Abstract
Developmental mechanisms play an important role in determining the costs, limits, and evolutionary consequences of phenotypic plasticity. One issue central to these claims is the hypothesis of developmental decoupling, where alternate morphs result from evolutionarily independent developmental pathways. We address this assumption through a microarray study that tests whether differences in gene expression between alternate morphs are as divergent as those between sexes, a classic example of developmental decoupling. We then examine whether genes with morph-biased expression are less conserved than genes with shared expression between morphs, as predicted if developmental decoupling relaxes pleiotropic constraints on divergence. We focus on the developing horns and brains of two species of horned beetles with spectacular sexual- and morph-dimorphism in the expression of horns and fighting behavior. We find that patterns of gene expression were as divergent between morphs as they were between sexes. However, overall patterns of gene expression were also highly correlated across morphs and sexes. Morph-biased genes were more evolutionarily divergent, suggesting a role of relaxed pleiotropic constraints or relaxed selection. Together these results suggest that alternate morphs are to some extent developmentally decoupled, and that this decoupling has significant evolutionary consequences. However, alternative morphs may not be as developmentally decoupled as sometimes assumed and such hypotheses of development should be revisited and refined.
Keywords: polyphenism, developmental decoupling, horned beetles, Onthophagus, microarray, sexual dimorphism, pleiotropy, relaxed selection
Introduction
Polyphenisms are an impressive form of phenotypic plasticity where a genotype expresses one of several discrete, alternative phenotypes appropriate to local conditions. Polyphenisms are adaptive, allowing organisms to survive in a range of environments that differ in climate, predation regime, or nutritional conditions (Kingsolver and Wiernasz 1991; McCollum and VanBuskirk 1996; Nice and Fordyce 2006). Furthermore, polyphenisms are thought to play important roles in the evolution of organismal diversity from speciation (Pfennig et al. 2007) to the origins of novel features (West-Eberhard 1989, 2003).
A knowledge of the developmental and genetic mechanisms underlying polyphenisms is crucial to understanding the costs, limits, and evolutionary consequences of phenotypic plasticity. For instance, in cases in which a polyphenism is mediated by gene expression specific to a particular morph or environment, several mechanisms may promote rapid divergence of these genes, relative to genes shared between morphs or environments. First, pleiotropic constraints on morph- or environment-specific genes are relaxed, permitting evolutionary diversification (Fisher 1930; Pal et al. 2006). This idea is similar to the observation that proteins specific to certain contexts show relatively greater evolutionary divergence, presumably due to reduced pleiotropic constraints (reviewed in Pal et al. 2006). For instance, evolution is accelerated in genes with sex-specific expression (Jagadeeshan and Singh 2005; Ellegren and Parsch 2007; Haerty et al. 2007; Larracuente et al. 2008) and tissue-specific expression (Hastings 1996; Duret and Mouchiroud 2000; Zhang and Li 2004; Liao et al. 2006), and in genes with protein products that execute a small range of functions or exhibit low connectivity or centrality in interaction networks (Hahn and Kern 2005; Salathe et al. 2006). Second, morph-specific gene expression may lead to relaxed selection (Kawecki 1994; Kawecki et al. 1997) which should result in even greater sequence divergence due to the increased chance of fixing slightly deleterious mutations (Snell-Rood et al. 2010; Van Dyken and Wade 2010).
Such decoupling of developmental pathways, where gene expression is specific to a particular morph or environment, has long been a central hypothesis in the evolution of plasticity and, more broadly, exaggerated and novel traits (West-Eberhard 1989; Nijhout 1994; Emlen and Nijhout 2000; Nijhout 2003; West-Eberhard 2003, 2005). By reducing pleiotropic constraints, decoupling is believed to allow alternative morphs to adapt to their specific selective environment independent of one another and to explore a wide phenotypic space, facilitating the origin of novel traits. A wide variety of studies that have investigated divergent patterns of gene expression suggest that polyphenic morphs may indeed be decoupled, at least to some degree, in their development (Hymenoptera: Evans and Wheeler 1999, 2001; Cash et al. 2005; Pereboom et al. 2005; Donnell and Strand 2006; Judice et al. 2006; Sumner et al. 2006; Barchuk et al. 2007; Hoffman and Goodisman 2007; Isoptera: Scharf et al. 2003; Hojo et al. 2005; social Hemiptera: Kutsukake et al. 2004; vertebrates: Aubin-Horth et al. 2005).
In this study we hoped to build on existing literature in several ways. First, we were interested in expanding the taxonomic survey of patterns of gene expression between polyphenic morphs by focusing on a non-social insect: horned beetles. Second, we were interested in testing whether the degree of developmental decoupling between polyphenic morphs is comparable to that between males and females, a commonly cited example of relative developmental (and evolutionary) independence (Bull 1983; West-Eberhard 2003; Williams and Carroll 2009). Finally, we were interested in explicitly addressing the developmental decoupling hypothesis by testing whether morph-biased genes are indeed under less evolutionary constraint than morph-shared genes. Population genetic models of relaxed constraint stress the importance of whether a gene is “on” or “off” in one morph or another (Kawecki 1994; Kawecki et al. 1997; Van Dyken and Wade 2010), even though more typically, gene expression is only biased across environments (Aubin-Horth and Renn 2009; Hodgins-Davis and Townsend 2009; Snell-Rood et al. 2010). We test whether morph-biased expression results in the same “freeing” of selection as morph-specific expression.
We focus on patterns of gene expression in beetles in the genus Onthophagus, which are famous for their intra- and inter-specific diversity in horns (Emlen et al. 2005a; Emlen et al. 2005b). Most male onthophagine beetles express horns, used in aggressive encounters, while females do not (but see Watson and Simmons 2010). However, horn expression in males is highly dependent on nutrition and body size (Emlen 1994; Moczek and Emlen 1999), resulting in an impressive polyphenism in both morphology and behavior. Large, horned males guard females and their tunnels (Emlen 1997). Provided competition with other males is relatively low, horned males will also help females in provisioning brood balls (Moczek 1999; Hunt and Simmons 2000, 2002), which support the entire larval development of their offspring. In contrast, small males, which express only rudimentary horns, sneak copulations with females by bypassing horned males and burrowing through side tunnels (Emlen 1997). These sneaker males also show increased investment in testes (Simmons and Emlen 2006) and sometimes ejaculates (Simmons et al. 1999). Rudimentary horn expression in small males favors increased maneuverability in tunnels (Moczek and Emlen 2000; Madewell and Moczek 2006), and reduces tradeoffs associated with the expression of large, costly horns (Emlen 2001; Moczek and Nijhout 2004). The range of differences between horned and sneaker morphs, and between male and female beetles, all of which vary widely across species, provides an opportunity to test the hypothesis of decoupling in polyphenic development. We use differences in tissue-specific transcription profiles between sexes to ask whether morph-biased patterns of expression carry with it gene expression differences similar to those detected across sexes. We also test the evolutionary significance of morph-biased expression by relating patterns of gene expression to measures of sequence divergence.
Methods
Study System and Husbandry
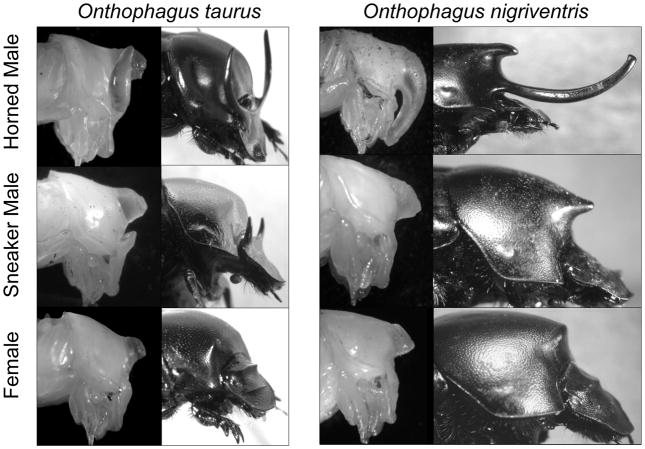
We chose to focus on developing horns in the beetle Onthophagus taurus (Fig. 1). A subset of arrays were used to contrast development with the related species, O. nigriventris (Fig. 1). Both species show a pronounced difference between male morphs in both morphology and behavior, as well as striking sexual dimorphism. In O. taurus, only large males develop paired, curved head horns, and fight for access to females; small males have two small residual horns and are more likely to sneak copulations (Moczek and Emlen 2000). Instead of horns, female O. taurus express a narrow ridge on their head. In addition, both sexes and both male morphs also develop a single medial prothoracic horn, which is clearly visible externally in pupae but becomes resorbed during the pupal stage in all individuals (Fig. 1).
Figure 1. Study System.
Shown are pupae (left) and corresponding adults (right) of horned (top) and sneaker (middle) male morphs and females (bottom) of O. taurus and O. nigriventris. For each treatment group, we harvested tissue from the developing thoracic horn epidermis, head horn epidermis, legs, and brains. We focused on an array design that compared expression profiles of these focal tissues to those of reference tissues (dorsal abdominal epidermis or ganglia).
O. nigriventris shows a somewhat similar polyphenism in mating behavior and horn development, but differs in the location of horn expression: large adult males bear a single long, curved thoracic horn, while small adult males develop only a small point on their prothorax. Adult females express only a small prothoracic ridge. In this species pupal resorption of thoracic horns is restricted to females (Moczek 2006) and, albeit to a lesser degree, small males (Moczek 2007), but is absent in large males.
O. taurus were collected from a population near Charlottesville, VA, while O. nigriventris were collected from established populations in Waimea, Hawaii. Beetles were maintained in laboratory colonies using established methods (described in Moczek and Nijhout 2003). Offspring were collected by setting up males and females in separate low-density containers (see Moczek and Nagy 2005). Second or early third instar larvae were transferred from their brood balls to fresh dung in 12-well plates where their developmental stage could be monitored (see Shafiei et al. 2001).
Tissue Collection
Beetles were sacrificed for tissue collection within 24 hours of pupation. We chose to focus on this stage of development primarily for two reasons. First, at this developmental stage, the basic horn structure is externally visible and the horn (and homologous areas in the female and sneaker males), including underlying epidermal tissue, can be easily and quickly harvested. Second, in early pupae, extensive changes are occurring within the developing horn, including differentiation of the horn epidermis, tissue remodeling and cell death, as well as growth of the adult cuticle (Moczek 2006).
