibrium analyses of lower-affinity interactions
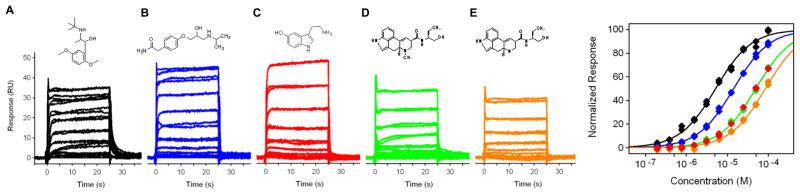
A powerful feature of Biacore technology is the ability to detect and quantitate relatively weak interactions (KD > 1 uM). To explore this possibility with the GPCR systems, we characterized the binding of five lower-affinity compounds against β1AR*. As shown in Figure 4, each of these compounds bound in a concentration-dependent manner and the responses were reproducible. Because they all have very fast dissociation rates, they reached equilibrium rapidly during the association phase. The responses at equilibrium all fit well to a simple 1:1 binding model as shown in Figure 4. The affinities for these compounds ranged from ~5 to ~85 uM (Table 1).
Figure 4.
Equilibrium analyses of five lower-affinity β1AR analytes. Each compound was tested in duplicate in a two-fold dilution series starting at 100uM. Responses at equilibrium (t = 10–20 sec) were plotted against compound concentration and fit to a simple binding isotherm. Analyte structures are shown in the insets; compound identities and affinities from the isotherms are listed in Table 1.
Discussion
We showed previously how Biacore could be used to study native GPCRs from both purified and crude preparations [4–9]. We developed assays to identify solubilization and purification conditions [4–6]. And we applied the technology to characterize the binding kinetics of antibodies, natural ligands and small molecules ranging in affinity from pM to mM [7–9].
In this report, we illustrate the advantages of using stabilized forms of GPCRs as ligands in Biacore experiments. Since a key requirement for the biosensor technology is that the ligand be active, a major advantage of the stabilized receptors is that they can be prepared with high overall activity and conformational homogeneity. (The β1AR* and A2AR* we used in this study were engineered to be in an antagonist conformation [10,13–15]). This high binding activity allows us to achieve relatively large binding responses even for small analytes. The conformational homogeneity likely contributes to the fact that the binding response data for all the compounds studied were reproducible and could be fit to a simple model.
However, even with active and stable starting material, we needed to optimize the receptor immobilization conditions by employing a capture-coupling approach. This method worked well with both the His-tagged β1AR* and A2aR* receptor systems, providing high capacity surfaces that were stable over time. However, we want to stress that with any new receptor system, it is important to run control experiments to ensure that the capture-coupling approach does not significantly affect receptor binding activity.
We developed a novel regeneration method that utilizes high concentrations of a weak binding analyte to passively dissociate a bound compound. This method will be most effective at regenerating analytes with fast association rates. These systems tend to be mass transport limited and can be easily displaced from the sensor surface by blocking rebinding as compared to a compound that has an inherently slow dissociation rate. Given the simplicity of this approach, we would encourage biosensor users to try displacement regeneration when they encounter slowly dissociating analytes with their own systems.
Using optimized immobilization and regeneration methods, we were able to characterize the binding of a range of small-molecule analytes to both β1AR* and A2aR*. This series of compounds displayed a wide range of association and dissociation rate constants that produced an almost 300-fold span in affinity. Having established the conditions for analysis of the stabilized receptors, we are in the process of transferring the methodology to study native forms of these same receptors. Biacore should provide an excellent method of assessing in detail if and how the stabilizing mutations change the recognition properties of the receptors.
Finally, we demonstrated that it is possible to characterize the affinity of relatively weak compounds (KD ~10 to 100 uM) binding to these receptors. These results hint at the potential of using Biacore to screen fragment libraries against GPCRs. One of our concerns, however, is that high concentrations of compounds used in fragment screening (100 to 500 uM) may show high levels of nonspecific binding to the receptor surfaces. We are currently investigating if this will be the case. But an additional advantage of utilizing stabilized receptors is that they can be crystallized [18] to confirm hits from a fragment library and identify binding modes for these compounds, which would be a useful in elaborating the hits.
Of course, engineering stabilized receptors requires a substantial amount of effort. But it is encouraging to see that these types of reagents are excellent tools for biosensor analysis. We are certain that the combination of stabilized receptors and biosensor technology will provide new insights into receptor structure/function and the high quality of direct binding data will positively impact drug discovery.
Acknowledgments
This work was funded by a grant (GM 071697) awarded to DGM by the National Institutes of Health.