In O. taurus, we collected six tissue types (see complete list of arrays, Table 1). First, three focal epidermal tissues were harvested: 1) the prothoracic epidermis, which included the developing horn and the surrounding prothorax, 2) the dorsal head epidermis, which included head horns in large males, 3) all six legs. These focal epidermal tissues were hybridized on arrays with dorsal abdominal epidermis, which served as a comparative epidermal tissue that does not produce any appendages or outgrowths similar to horns (we avoided small lateral projections common in onthophagine pupae, see Moczek 2006). Second, we harvested developing neural tissue, including the developing central brain and optic lobes, which were hybridized on arrays with ganglionic tissue, including the subesophageal ganglion and the thoracic ganglia. In O. nigriventris, we focused only on the prothoracic epidermis and the dorsal abdominal epidermis.
Table 1. List of Microarray Hybridizations.
For the majority of microarrays, different tissues were compared within the same morph or sex – for instance, hybridization between head horn epidermis and abdominal epidermis of horned males. For a subset of arrays (N = 4), the same tissues were compared between different morphs. For each microarray (total N = 71), tissue samples originated from four individual beetles. Abbreviations: HM = horned male; SM = sneaker male; F = female; OT = Onthophagus taurus; ON = Onthophagus nigriventris. All processed and raw microarray data are available at NCBI’s Gene Expression Omnibus http://www.ncbi.nlm.nih.gov/geo/, series accession number GSE23425.
| Sample 1 | Sample 2 | Morph, Sex | Species | Total Arrays |
|---|---|---|---|---|
| Head horn Epidermis | Abdominal Epidermis | HM/SM/F | OT | 12 |
| Thoracic Horn Epidermis | Abdominal Epidermis | HM/SM/F | OT | 12 |
| Legs | Abdominal Epidermis | HM/SM/F | OT | 12 |
| Brain | Ganglia | HM/SM/F | OT | 12 |
| Thoracic Horn Epidermis | Abdominal Epidermis | HM/SM/F | ON | 19 |
| Head horn (HM) | Head Horn (SM) | HM -- SM | OT | 4 |
All dissections were performed in 1x RNase-free PBS (Ambion), under RNase-free conditions: all dissecting scissors, forceps, and containers were treated with RNase-Zap (Ambion). All tissues were rinsed with 1x RNase-free PBS (Ambion) to remove as much non-epidermal tissue as possible. Immediately after removal, tissue was placed in 350 μl Buffer RLT 1% v/v BME (RNeasy Mini Kit, Qiagen). Tissue was ground using a sterile, RNase-free pestle fit to the 1.5 μl microcentrifuge tube (Kontes grinders, Kimble Chase, VWR) and immediately frozen in liquid nitrogen before being stored at −80 C until RNA extraction.
RNA extraction and amplification
Total RNA was extracted from tissue samples using the RNeasy Mini Kit (Qiagen), following standard kit protocols. RNA was eluted in 50 μl RNase-free water (Qiagen), and quantified using a Nanodrop (Thermo Scientific). Extracted RNA had an average purity of 260/230 = 2.11, 260/280 = 2.06. For a subset of samples, we verified the quality of the RNA using an Agilent 2100 BioAnalyzer. Total RNA was stored at −80C until amplification.
We amplified RNA using a protocol developed by A. Cash and J. A. Andrews, which drew from previously developed protocols (Vangelder et al. 1990; Klebes et al. 2002; Kijimoto et al. 2009). Briefly, we reverse-transcribed total RNA using a T7 Oligo (dT) primer (Ambion) and Super Script III (Invitrogen). Following second-strand synthesis (using DNA polymerase and RNase H (Invitrogen)), we converted the cDNA to anti-sense RNA using the MEGAscript T7 In Vitro Transcription Kit (Ambion). The final, amplified antisense RNA was purified using the RNeasy Mini Kit (Qiagen), eluting the sample in 50 μl RNase-free water (Qiagen). The final amplified antisense RNA product was quantified using a Nanodrop (Thermo Scientific). We used Microcon-30 centrifugal columns (Millipore) to purify any extracted or amplified samples with 260/230 or 260/280 ratios less than 1.80. Amplified RNA had an average purity of 260/230 = 2.76, 260/280 = 2.19.
Microarray Design
Overview
To assay gene expression, we used a cDNA microarray custom-built for Onthophagus taurus (Kijimoto et al. 2009). We assayed gene expression between tissue types to determine the similarity of overall patterns of expression between morphs and sexes (Fig. 1, Table 1). Given the differences between morphs in morphology and behavior, we focused on epidermal and neural tissues. The “morphology arrays,” which included contrasts between epidermal tissues, allowed us to identify the level of gene expression in developing horns relative to that in abdominal epidermal cells. Specifically, we were able to identify genes whose expression was consistently (i.e., significantly) higher or lower in horn epidermal cells compared to abdominal epidermis. By comparing horn epidermis to abdominal epidermis, we could identify genes biased in expression to developing horns, as opposed to more general genes involved in epidermis development, such as housekeeping genes. The neural tissue arrays allowed us to identify the level of gene expression in developing brains (and eyes) relative to that in developing ganglia. Brain gene expression has been shown to covary with morph differences in behavior in other systems (Aubin-Horth et al. 2005; Whitfield et al. 2006; Toth et al. 2007; Alaux et al. 2009). Moreover, given the importance of metamorphosis in beetle brain development, we hypothesized neural tissue would show morph-biased patterns of gene expression as soon as first day pupae (Paspalas et al. 1993; Wegerhoff 1999). By replicating this approach across sexes and male morphs we could therefore gain a better understanding of both the similarities and differences in gene expression between morphs and sexes across tissue types. All processed and raw microarray data (N = 71 arrays) are available at NCBI’s Gene Expression Omnibus http://www.ncbi.nlm.nih.gov/geo/, series accession number GSE23425.
Experimental procedures
We focused on array comparisons in O. taurus, the species for which the arrays were developed. We executed 48 arrays of four comparison types (Table 1), head horn – abdomen, thoracic horn – abdomen, leg – abdomen, and brain – ganglia (N = 48 arrays). We performed an additional set of arrays (N = 4), directly hybridizing horn tissue of each morph, to validate measures of morph-biased expression. In O. nigriventris, we focused only on thoracic horn – abdomen arrays (N = 19 arrays). Within each tissue comparison, we included 4–7 independent biological replicates (Table 1), each of which included tissue pooled from four individuals, with balanced dye flips. While the interpretation of cross-species microarrays must be treated with caution, we believe that broad comparisons are justified because overall patterns of expression were highly correlated between species (e.g., M values for thorax-abdomen arrays of horned males: R2 = 0.71, F1,446 = 1113.23, P < 0.0001, bST = 0.77, Supplementary Fig. 1).
The cDNA microarrays used in the present study were developed for O. taurus using an EST library described in detail in Kijimoto et al (2009). Briefly, these arrays included 3,756 cDNA spots, where each spot represented an EST from two normalized libraries developed from 16 beetles (male and female) harvested over eight developmental stages (4 time points in the larval stage; 4 time points in the pupal stage). High quality sequence reads were generated for 3,488 of these ESTs (GenBank accession numbers FG539013-FG542500); these sequences were assembled using ESTPiper (Tang et al. 2009) into 451 contigs (2.6 ESTs per contig) and 2,330 singletons (N = 2781 non-redundant sequences, see Kijimoto et al. 2009). Seventy one percent of the non-redundant sequences were annotated using the UniProtKB/TrEMBL protein sequence database (E value < 10−5; median e value = 10−50). The cDNA microarrays were printed by the Center for Genomics and Bioinformatics at Indiana University on GAPSII Microarray Slides (Corning) using an Omnigrid 300 Printing Robot and developed protocols (Andrews et al. 2006; Kijimoto et al. 2009). Each microarray included 564 control spots (GAPDH, actin-5c, and spotting buffer only). The gene list and platform description is available at Gene Expression Omnibus http://www.ncbi.nlm.nih.gov/geo/ accession number GPL7555.
We followed an RNA labeling and microarray hybridization protocol developed by A Cash and J Andrews (Kijimoto et al. 2009), based closely on the protocol from the Kreatech ULS-Cy 3/5 aRNA fluorescent labeling kit (Biomicrosystems). We labeled 2 ug of our aRNA with the Kreatech Cy3 or Cy 5-ULS (Biomicrosystems). For the hybridization, samples were balanced for labeling efficiency: we added enough labeled solution such that each sample contributed 60 pmol of dye (for samples with lower labeling efficiency we matched for the maximum amount possible). In general, this resulted in not only dye balance, but also sample (in total RNA) balance. Microarrays were pre-hybridized for 1 hour at 55 C in a solution of 5XSSC (Ambion), 0.1% 10% SDS (Ambion), and 1% I-block (Applied Biosystems). Labeled samples were mixed with KREAblock (ULS aRNA fluorescent labeling kit) and 2x Enhanced cDNA hybridization buffer (Genisphere); hybridization occurred at 55 C water for 16–18 hours. Slides were rinsed in 2x SSC 0.2% SDS at 55 C for 10 min, 2X SSC RT for 10 min, and 0.2X SSC RT for 10 min. Microarrays were scanned using a GenePix Scanner 4200 (Molecular Devices; PMTs were balanced using the Set PMT Gain function) and spot intensity quantified (after manual inspection) using GenePix Pro 5.0 software. Slide quality (spot # and foreground/background ratio for each dye) was comparable across all treatment groups.
Microarray Analyses
Microarrays were analyzed through several steps. First, we performed a basic quality control step to ensure dyes were balanced and signal to noise ratio was adequate (see https://dgrc.cgb.indiana.edu/microarrays/support/bha.html). Second, we quantified differential gene expression between tissues – for each EST (spot) on the array – using a custom R program developed by J Costello and J Andrews that employed the “biobase” and “marray” bioconductor packages (Yang et al. 2002), the OLIN normalization package (Futschik and Crompton 2004), and the limma differential expression package (Smyth 2004). In our analysis we performed OLIN normalization using the background correction “normexp” which produces only positive adjusted intensities. We set a threshold of inclusion for intensities of at least 150, and included only spots that were present in at least 70% of arrays for a treatment group. These analyses employ standard t tests, adjusted for multiple testing, to determine whether a gene is consistently (i.e., across 4 arrays) more highly expressed in one tissue relative to another (i.e., between the two florescent dyes on the array).
Third, we adjusted for the fact that some ESTs (spots) on the array represented the same gene; specifically, in the prior analysis of the EST library, 1158 EST sequence reads assembled into 451 contigs (Kijimoto et al. 2009; Tang et al. 2009). We combined data across all spots within a contig. Expression intensity (A) was quantified as average intensity across all spots within a contig. We combined differential expression (generally, “M,” the log2[focal tissue expression/comparison tissue expression]), by first converting M value to fold change (2M), averaging these values, and then taking the log2 to convert back to M value. We combined adjusted P values (pi) using Fisher’s method for combined P values, where the product −2* [Σloge (pi)] is distributed as a Χ2 with degrees of freedom equal to two times the number of ESTs in a given contig.
After processing microarrays in this way, we focused subsequent clustering analyses on a subset of genes that passed a set of inclusion criteria. First, we considered only genes with an adjusted P value of 0.05 for at least one of the treatment or tissue groups included in an analysis. This P value is adjusted for the false-discovery rate of Benjamini and Hochberg (1995) such that a threshold of 0.05 corresponds to less that 5% false discoveries (Smyth 2005). Second, we considered only genes with a fold-expression difference of at least two (|M| > 1) in at least one of the treatment of tissue groups in a given analysis. Analyses without this latter filtering step were comparable to those with the filtering step, but the filtering helped to make data visualization more manageable.
We compared microarrays from different tissues, sexes, and morphs using clustering analyses. We used the TM4 Microarray Experiment Viewer (Saeed et al. 2003) for all genes that fit the above criteria (Adjusted P < 0.05, |M| > 1) and additionally met the intensity threshold for each treatment group considered. This clustering approach allowed us to assess overall patterns of gene expression and determine both shared and biased patterns of gene expression between morphs and sexes. We inspected clustering patterns of treatment groups using Euclidian distance; confidence in the observed clustering was evaluated through a bootstrapping support tree (100 replicates). We measured the similarity of expression patterns by calculating the Pearson correlation between groups.
Sequence Divergence
We calculated sequence divergence using previously reported sequence data for the ESTs used to build the microarrays (Kijimoto et al. 2009). Tribolium castaneum is the closest sequenced genome to Onthophaus taurus, but the species diverged from a common ancestor over 150 million years ago, making sequence alignments and calculations of standard metrics (e.g., dN/dS) difficult. We used amino acid distance (dA) as estimates of divergence. We identified orthologous gene pairs between Tribolium and Onthophagus as genes with the best BLASTX hit (e value cutoff = 1e-10). We filtered out the proteins with multiple homologs, and only retained one-to-one pairs for subsequent analyses. We then used ESTwise (Birney et al. 2004) to identify the frame and structure of the alignment between the EST sequence and the Tribolium protein. Frameshifts predicted by ESTwise were fixed manually, assuming they resulted from sequencing error. We believe this assumption is reasonable, given that EST sequences are generally prone to small sequencing errors. The dA set was further filtered to include only the pairs with predicted peptide alignments that exceeded 50 amino acids and 50% of the total length of both the EST sequence and the Tribolium protein sequence. We extracted the protein sequences predicted by ESTwise and aligned them again using MUSCLE (Edgar 2004). Amino acid distance was estimated in PAML4 (Yang 2007) as the maximum likelihood estimates of number of amino acid replacements per site based on the empirical substitution model WAG (Whelan and Goldman 2001).
We tested whether patterns of divergence were related to patterns of gene expression. We limited these analyses to only O. taurus array data, and included all genes with significant expression (Adjusted P value < 0.05) in at least one tissue in females, sneaker males or horned males. Morph-biased expression was quantified as the absolute difference in expression (M value) between horned and sneaker morphs, averaged across all tissues (brain, head horn, thoracic horn, legs). We validated this measure of morph-biased expression by comparing the difference in M values between horn-abdomen comparisons of each morph with a more direct measure of morph-biased expression: direct comparisons between the head horn tissue of two morphs (see results). We also tested for effects of sex-biased and tissue-biased expression, total expression level and sequence length, all of which have been shown to have effects on sequence divergence (Pal et al. 2006). Sex-biased expression was measured as the absolute difference in expression (M value) between females and the average expression of the two male morphs, averaged across all tissues. Tissue bias was quantified as the number of tissue-specific arrays in which a gene was significantly expressed (in females, horned males or sneaker males). Total expression was the average expression (A value) of a gene across all arrays. We transformed all non-normally distributed data (log transformation for morph- and sex-specific expression; arc-sin square root transformation for dA).
Results
Onthophagus taurus: patterns of expression in developing horns
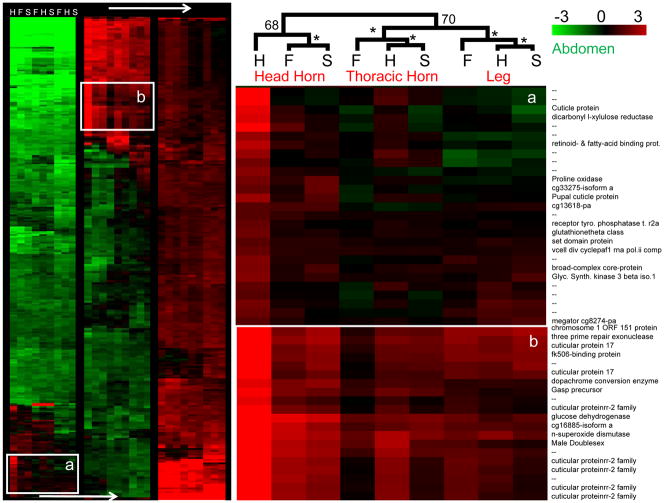
We first present our results for O. taurus patterns of gene expression. Recall that in this species only large males develop paired head horns, while all males and females transiently express prothoracic horns which are subsequently resorbed during the pupal stage prior to the adult molt (Fig. 1). For the epidermal tissue arrays, 794 genes fit our criteria for inclusion in the hierarchical clustering analysis (P < 0.05 and |M| >1 for at least one treatment category). We found that in prothoracic horn-abdomen and leg-abdomen arrays – tissues with no obvious differences between the morphs – patterns of expression in the two male morphs were more similar to each other than to that in females (Fig 2, Table 2). In contrast, in the developing head epidermis, where morph dimorphism is pronounced, patterns of gene expression in the developing small, sneaker males were more similar to expression in the females than in the horned male morph (Fig. 2, Table 2). Overall, morph- and sex-specific patterns of expression were more similar within tissue types than between tissue types (Fig. 2, Table 3).
Figure 2. Morphology Arrays for O. taurus.
Shown are the results of a clustering analysis of head horn- abdomen, thoracic horn-abdomen and leg-abdomen arrays for large, horned males (H), small, sneaker males (S) and females (F). Genes included in the analysis were significantly differentially expressed (P < 0.05) at least two-fold between tissues (|M| >1) in at least one treatment category. We identified two clusters of genes with biased expression in the developing head epidermis of horned males. Bootstrapping values are indicated (* = 100%).
Table 2. Results of Clustering for O. taurus arrays.
Each clustering analysis was executed independently for each array (tissue hybridization) type (versus the consolidated clustering analysis shown in Fig. 2). Genes included in this clustering analysis (N = 794 for O. taurus morphology arrays; 189 for O. taurus brain arrays; 448 for O. nigriventris arrays) were significant (adjusted P < 0.05) with a threshold differential expression (|M| >1) in at least one of the three categories (females (F), horned male (H) or sneaker male (S)). Shown are Pearson correlation coefficients between each treatment group. Bold values represent highly correlated arrays (> 0.95). Abbreviations: OT = Onthophagus taurus; ON = Onthophagus nigriventris
| Array Type | Female-Horned Male | Female-Sneaker Male | Sneaker-Horned Male | |
|---|---|---|---|---|
| OT | Head-Ab | 0.9244 | 0.9739 | 0.9244 |
| Thorax-Ab | 0.9386 | 0.9632 | 0.9700 | |
| Legs-Ab | 0.9705 | 0.9706 | 0.9769 | |
| Brain-Gan. | 0.958 | 0.937 | 0.871 | |
| ON | Thorax-Ab | 0.941 | 0.978 | 0.944 |
Table 3. Correlations across all O. taurus morphology arrays.
Shown are Pearson correlation coefficients across arrays for head horn epidermis-abdomen (head), thoracic horn-abdomen (thorax) and leg-abdomen (leg) arrays for sneaker males (S), horned males (H) and females (F). Bold values represent highly correlated arrays (> 0.95).
| Head | Thorax | Leg | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| F | H | S | F | H | S | F | H | S | ||
| Head | F | 0.924 | 0.974 | 0.895 | 0.872 | 0.869 | 0.881 | 0.845 | 0.836 | |
| H | 0.926 | 0.819 | 0.881 | 0.845 | 0.832 | 0.825 | 0.809 | |||
| S | 0.895 | 0.884 | 0.888 | 0.875 | 0.844 | 0.839 | ||||
| Thorax | F | 0.939 | 0.963 | 0.877 | 0.851 | 0.835 | ||||
| H | 0.970 | 0.872 | 0.877 | 0.864 | ||||||
| S | 0.865 | 0.857 | 0.859 | |||||||
| Leg | F | 0.970 | 0.971 | |||||||
| H | 0.977 | |||||||||
| S | ||||||||||
We were interested in genes or pathways that exhibited morph-biased patterns of gene expression, in particular genes with expression patterns unique to the head epidermis (relative to the abdomen) in the horned male morph, but not in the other treatments or tissues. Several clusters fit these criteria (Fig. 2). Nineteen percent of these genes were involved in cuticle formation, and 36% of these genes were un-annotated (Fig. 2, genes marked with “--”). A complete list of O. taurus head-epidermis genes with the most divergent expression patterns between morphs and sexes is presented in Appendix 1.
Appendix 1. Genes of the developing head epidermis of O. taurus with the most divergent expression patterns between morphs and sexes.