Footnotes
Abbreviations used: A2AR, A2A adenosine receptor; β1AR, β1 adrenergic receptor; DMSO, dimethyl sulfoxide; DPCPX, 8-cyclopentyl-1,3-dipropylxanthine; EDC, 1-ethyl-3-(3-dimethylaminpropyl)-carbodiimde hydrochloride; GPCR, G-protein-coupled receptor; HBS, HEPES-buffered saline; HEPES, N-(2-hydroxyethyl)piperazine-N′-(2-ethanesulfonic acid); RU, resonance units; sulfo-NHS, sulfo-N-hydroxysuccinimide; SPR, surface plasmon resonance; XAC, xanthine amine congener.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Löfås S, Johnsson B. A novel hydrogel matrix on gold surfaces in surface plasmon resonance sensors for fast and efficient covalent immobilization of ligands. J Chem Soc, Chem Comm. 1990:1526–1528. [Google Scholar]; Löfås S, Malmqvist M, Rönnberg I, Stenberg E, Liedberg B, Lundström I. Bioanalysis with surface plasmon resonance. Sensors Actuat B. 1991;5:79–84. [Google Scholar]; Karlsson R, Michaelsson A, Mattsson L. Kinetic analysis of monoclonal antibody-antigen interactions with a new biosensor based analytical system. J Immunol Methods. 1991:229–240. doi: 10.1016/0022-1759(91)90331-9. [DOI] [PubMed] [Google Scholar]
- 2.Rich RL, Myszka DG. Survey of the year 2007 commercial optical biosensor literature. J Mol Recog. 2008;21:355–400. doi: 10.1002/jmr.928. [DOI] [PubMed] [Google Scholar]
- 3.Rich RL, Myszka DG. Grading the commercial optical biosensor literature – Class of 2008: “The Mighty Binders”. J Mol Recog. 2010;23:1–64. doi: 10.1002/jmr.1004. [DOI] [PubMed] [Google Scholar]
- 4.Navratilova I, Sodroski J, Myszka DG. Solubilization, stabilization, and purification of chemokine receptors using Biacore technology. Anal Biochem. 2005;339:271–281. doi: 10.1016/j.ab.2004.12.017. [DOI] [PubMed] [Google Scholar]
- 5.Rich RL, Miles AR, Gale BK, Myszka DG. Detergent screening of a GPCR using serial and array biosensor technologies. Anal Biochem. 2009;386:98–104. doi: 10.1016/j.ab.2008.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Navratilova I, Pancera M, Wyatt RT, Myszka DG. A biosensor-based approach toward purification and crystallization of G protein-coupled receptors. Anal Biochem. 2006;353:278–273. doi: 10.1016/j.ab.2006.03.049. [DOI] [PubMed] [Google Scholar]
- 7.Navratilova I, Dioszegi M, Myszka DG. Analyzing ligand and small molecule binding activity of solubilized GPCRs using biosensor technology. Anal Biochem. 2006;355:132–139. doi: 10.1016/j.ab.2006.04.021. [DOI] [PubMed] [Google Scholar]
- 8.Navratilova I, Myszka DG, Rich RL. Probing membrane protein interactions with real-time biosensor technology. In: Pebay-Peroula E, editor. Biophysical Analysis of Membrane Proteins. 2008. pp. 121–140. [Google Scholar]
- 9.Congreve M, Rich RL, Myszka DG, Siegal G, Marshall F. Fragment screening of stabilized G-protein coupled receptors using biophysical methods. Methods Enzymol. 2010 doi: 10.1016/B978-0-12-381274-2.00005-4. In press. [DOI] [PubMed] [Google Scholar]
- 10.Warne T, Serrano-Vega MJ, Tate CG, Schertler GF. Development and crystallization of a minimal thermostabilised G protein-coupled receptor. Protein Expr Purif. 2009;65:204–213. doi: 10.1016/j.pep.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 11.Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Kuhn P, Weis WI, Kobilka BK, Stevens RC. High-resolution crystal structure of an engineered human β2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jaakola VP, Griffith MT, Hanson MA, Cherezov V, Chien EY, Lane JR, Ijzerman AP, Stevens RC. The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science. 2008;322:1211–1217. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Serrano-Vega MJ, Magnani F, Shibata Y, Tate CG. Conformational thermostabilization of the β1-adrenergic receptor in a detergent-resistant form. Proc Natl Acad Sci USA. 2008;105:877–882. doi: 10.1073/pnas.0711253105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Serrano-Vega MJ, Tate CG. Transferability of thermostabilizing mutations between β-adrenergic receptors. Mol Membr Biol. 2009;26:385–396. doi: 10.3109/09687680903208239. [DOI] [PubMed] [Google Scholar]
- 15.Magnani F, Shibata Y, Serrano-Vega MJ, Tate CG. Co-evolving stability and conformational homogeneity of the human adenosine A2A receptor. Proc Natl Acad Sci USA. 2008;105:10744–10749. doi: 10.1073/pnas.0804396105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Myszka DG. Improving biosensor analysis. J Mol Recog. 1999;12:279–284. doi: 10.1002/(SICI)1099-1352(199909/10)12:5<279::AID-JMR473>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 17.Papalia GA, Giannetti AM, Arora N, Myszka DG. Thermodynamic characterization of pyrazole and azaindole derivatives binding to p38 MAP kinase using Biacore T100 technology and van’t Hoff analysis. Anal Biochem. 2008;383:255–264. doi: 10.1016/j.ab.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 18.Warne T, Serrano-Vega MJ, Baker JG, Moukhametzianov R, Edwards PC, Henderson R, Leslie AG, Tate CG, Schertler GF. Structure of a β1-adrenergic G-protein-coupled receptor. Nature. 2008;454:486–491. doi: 10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]