Genes were filtered as follows: 1) genes were significantly differentially expressed in the head epidermis relative to the abdomen (M > 0) or in the abdomen relative to the head (M < 0) in at least one treatment category; 2) absolute difference in expression (M value) was greater than 1 when horned males were compared to both females and sneaker males. Shown are M values for genes fitting these criteria in head epidermis-abdomen arrays for tissue from females, horned males and sneaker males. Gene annotation is according to (Kijimoto et al. 2009).
| Gene | Female | Horned | Sneaker |
|---|---|---|---|
| ---NA--- | −2.61264 | −4.26973 | −2.70081 |
| ---NA--- | −2.37502 | −3.73505 | −2.3695 |
| ---NA--- | −1.32496 | −3.67555 | −1.73222 |
| cg3960-isoform d | −1.63447 | −2.95738 | −1.63701 |
| ---NA--- | −1.66075 | −2.69271 | −1.65682 |
| ---NA--- | −0.19619 | −1.64336 | −0.48173 |
| von willebrand factor | −0.31834 | −1.57492 | −0.50605 |
| −2.79414 | −1.50176 | −2.60504 | |
| hemolymph proteinase 19 | −0.01858 | −1.47872 | −0.23046 |
| cuticular proteinrr-1 family (agap009868-pa) | −2.69777 | −1.37197 | −2.55881 |
| cuticular proteinrr-1 family (agap009868-pa) | −2.76351 | −1.32027 | −2.42512 |
| translationally controlled tumor protein | 0.034448 | −1.09268 | −0.08592 |
| ---NA--- | 0.577938 | −0.64285 | 0.464805 |
| cg7759-isoform a | −3.12349 | −0.57699 | −2.50153 |
| ---NA--- | 1.009491 | −0.54194 | 0.573985 |
| ---NA--- | −1.55917 | −0.42465 | −1.51986 |
| cg16973-isoform c | −1.53556 | −0.1353 | −1.3262 |
| cg17816-isoform d | 1.306045 | 0.208671 | 1.382103 |
| cg10576-isoform a | −1.62996 | 0.609209 | −0.82538 |
| tollo cg6890-pa | −1.07589 | 0.613 | −0.52011 |
| smooth muscle | −0.15391 | 0.992315 | −1.22664 |
| ---NA--- | 0.055575 | 1.134956 | 0.073985 |
| ---NA--- | −0.02831 | 1.343717 | 0.004151 |
| ---NA--- | 0.29424 | 1.396849 | 0.19818 |
| ---NA--- | 0.433958 | 1.517664 | 0.468252 |
| ---NA--- | 0.421735 | 1.593478 | 0.320868 |
| ---NA--- | 0.313366 | 1.776955 | 0.436652 |
| ---NA--- | −0.01622 | 1.781616 | 0.37655 |
| pupal cuticle | −0.11425 | 2.05139 | 0.16826 |
| retinoid- and fatty-acid binding protein cg11064-pa isoform 1 | 0.637634 | 2.181204 | 0.327091 |
| ---NA--- | 1.019918 | 2.189003 | 0.530595 |
| golgi transport 1 homolog b | −0.13215 | 2.32078 | −0.23713 |
| cytochrome p450 | 3.899183 | 2.329793 | 4.20731 |
| pupal cuticle | 0.031307 | 2.484786 | −0.04927 |
| transcriptional regulator | 0.617591 | 2.505465 | 1.265079 |
| dicarbonyl l-xylulose reductase | 0.654662 | 2.569478 | 0.292406 |
| dopachrome conversion enzyme | 1.43119 | 2.666762 | 1.648458 |
| cuticle protein | 0.532201 | 2.707232 | 0.018046 |
| cg15527 | 1.603184 | 2.802856 | 1.588836 |
| gasp precursor | 1.429647 | 2.944655 | 1.202601 |
| ---NA--- | 0.008141 | 3.119834 | −0.24211 |
| cuticular protein 17 from low complexity family (agap006149-pa) | 0.849878 | 3.175253 | 1.237865 |
| ---NA--- | 1.420134 | 3.286538 | 1.896568 |
| ---NA--- | −0.09458 | 3.302558 | −0.26646 |
| ---NA--- | 1.076751 | 3.33122 | 0.29308 |
| male doublesex | 2.087831 | 3.378044 | 1.742706 |
| cg16885-isoform a | 2.342864 | 3.453355 | 1.745159 |
| collagen adhesin protein | 0.652254 | 3.526428 | 1.333105 |
| cuticular protein 17 from low complexity family (agap006149-pa) | 1.278416 | 3.564511 | 1.6851 |
| fk506-binding protein | 1.348947 | 3.638417 | 1.577823 |
| cuticular proteinrr-2 family (agap001668-pa) | 1.386482 | 3.799004 | 1.139587 |
| cuticular protein | 2.096998 | 3.855633 | 2.768874 |
| cuticle | 2.88288 | 3.921788 | 2.629512 |
| ---NA--- | −0.49092 | 3.938912 | −0.13293 |
| three prime repair exonuclease | 1.652532 | 3.962492 | 1.983267 |
| chromosome 1 open reading frame 151 protein | 1.40921 | 4.182193 | 1.924277 |
| cuticular proteinrr-2 family (agap001668-pa) | 1.791403 | 4.533675 | 1.469891 |
| cuticle | 3.104258 | 4.573813 | 3.216351 |
| cuticular proteinrr-2 family (agap010095-pa) | 1.880374 | 4.60833 | 1.604438 |
| cuticle | 3.124514 | 4.616544 | 3.180159 |
| cuticle | 2.979066 | 4.70642 | 2.795767 |
| cuticular proteinrr-2 family (agap001668-pa) | 1.841373 | 4.717938 | 1.726234 |
| cuticular proteinrr-2 family (agap001668-pa) | 1.760505 | 4.744734 | 1.418281 |
| major intrinsic protein of eye lens fiber | 3.72832 | 5.456043 | 4.260327 |
| cuticle | 3.133595 | 5.498162 | 4.136754 |
| cuticle | 3.476786 | 5.506462 | 2.983314 |
| cuticular proteinrr-2 family (agap003390-pa) | 3.90834 | 5.740922 | 4.431792 |
We performed an additional set of microarray comparisons to validate our estimate of morph-biased patterns of expression (used in later analyses of sequence divergence): head horn tissue of horned males was directly hybridized against head epidermis tissue of sneaker males. We found 38 genes significantly over-expressed in the horned male relative to the sneaker male, and 19 genes significantly over-expressed in the sneaker male relative to the horned male (Appendix 2). Additionally, our measure of morph-biased patterns of expression (e.g., the absolute difference in M value between head-abdomen arrays of horned and sneaker males) was highly correlated with the direct comparison of morph-biased expression in head epidermal tissue (i.e., the M value from horned-sneaker arrays; Supplementary Figure 2; R2 = 0.73, F1,1522 = 4160, P < 0.00001).
Appendix 2. Morph-biased genes as revealed by direct head tissue comparisons between horned and sneaker morphs.
Shown are genes that are significantly differentially expressed (adjusted P value < 0.05) in male morphs when head horn tissue was directly hybridized. Negative M values indicate a gene is more highly expressed in sneaker males relative to horned males; positive M values indicate a gene is more highly expressed in horned males relative to sneaker males. Gene annotation is according to (Kijimoto et al. 2009).
| Gene Name | M value |
|---|---|
| myosin light chain | −2.79951 |
| cg5939-isoform a | −2.65597 |
| muscle lim protein | −2.46179 |
| troponin i | −2.29138 |
| ---NA--- | −2.17178 |
| ---NA--- | −2.14284 |
| cg2867 | −1.85051 |
| tubulin beta chain | −1.70499 |
| ---NA--- | −1.24708 |
| ---NA--- | −1.19598 |
| ---NA--- | −0.94248 |
| ---NA--- | −0.85905 |
| hemolymph proteinase 19 | −0.78203 |
| homeobox protein araucan | −0.75062 |
| catalase | −0.74422 |
| cellular repressor of e1a-stimulated genes 1 | −0.74029 |
| cg14681-isoform b | −0.71791 |
| ---NA--- | −0.67913 |
| tubulin alpha chain | −0.48906 |
| lipoma hmgic fusion partner-like 3 | 0.858505 |
| leucine-rich ppr-motif containing | 0.923889 |
| cg30463-isoform a | 0.95565 |
| guanine nucleotide-binding protein galpha subunit | 1.020867 |
| ---NA--- | 1.024681 |
| ---NA--- | 1.030074 |
| ---NA--- | 1.062299 |
| serine threonine kinase 23 | 1.099674 |
| hypoxia up-regulated 1 | 1.1402 |
| cg8927-isoform a | 1.140227 |
| n-superoxide dismutase | 1.172646 |
| tollo cg6890-pa | 1.180807 |
| ---NA--- | 1.191761 |
| ---NA--- | 1.201925 |
| cg8384-isoform e | 1.228672 |
| vcell division cyclepaf1 rna polymerase ii complexhomolog | 1.255519 |
| nonmuscle myosin-ii heavy chain | 1.315133 |
| ---NA--- | 1.357758 |
| la ribonucleoprotein domainmember 1 | 1.363963 |
| cg1386-pa | 1.420593 |
| ---NA--- | 1.501933 |
| male doublesex | 1.631273 |
| golgi transport 1 homolog b | 1.707278 |
| transcriptional regulator | 1.723254 |
| cg10576-isoform a | 1.728858 |
| ---NA--- | 1.860702 |
| ---NA--- | 1.91156 |
| cuticular proteinrr-2 family (agap001668-pa) | 2.159434 |
| collagen adhesin protein | 2.547277 |
| cg7759-isoform a | 2.615693 |
| cuticular proteinrr-2 family (agap001668-pa) | 2.759644 |
| cuticular proteinrr-2 family (agap001668-pa) | 2.761131 |
| cuticular proteinrr-2 family (agap001668-pa) | 2.862221 |
| cuticular proteinrr-2 family (agap010095-pa) | 2.877466 |
| cuticle | 3.189423 |
| ---NA--- | 3.249518 |
| ---NA--- | 3.43462 |
| ---NA--- | 3.515686 |
Overall patterns of gene expression were highly correlated between morphs, sexes, and across tissue types (Pearson’s correlation > 0.80, Table 2, 3). Furthermore, for a given gene, the polarity of differential expression (i.e., higher in tissue A versus tissue B) was generally the same across morphs and sexes (Fig. 2).
Onthophagus taurus: patterns of expression in developing brains
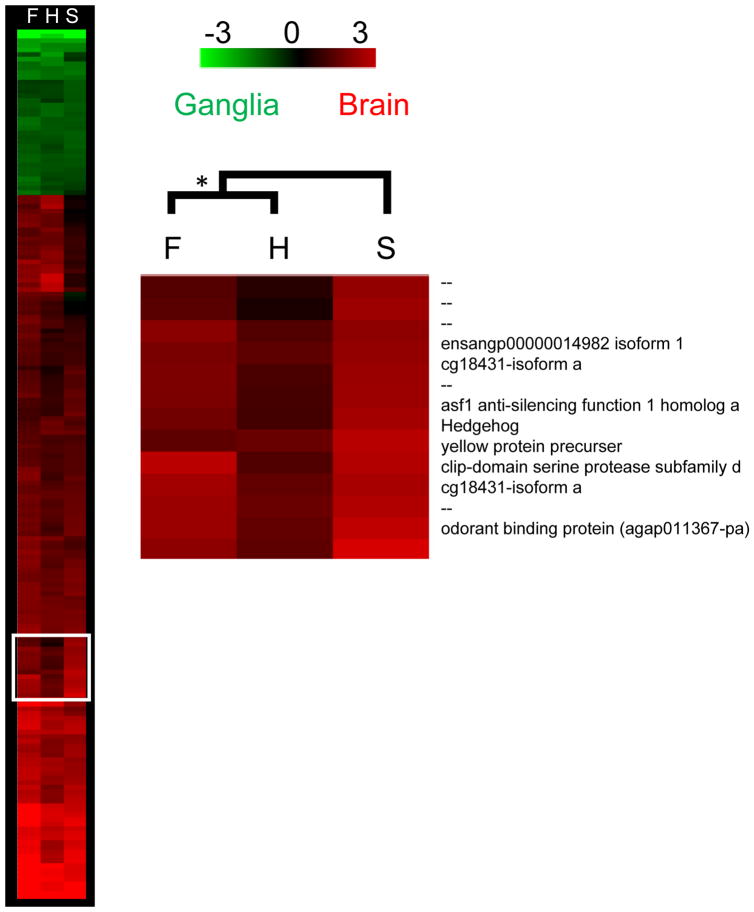
For the neural tissue arrays, 189 genes fit our criteria for inclusion in the hierarchical clustering analysis (P < 0.05 and |M| >1 for at least one treatment category). Overall patterns of expression in the developing brain of O. taurus were more similar between horned males and females than between the two male morphs (Fig. 3, Table 2). Moreover, patterns of gene expression in the brain and ganglia were remarkably dissimilar from the developing epidermis when brain and morphology arrays were compared directly (overall Pearson correlation mean (SD) = 0.07 (0.03) in N = 27 array comparisons).
Figure 3. Brain Arrays for O. taurus.
Shown are the results of a clustering analysis for the brain-ganglia arrays for O. taurus large, horned males (H), small, sneaker males (S) and females (F). Genes included in the analysis were significantly differentially expressed (P < 0.05) at least two-fold between tissues (|M| >1) in at least one treatment category (H, S, or F). We identified one cluster of genes with biased expression in the developing brain of sneaker males. Bootstrapping is 100%.
We were interested in genes and pathways with divergent expression between morphs, in particular with patterns of expression unique to the most different morph, in this case sneaker males, relative to horned males and females. One cluster fit these criteria (Fig. 3). Twenty-eight percent of the genes in this cluster were un-annotated. A complete list of O. taurus neural tissue genes with the most divergent expression patterns between morphs and sexes is presented in Appendix 3.
Appendix 3. Genes of the developing brain of O. taurus with the most divergent expression patterns between morphs and sexes.
Genes were filtered as follows: 1) genes were significantly differentially expressed in the brain relative to the ganglia (M > 0) or in the ganglia relative to the brain (M < 0) in at least one treatment category; 2) absolute difference in expression (M value) was greater than 0.75 when sneaker males were compared to both females and horned males. Shown are M values for genes fitting these criteria in brain-ganglia arrays for tissue from females, horned males and sneaker males. Gene annotation is according to (Kijimoto et al. 2009).
| Gene | Female | Horned | Sneaker |
|---|---|---|---|
| glucose dehydrogenase | −1.7085 | −1.52307 | −0.5793 |
| cuticle | 1.294428 | 1.209115 | −0.32958 |
| cuticle | 1.116113 | 0.964331 | −0.17994 |
| cuticle | 0.78352 | 0.849632 | −0.13251 |
| cuticular proteinrr-2 family (agap001668-pa) | 1.105493 | 0.744713 | −0.02102 |
| cuticle | 1.196771 | 1.432571 | 0.042271 |
| chromosome 1 open reading frame 151 protein | 1.29145 | 1.870667 | 0.121638 |
| fk506-binding protein | 1.399322 | 1.190266 | 0.1529 |
| lethalessential for lifel2efl | 1.398837 | 1.168393 | 0.161068 |
| cg33205-isoform f | 1.263757 | 1.852392 | 0.271971 |
| three prime repair exonuclease | 1.50772 | 1.158483 | 0.358808 |
| troponin i | 1.555121 | 2.373698 | 0.478137 |
| troponin i | 1.707857 | 2.417993 | 0.488219 |
| troponin i | 1.375902 | 2.190387 | 0.492013 |
| cg5939-isoform a | 1.541546 | 1.782398 | 0.60671 |
| paramyosin | 1.63252 | 1.800172 | 0.865861 |
| cg5939-isoform a | 1.759214 | 2.223351 | 0.972434 |
| ---NA--- | 2.475091 | 2.458102 | 1.307608 |
| cuticle | 2.97928 | 2.649723 | 1.759408 |
| ---NA--- | 1.064104 | 0.313479 | 1.858458 |
| sonic hedgehog b | 1.115904 | 1.253558 | 2.181145 |
| odorant binding protein (agap011367-pa) | 1.667497 | 1.117349 | 2.526956 |
Onthophagus nigriventris: patterns of expression in developing horns
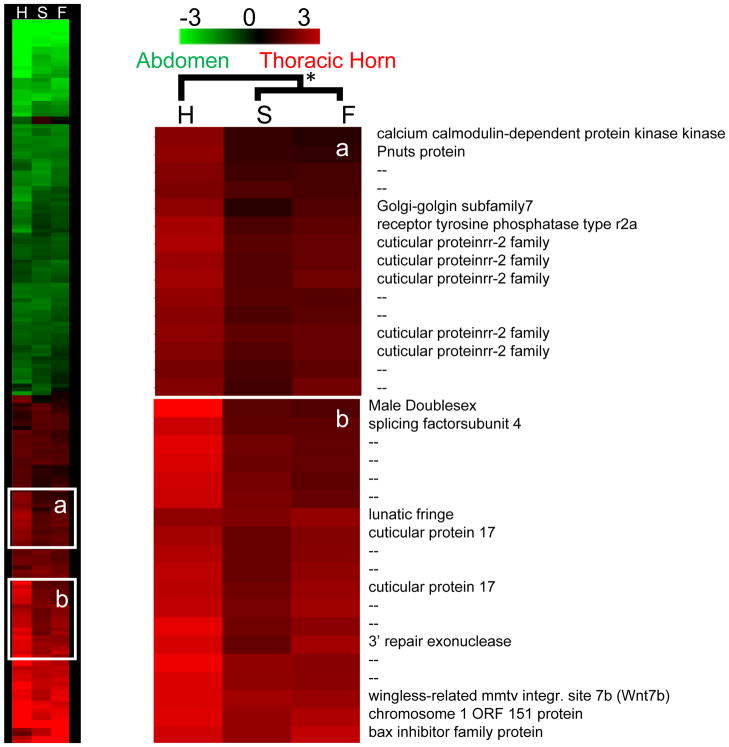
We replicated our approach for the prothoracic horns found in the congener O. nigriventris. Recall that in this species horn expression is confined to the prothorax. Only large adult males bear a single long, curved prothoracic horn, while small adult males and females develop only a small point or ridge on their prothorax, respectively. Also recall that in contrast to O. taurus, pupal resorption of thoracic horns is restricted to females and small males. For the O. nigriventris arrays, 448 genes fit our criteria for inclusion in the hierarchical clustering analysis (P < 0.05 and |M| >1 for at least one treatment category). While the present microarrays were developed for O. taurus, patterns of thoracic epidermal gene expression were highly correlated between species (Supplementary Fig. 1), suggesting limited use of these arrays in cross-species analyses was valid, although results should be interpreted with caution.
In contrast to the thoracic horns of O. taurus, overall patterns of expression in the thoracic horns of O. nigriventris were more similar between sneaker males and females than between the two male morphs (Fig. 4, Table 2). Two clusters of genes showed patterns of expression biased to the horned male (Fig. 4). Between these clusters, 47% of the genes were un-annotated and 21% were involved in cuticle development. A complete list of O. nigriventris thoracic-epidermis genes with the most divergent expression patterns between morphs and sexes is presented in Appendix 4.
Figure 4. Thoracic horn Arrays for O. nigriventris.
Shown are the results of a clustering analysis of thoracic horn-abdomen arrays for large, horned males (H), small, sneaker males (S) and females (F). Genes included in the analysis were significantly differentially expressed (P < 0.05) at least two-fold between tissues (|M| >1) in at least one treatment category (H, S, or F) We identified two clusters of genes with biased expression in the developing thoracic horn epidermis of horned males. Bootstrapping is 100%.
Appendix 4. Genes of the developing thoracic epidermis of O. nigriventris with the most divergent expression patterns between morphs and sexes.
Genes were filtered as follows: 1) genes were significantly differentially expressed in the thoracic epidermis relative to the abdomen (M > 0) or in the abdomen relative to the thorax (M < 0) in at least one treatment category; 2) absolute difference in expression (M value) was greater than 0.75 when horned males were compared to both females and sneaker males. Shown are M values for genes fitting these criteria in thoracic epidermis-abdomen arrays for tissue from females, horned males and sneaker males. Gene annotation is according to (Kijimoto et al. 2009).
| Gene | Female | Horned | Sneaker |
|---|---|---|---|
| myofilin protein | −3.36636 | −4.28976 | −3.52464 |
| leucine rich repeat containing 20 | −2.82641 | −3.95324 | −3.14418 |
| cg5939-isoform a | −2.20127 | −3.50905 | −2.13895 |
| cg33205-isoform f | −2.43331 | −3.49714 | −2.32824 |
| cg5939-isoform a | −2.44679 | −3.48479 | −2.34693 |
| cg5939-isoform a | −2.33613 | −3.3169 | −2.10347 |
| paramyosin | −2.43609 | −3.25782 | −2.00963 |
| muscular protein 20 | −1.84373 | −2.97949 | −1.89135 |
| myosin light chain | −1.89345 | −2.97087 | −2.21614 |
| myosin heavynonmuscle or smooth muscle | −1.74439 | −2.4968 | −1.52148 |
| muscle lim protein | −1.4099 | −2.47528 | −1.33804 |
| muscle lim protein | −0.84231 | −1.98286 | −1.02165 |
| ---NA--- | −0.632 | −1.87506 | −0.71674 |
| troponin i | −1.03083 | −1.86206 | −0.9607 |
| f-box leucine rich repeat protein | 0.290666 | −1.80495 | −0.93902 |
| slc39a9 protein | −0.81465 | −1.58076 | −0.53927 |
| 205 kda microtubule-associated protein | 0.608656 | −0.25735 | 0.56965 |
| cg2867 | 0.847172 | −0.22563 | 0.782536 |
| translocase of inner mitochondrial membrane 23 homolog | 3.332519 | 1.213426 | 2.200428 |
| obstractor b | 0.30255 | 1.237716 | 0.096169 |
| ---NA--- | 0.45308 | 1.440766 | −0.16057 |
| calcium calmodulin-dependent protein kinase kinase | 0.490072 | 1.621284 | 0.662207 |
| pnuts protein | 0.606768 | 1.699443 | 0.675363 |
| receptor tyrosine phosphatase type r2a | 1.112783 | 1.973167 | 0.9052 |
| cuticular proteinrr-2 family (agap010095-pa) | 1.208615 | 2.002869 | 1.048826 |
| smooth muscle | 3.673561 | 2.137674 | 3.510956 |
| ---NA--- | 1.201252 | 2.378341 | 1.442603 |
| splicing factorsubunit 4 | 1.132123 | 2.37955 | 1.121552 |
| ---NA--- | 1.125744 | 2.429575 | 1.399506 |
| cg15920-isoform a | 4.215076 | 2.45339 | 3.798524 |
| ---NA--- | 1.194697 | 2.576861 | 1.281907 |
| ---NA--- | 1.163802 | 2.661204 | 1.335176 |
| ---NA--- | 1.635242 | 2.708232 | 1.342403 |
| male doublesex | 0.953268 | 3.289013 | 1.081923 |
| cuticular proteinrr-3 family (agap006931-pa) | 2.115994 | 3.49637 | 2.236123 |
| cuticle | 2.428872 | 3.518445 | 2.646961 |
| ---NA--- | 1.936041 | 3.644659 | 2.425459 |
Patterns of Sequence Divergence
Morph-biased expression of genes was positively related to evolutionary divergence at the amino acid level (F1 = 3.82, P < 0.05, bst = 0.05) in a model that controlled for overall expression level, sex-biased expression, and the number of tissues in which the gene was expressed (overall model: F4,336 = 12.96, P < 0.0001, N = 341 genes). Overall expression level was negatively related to divergence (F1 = 48.8, P < 0.0001, bst = − 0.06), but there was no effect of sex-biased expression (F1 = 0.08, P = 0.78) or tissue-biased expression (F1 = 0.02, P = 0.88).
Discussion
Identifying the developmental genetic mechanisms underlying plasticity is critical for our understanding of the evolutionary origins and consequences of plasticity. The hypothesis of developmental decoupling views alternate phenotypes as resulting from switching on or off independent developmental pathways and suggests that alternative phenotypes have the capacity to be honed by selection independent of each other, similar to the positive effect of tissue- and sex-specific expression on sequence divergence (Hastings 1996; Duret and Mouchiroud 2000; Zhang and Li 2004; Jagadeeshan and Singh 2005; Liao et al. 2006; Ellegren and Parsch 2007; Haerty et al. 2007; Larracuente et al. 2008). In this work, we take steps to quantify the degree of developmental decoupling between morphs – with reference to classic examples of decoupling – and determine the evolutionary consequences of the observed morph-biased patterns of gene expression.
Decoupling in Polyphenic Development
We found that, at least for several tissues types, morph-biased expression is as divergent as sex-biased expression, a classical example of developmental decoupling (Bull 1983; Williams and Carroll 2009). We found that patterns of expression in the horn epidermis and neural tissue of developing beetle morphs were just as biased as expression patterns between the sexes (as supported by bootstrapping our clustering analysis). For instance, in the developing head epidermis of O. taurus (relative to the abdominal epidermis), overall patterns of gene expression were more similar between female and sneaker males than between the two male morphs (Fig. 2, Table 2), which matches morphological differences in horn expression. However, the development of the sneaker morph is not as simple as feminizing patterns of gene expression: in the developing brain of O. taurus (relative to the ganglia), patterns of expression in the horned male were more similar to those in the female than to the sneaker male (Fig. 3, Table 2). While our array results suggest that morph-biased expression is as divergent as sex-biased expression, it is important to note that our results apply solely to somatic tissue comparisons. Sex-specific expression is extensive when gonadal tissues are directly compared to each other, at times exceeding 30–50% of the expressed genome (Parisi et al. 2004) and it is presently unclear whether morph-biased gonadal expression in horned beetles would be as divergent as sex-specific expression.
We also observed that, across species, morph-biased expression was associated with polyphenic development; that is, morph-biased expression is as divergent as sex-biased expression only when a given trait shows differences between morphs. In the developing thoracic horn of O. taurus, which is re-absorbed in pupae such that neither male morph expresses an adult thoracic horn (Fig. 1), overall patterns of gene expression are more similar between male morphs than between males and females (Fig. 2), which show significant differences in thoracic horn allometry (Moczek 2006). In contrast, in the related O. nigriventris, where male morphs show pronounced differences in adult thoracic horn expression (Fig. 1), overall patterns of gene expression in the developing thoracic horn are more similar between sneaker males and females than between the two male morphs (Fig. 4), paralleling the results in O. taurus head horns (Fig. 2).
It is important to note that our study was restricted to one (24-hour) time point in development. To accurately determine the proportion of development that is decoupled between morphs, a more complete survey of gene expression over development time would be needed. It is possible that patterns of expression would be less biased between morphs if more time points were considered. Heterochronic shifts in gene expression are common (Abzhanov et al. 2004; Badyaev et al. 2008; Carleton et al. 2008) and such a shift in the developmental timing of one morph relative to another could account for our observed differences. On the other hand, surveying other time points in development – for instance during the horn proliferation phase in larvae or the horn scleritization phase in late pupae – could reveal even stronger patterns of morph-biased expression. Given the fact that relatively minor changes in upstream networks can lead to diverse changes in traits (Brakefield et al. 1996; Abouheif and Wray 2002; Moczek and Nagy 2005), it is possible that the most pronounced decoupling of morphs will actually be later in development, when more downstream genes have been turned on or off. Continuing research will help clarify these possibilities, in particular whether the same genes are employed over and over throughout development to produce different alternate phenotypes.
Alternatives to the Decoupling Hypothesis
The developmental decoupling hypothesis is in many ways a metaphor in science: a powerful tool by which we can summarize and apply complex processes, but also a hypothetical model, rather than biological reality (Nijhout 1990). The developmental decoupling hypothesis can be juxtaposed with the idea that phenotypic differences between morphs can come about through very minor changes in gene regulation, such that a common developmental program may be sufficient to govern the expression of alternative forms, as suggested by a range of allometric modeling studies (Nijhout and Wheeler 1996; Tomkins et al. 2005; Tomkins and Moczek 2009). This notion parallels an emerging theme in evo-devo that much intra- and inter-specific diversity can arise from the re-deployment of, and subtle changes in, the same developmental genes and networks over developmental time and space (Brakefield et al. 1996; Abouheif and Wray 2002; Moczek and Nagy 2005;Khila and Abouheif 2008).
Our data are consistent with both developmental decoupling and this “alternate” view of development. While our clustering analysis showed that morph-biased expression was as divergent as sex-biased expression, overall patterns of gene expression, as measured by Pearson correlation, were highly correlated between morphs and sexes (Table 2, 3). For instance, differences between morphs were much less than differences between tissues (e.g., 0.89–0.95 for morph comparisons versus 0.05–0.15 for correlation between brain and morphology arrays). Furthermore, few, if any, genes were entirely “on” in one morph and “off” in the other morph. These results are consistent with those from countless microarray studies which suggest that morph- or environment-biased, but not necessarily morph- or environment-specific, gene expression is common (rev. in Aubin-Horth and Renn 2009; Snell-Rood et al. 2010).
This study suggests that components of both the developmental decoupling hypothesis and the gene regulation hypothesis apply to polyphenic development. This juxtaposition of hypotheses explaining intraspecific diversity recalls a similar debate in the study of interspecific diversity. In discussing the relative importance of changes in regulatory regions versus protein coding regions (Hoekstra and Coyne 2007; Carroll 2008; Lynch and Wagner 2008; Stern and Orgogozo 2008), it seems that in reality, both processes contribute to generating diversity (Steiner et al. 2007). Such debates highlight the need for integrative models of development that account for inter- and intraspecific diversity.
Our data suggest that alternate morphs are to some extent developmentally decoupled, but not to an extreme extent, such as when organs arise from stem cells (sensu Morgan et al. 2005). Because gene expression between morphs is not entirely independent, alternative morphs may not be as free from pleiotropic constraints on divergence as is sometimes assumed. If 3–10% of development is decoupled, as suggested by our Pearson correlations (Table 2, 3), what effect does this have on the evolution of plasticity? If much of this presumed decoupling is due to morph-biased expression and not morph-specific expression, do the same evolutionary consequences hold? An analysis of the effects of such gene expression on sequence divergence allows a first step in answering these questions.
The effects of morph-biased expression on sequence divergence
Our results suggest that genes with morph-biased expression are more evolutionarily divergent than those with morph-shared expression. This effect is independent of other factors known to influence sequence divergence, such as sex-biased expression (Jagadeeshan and Singh 2005; Eads et al. 2007; Ellegren and Parsch 2007; Haerty et al. 2007; Larracuente et al. 2008), tissue-biased expression (Hastings 1996; Duret and Mouchiroud 2000; Zhang and Li 2004; Liao et al. 2006), or overall levels of gene expression (Drummond et al. 2005; Drummond and Wilke 2008). These data suggest that even when a small proportion of development is decoupled between morphs, there can be evolutionary consequences. Furthermore, it suggests that models of relaxed constraint that rely on morph-specific expression (Kawecki 1994; Kawecki et al. 1997; Van Dyken and Wade 2010) may also apply to morph-biased expression. This is an important implication because morph- or environment-biased expression is widespread (Aubin-Horth and Renn 2009; Hodgins-Davis and Townsend 2009; Snell-Rood et al. 2010) and is likely to be a far more general phenomenon than morph- and environment-specific gene expression.
Two non-mutually exclusive mechanisms can explain the observation that morph-biased genes are less conserved than morph-shared genes. First, morph-biased expression should relax pleiotropic constraints, “freeing” genes to adapt to the unique selective environment of either a sneaker male or a horned male (West-Eberhard 1989, 2003). Second, morph-biased expression increases the potential for relaxed selection because genes specific to one morph are hidden from selection when they are unexpressed in the alternate morph (Kawecki 1994; Kawecki et al. 1997; Snell-Rood et al. 2010; Van Dyken and Wade 2010). Thus, the probability of fixing deleterious mutations is higher and sequence divergence between species should be greater for morph-specific genes (Van Dyken and Wade 2010). It is likely that both mechanisms are playing a role in this system. Future, more thorough expression and sequence data may allow us to tease apart these separate mechanisms. For instance, because the frequency of each morph varies between species (Simmons et al. 2007), the degree of relaxed selection and thus sequence divergence of morph-biased genes should also vary between species (similar to analyses of Brisson and Nuzhdin 2008). In addition, more complete genomic sampling in this taxon will allow precise estimates of sequence divergence, signatures of the strength of purifying selection, and measures of genetic variation within species. In the future we will be able to more precisely determine the relationship between morph-biased expression and relaxed selection, reduced pleiotropic constraint, and other factors such as differences in positive selection between morphs.
Conclusions and Future Directions
The developmental mechanisms underlying phenotypic plasticity play a critical role in determining costs, limits and evolutionary consequences of plasticity. The hypothesis of developmental decoupling carries with it important implications about how plasticity may affect organismal diversification and the origin of novel traits, and yet we have a highly incomplete empirical picture of the proportion of development that is decoupled and the evolutionary effects of such decoupling. We present a first step in addressing these issues by comparing patterns of gene expression in beetle morphs. We found that morph-biased expression is comparable to other examples of developmental decoupling, in particular, the development of different sexes. However, gene expression was highly correlated across morphs (and sexes), suggesting that only a small proportion of development is decoupled and alternate developmental pathways are not as independent as often assumed. Nevertheless, the observed degree of decoupling had important evolutionary consequences. We found that morph-biased genes were more divergent than those with shared patterns of expression between morphs. This effect of morph-biased expression on sequence divergence was independent of sex-biased expression, tissue-biased expression, and overall levels of expression.
Future work is necessary to distinguish between the importance of relaxed selection and reduced pleiotropic constraints as consequences of morph-biased expression. A more thorough survey of patterns of gene expression across different tissues, developmental time points, and diverse species can distinguish between these hypotheses and further quantify the degree of decoupling in polyphenic development. Our results strongly suggest that the study of plasticity would also benefit from models of development that could incorporate complex interactions between decoupling and alternate regulation of the same pathways over developmental time and space. Finally, our data suggest interesting candidate genes and pathways for future studies of the developmental genetics of plasticity. For instance, the gene doublesex, a major regulator of sex differentiation (Christiansen et al. 2002; Estrada et al. 2003; Billeter et al. 2006; Camara et al. 2008; Sanchez 2008), was highly morph-biased in regions of horn development of both species. Future functional work will yield insights into developmental changes underlying this nutritional polyphenism and the diversification of horns more generally.
Supplementary Material
Each axis shows the M value for thoracic horn-abdominal epidermis comparisons, where positive values indicate greater expression in the thoracic horn relative to the abdomen. Shown is the correlation between such M values for microarrays conducted with O. taurus horned male tissue (X axis) and O. nigriventris horned male tissue (Y axis). Correlations are similarly significant for cross-species comparisons between sneaker male and female tissue. This figure illustrates only genes that were significantly expressed in at least one O. nigriventris array comparison (horned males, sneaker males, females).
Each axis shows a measure of morph-biased patterns of gene expression where positive values indicate greater expression in the head horn tissue of horned males relative to sneaker males. The X axis shows a direct measure of this value, where tissue from one male morph was directly hybridized from tissue with the other male morph. The Y axis shows an indirect measure of this value, the difference in M values between head-abdomen arrays of horned males and sneaker males. This figure illustrates only genes that were significantly differentially expressed in the head-abdomen arrays for either sneaker or horned male morphs.
Acknowledgments
The Center for Genomics and Bioinformatics at Indiana University and its staff, especially John Colbourne, Jacqueline Lopez, and James Ford, provided essential help and expertise during this work. We are also grateful to Matt Hahn for providing advice on measures of sequence divergence and to Tami Cruickshank, David Van Dyken and Mike Wade for discussions on relaxed selection and the evolution of plasticity. The Center for Genomics and Bioinformatics is funded in part by the METACyt Initiative of Indiana University, which is funded in part through a major grant from the Lilly Endowment. Work presented here was carried out while ECSR was supported by NIH NRSA F32GM083830 and AC was supported NSF IGERT training grant 0504627--206251A. Additional support was provided by National Science Foundation grant IOS 0820411 to JA and APM. The content of this paper does not necessarily represent the official views of the National Institutes of Health or the National Science Foundation.
Literature Cited
- Abouheif E, Wray GA. Evolution of the gene network underlying wing polyphenism in ants. Science. 2002;297:249–252. doi: 10.1126/science.1071468. [DOI] [PubMed] [Google Scholar]
- Abzhanov A, Protas M, Grant BR, Grant PR, Tabin CJ. Bmp4 and morphological variation of beaks in Darwin’s finches. Science. 2004;305:1462–1465. doi: 10.1126/science.1098095. [DOI] [PubMed] [Google Scholar]
- Alaux C, Sinha S, Hasadsri L, Hunt GJ, Guzman-Novoa E, DeGrandi-Hoffman G, Uribe-Rubio JL, Southey BR, Rodriguez-Zas S, Robinson GE. Honey bee aggression supports a link between gene regulation and behavioral evolution. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:15400–15405. doi: 10.1073/pnas.0907043106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews J, Bogart K, Burr A, Conaty J. Fabrication of DGRC cDNA microarrays. CGB Technical Report 2006. 2006:11. [Google Scholar]
- Aubin-Horth N, Landry CR, Letcher BH, Hofmann HA. Alternative life histories shape brain gene expression profiles in males of the same population. Proceedings of the Royal Society B-Biological Sciences. 2005;272:1655–1662. doi: 10.1098/rspb.2005.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubin-Horth N, Renn SCP. Genomic reaction norms: using integrative biology to understand molecular mechanisms of phenotypic plasticity. Molecular Ecology. 2009;18:3763–3780. doi: 10.1111/j.1365-294X.2009.04313.x. [DOI] [PubMed] [Google Scholar]
- Badyaev AV, Young RL, Oh KP, Addison C. Evolution on a local scale: Developmental, functional, and genetic bases of divergence in bill form and associated changes in song structure between adjacent habitats. Evolution. 2008;62:1951–1964. doi: 10.1111/j.1558-5646.2008.00428.x. [DOI] [PubMed] [Google Scholar]
- Barchuk AR, Cristino AS, Kucharski R, Costa LF, Simoes ZLP, Maleszka R. Molecular determinants of caste differentiation in the highly eusocial honeybeeApis mellifera. BMC Developmental Biology. 2007;7 doi: 10.1186/1471-213X-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate - a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B-Methodological. 1995;57:289–300. [Google Scholar]
- Billeter JC, Rideout EJ, Dornan AJ, Goodwin SF. Control of male sexual behavior in Drosophila by the sex determination pathway. Current Biology. 2006;16:R766–R776. doi: 10.1016/j.cub.2006.08.025. [DOI] [PubMed] [Google Scholar]
- Birney E, Clamp M, Durbin R. GeneWise and genomewise. Genome Research. 2004;14:988–995. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brakefield PM, Gates J, Keys D, Kesbeke F, Wijngaarden PJ, Monteiro A, French V, Carroll SB. Development, plasticity and evolution of butterfly eyespot patterns. Nature. 1996;384:236–242. doi: 10.1038/384236a0. [DOI] [PubMed] [Google Scholar]
- Brisson JA, Nuzhdin SV. Rarity of males in pea aphids results in mutational decay. Science. 2008;319:58–58. doi: 10.1126/science.1147919. [DOI] [PubMed] [Google Scholar]
- Bull JJ. Advanced Book Program. Benjamin/Cummings Pub. Co; Menlo Park, Calif: 1983. Evolution of sex determining mechanisms. [Google Scholar]
- Camara N, Whitworth C, Van Doren M. The creation of sexual dimorphism in the Drosophila soma. Sex Determination and Sexual Development. 2008;83:65. doi: 10.1016/S0070-2153(08)00403-1. [DOI] [PubMed] [Google Scholar]
- Carleton KL, Spady TC, Streelman JT, Kidd MR, McFarland WN, Loew ER. Visual sensitivities tuned by heterochronic shifts in opsin gene expression. BMC Biology. 2008;6:14. doi: 10.1186/1741-7007-6-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB. Evo-devo and an expanding evolutionary synthesis: A genetic theory of morphological evolution. Cell. 2008;134:25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- Cash AC, Whitfield CW, Ismail N, Robinson GE. Behavior and the limits of genomic plasticity: power and replicability in microarray analysis of honeybee brains. Genes Brain and Behavior. 2005;4:267–271. doi: 10.1111/j.1601-183X.2005.00131.x. [DOI] [PubMed] [Google Scholar]
- Christiansen AE, Keisman EL, Ahmad SM, Baker BS. Sex comes in from the cold: the integration of sex and pattern. Trends in Genetics. 2002;18:510–516. doi: 10.1016/s0168-9525(02)02769-5. [DOI] [PubMed] [Google Scholar]
- Donnell DM, Strand MR. Caste-based differences in gene expression in the polyembryonic wasp Copidosoma floridanum. Insect Biochemistry and Molecular Biology. 2006;36:141–153. doi: 10.1016/j.ibmb.2005.11.009. [DOI] [PubMed] [Google Scholar]
- Drummond DA, Bloom JD, Adami C, Wilke CO, Arnold FH. Why highly expressed proteins evolve slowly. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:14338–14343. doi: 10.1073/pnas.0504070102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DA, Wilke CO. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell. 2008;134:341–352. doi: 10.1016/j.cell.2008.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L, Mouchiroud D. Determinants of substitution rates in mammalian genes: Expression pattern affects selection intensity but not mutation rate. Molecular Biology and Evolution. 2000;17:68–74. doi: 10.1093/oxfordjournals.molbev.a026239. [DOI] [PubMed] [Google Scholar]
- Eads BD, Colbourne JK, Bohuski E, Andrews J. Profiling sex-biased gene expression during parthenogenetic reproduction in Daphnia pulex. BMC Genomics. 2007;8:13. doi: 10.1186/1471-2164-8-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:1–19. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H, Parsch J. The evolution of sex-biased genes and sex-biased gene expression. Nature Reviews Genetics. 2007;8:689–698. doi: 10.1038/nrg2167. [DOI] [PubMed] [Google Scholar]
- Emlen DJ. Environmental control of horn length dimorphism in the beetle Onthophagus acuminatus (Coleoptera, Scarabidae) Proceedings of the Royal Society of London Series B-Biological Sciences. 1994;256:131–136. [Google Scholar]
- Emlen DJ. Alternative reproductive tactics and male-dimorphism in the horned beetle Onthophagus acuminatus (Coleoptera:Scarabaeidae) Behavioral Ecology and Sociobiology. 1997;41:335–341. [Google Scholar]
- Emlen DJ. Integrating development with evolution: A case study with beetle horns. Bioscience. 2000;50:403–418. [Google Scholar]
- Emlen DJ. Costs and the diversification of exaggerated animal structures. Science. 2001;291:1534–1536. doi: 10.1126/science.1056607. [DOI] [PubMed] [Google Scholar]
- Emlen DJ, Nijhout HF. The development and evolution of exaggerated morphologies in insects. Annual Review of Entomology. 2000;45:661–708. doi: 10.1146/annurev.ento.45.1.661. [DOI] [PubMed] [Google Scholar]
- Emlen DJ, Hunt J, Simmons LW. Evolution of sexual dimorphism and male dimorphism in the expression of beetle horns: Phylogenetic evidence for modularity, evolutionary lability, and constraint. American Naturalist. 2005a;166:S42–S68. doi: 10.1086/444599. [DOI] [PubMed] [Google Scholar]
- Emlen DJ, Marangelo J, Ball B, Cunningham CW. Diversity in the weapons of sexual selection: Horn evolution in the beetle genus Onthophagus (Coleoptera: Scarabaeidae) Evolution. 2005b;59:1060–1084. [PubMed] [Google Scholar]
- Estrada B, Casares F, Sanchez-Herrero E. Development of the genitalia in Drosophila melanogaster. Differentiation. 2003;71:299–310. doi: 10.1046/j.1432-0436.2003.03017.x. [DOI] [PubMed] [Google Scholar]
- Evans JD, Wheeler DE. Differential gene expression between developing queens and workers in the honey bee, Apis mellifera. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:5575–5580. doi: 10.1073/pnas.96.10.5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans JD, Wheeler DE. Gene expression and the evolution of insect polyphenisms. Bioessays. 2001;23:62–68. doi: 10.1002/1521-1878(200101)23:1<62::AID-BIES1008>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Fisher RA. The genetical theory of natural selection. Clarendon Press; Oxford [Eng.]: 1930. [Google Scholar]
- Futschik M, Crompton T. Model selection and efficiency testing for normalization of cDNA microarray data. Genome Biology. 2004;5 doi: 10.1186/gb-2004-5-8-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haerty W, Jagadeeshan S, Kulathinal RJ, Wong A, Ram KR, Sirot LK, Levesque L, Artieri CG, Wolfner MF, Civetta A, Singh RS. Evolution in the fast lane: Rapidly evolving sex-related genes in Drosophila. Genetics. 2007;177:1321–1335. doi: 10.1534/genetics.107.078865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW, Kern AD. Comparative genomics of centrality and essentiality in three eukaryotic protein-interaction networks. Molecular Biology and Evolution. 2005;22:803–806. doi: 10.1093/molbev/msi072. [DOI] [PubMed] [Google Scholar]
- Hastings KEM. Strong evolutionary conservation of broadly expressed protein isoforms in the troponin I gene family and other vertebrate gene families. Journal of Molecular Evolution. 1996;42:631–640. doi: 10.1007/BF02338796. [DOI] [PubMed] [Google Scholar]
- Hodgins-Davis A, Townsend JP. Evolving gene expression: from G to E to GxE. Trends Ecol Evol. 2009;24:649–658. doi: 10.1016/j.tree.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra HE, Coyne JA. The locus of evolution: Evo devo and the genetics of adaptation. Evolution. 2007;61:995–1016. doi: 10.1111/j.1558-5646.2007.00105.x. [DOI] [PubMed] [Google Scholar]
- Hoffman EA, Goodisman MAD. Gene expression and the evolution of phenotypic diversity in social wasps. BMC Biology. 2007;5 doi: 10.1186/1741-7007-5-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hojo M, Koshikawa S, Cornette R, Matsumoto T, Miura T. Identification of soldier-specific genes in the nasute termite Nasutitermes takasagoensis (Isoptera: Termitidae) Entomological Science. 2005;8:379–387. [Google Scholar]
- Hunt J, Simmons LW. Maternal and paternal effects on offspring phenotype in the dung beetle Onthophagus taurus. Evolution. 2000;54:936–941. doi: 10.1111/j.0014-3820.2000.tb00093.x. [DOI] [PubMed] [Google Scholar]
- Hunt J, Simmons LW. Confidence of paternity and paternal care: covariation revealed through the experimental manipulation of the mating system in the beetle Onthophagus taurus. Journal of Evolutionary Biology. 2002;15:784–795. [Google Scholar]
- Jagadeeshan S, Singh RS. Rapidly evolving genes of Drosophila: Differing levels of selective pressure in testis, ovary, and head tissues between sibling. Molecular Biology and Evolution. 2005;22:1793–1801. doi: 10.1093/molbev/msi175. [DOI] [PubMed] [Google Scholar]
- Judice CC, Carazzole MF, Festa F, Sogayar MC, Hartfelder K, Pereira GAG. Gene expression profiles underlying alternative caste phenotypes in a highly eusocial bee, Melipona quadrifasciata. Insect Molecular Biology. 2006;15:33–44. doi: 10.1111/j.1365-2583.2005.00605.x. [DOI] [PubMed] [Google Scholar]
- Kawecki TJ. Accumulation of deleterious mutations and the evolutionary cost of being a generalist. American Naturalist. 1994;144:833–838. [Google Scholar]
- Kawecki TJ, Barton NH, Fry JD. Mutational collapse of fitness in marginal habitats and the evolution of ecological specialisation. Journal of Evolutionary Biology. 1997;10:407–429. [Google Scholar]
- Khila A, Abouheif E. Reproductive constraint is a developmental mechanism that maintains social harmony in advanced ant societies. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:17884–17889. doi: 10.1073/pnas.0807351105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kijimoto T, Costello J, Tang Z, Moczek A, Andrews J. EST and micorarray analysis of horn development in Onthophagus beetles. BMC Genomics. 2009;10:504. doi: 10.1186/1471-2164-10-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsolver JG, Wiernasz DC. Seasonal polyphenism in wing-melanin pattern and thermoregulatory adaptation in Pieris butterflies. American Naturalist. 1991;137:816–830. [Google Scholar]
- Klebes A, Biehs B, Cifuentes F, Kornberg TB. Expression profiling of Drosophila imaginal discs. Genome Biol. 2002;3:0038. doi: 10.1186/gb-2002-3-8-research0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutsukake M, Shibao H, Lee J-M, Fukatsu T. Soldier-specific genes expressed in the second instar sterile soldier of Tuberaphis styraci. In: Simon J-C, Dedryver C, Rispe C, Hullé M, editors. Aphids in a New Millennium. INRA Editions; Versailles: 2004. pp. 233–237. [Google Scholar]
- Larracuente AM, Sackton TB, Greenberg AJ, Wong A, Singh ND, Sturgill D, Zhang Y, Oliver B, Clark AG. Evolution of protein-coding genes in Drosophila. Trends in Genetics. 2008;24:114–123. doi: 10.1016/j.tig.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Liao BY, Scott NM, Zhang JZ. Impacts of gene essentiality, expression pattern, and gene compactness on the evolutionary rate of mammalian proteins. Molecular Biology and Evolution. 2006;23:2072–2080. doi: 10.1093/molbev/msl076. [DOI] [PubMed] [Google Scholar]
- Lynch VJ, Wagner GP. Resurrecting the role of transcription factor change in developmental evolution. Evolution. 2008;62:2131–2154. doi: 10.1111/j.1558-5646.2008.00440.x. [DOI] [PubMed] [Google Scholar]
- Madewell R, Moczek AP. Horn possession reduces maneuverability in the horn-polyphenic beetle, Onthophagus nigriventris. Journal of Insect Science. 2006;6:10. doi: 10.1673/2006_06_21.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCollum SA, VanBuskirk J. Costs and benefits of a predator-induced polyphenism in the gray treefrog Hyla chrysoscelis. Evolution. 1996;50:583–593. doi: 10.1111/j.1558-5646.1996.tb03870.x. [DOI] [PubMed] [Google Scholar]
- Moczek AP. Facultative paternal investment in the polyphenic beetle Onthophagus taurus: the role of male morphology and social context. Behavioral Ecology. 1999;10:641–647. [Google Scholar]
- Moczek AP. Pupal remodeling and the development and evolution of sexual dimorphism in horned beetles. American Naturalist. 2006;168:711–729. doi: 10.1086/509051. [DOI] [PubMed] [Google Scholar]
- Moczek AP. Pupal remodeling and the evolution and development of alternative male morphologies in horned beetles. BMC Evolutionary Biology. 2007;7 doi: 10.1186/1471-2148-7-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moczek AP, Emlen DJ. Proximate determination of male horn dimorphism in the beetle Onthophagus taurus (Coleoptera: Scarabaeidae) Journal of Evolutionary Biology. 1999;12:27–37. [Google Scholar]
- Moczek AP, Emlen DJ. Male horn dimorphism in the scarab beetle, Onthophagus taurus: do alternative reproductive tactics favor alternative phenotypes? Animal Behaviour. 2000;59:459–466. doi: 10.1006/anbe.1999.1342. [DOI] [PubMed] [Google Scholar]
- Moczek AP, Nagy LM. Diverse developmental mechanisms contribute to different levels of diversity in horned beetles. Evolution & Development. 2005;7:175–185. doi: 10.1111/j.1525-142X.2005.05020.x. [DOI] [PubMed] [Google Scholar]
- Moczek AP, Nijhout HF. Rapid evolution of a polyphenic threshold. Evolution & Development. 2003;5:259–268. doi: 10.1046/j.1525-142x.2003.03033.x. [DOI] [PubMed] [Google Scholar]
- Moczek AP, Nijhout HF. Trade-offs during the development of primary and secondary sexual traits in a horned beetle. American Naturalist. 2004;163:184–191. doi: 10.1086/381741. [DOI] [PubMed] [Google Scholar]
- Morgan HD, Santos F, Green K, Dean W, Reik W. Epigenetic reprogramming in mammals. Human Molecular Genetics. 2005;14:R47–R58. doi: 10.1093/hmg/ddi114. [DOI] [PubMed] [Google Scholar]
- Nice CC, Fordyce JA. How caterpillars avoid overheating: behavioral and phenotypic plasticity of pipevine swallowtail larvae. Oecologia. 2006;146:541–548. doi: 10.1007/s00442-005-0229-7. [DOI] [PubMed] [Google Scholar]
- Nijhout HF. Metaphors and the role of genes in development. Bioessays. 1990;12:441–446. doi: 10.1002/bies.950120908. [DOI] [PubMed] [Google Scholar]
- Nijhout HF. Insect hormones. Princeton University Press; Princeton, N.J.: 1994. [Google Scholar]
- Nijhout HF. Development and evolution of adaptive polyphenisms. Evolution & Development. 2003;5:9–18. doi: 10.1046/j.1525-142x.2003.03003.x. [DOI] [PubMed] [Google Scholar]
- Nijhout HF, Wheeler DE. Growth models of complex allometries in holometabolous insects. American Naturalist. 1996;148:40–56. [Google Scholar]
- Pal C, Papp B, Lercher MJ. An integrated view of protein evolution. Nature Reviews Genetics. 2006;7:337–348. doi: 10.1038/nrg1838. [DOI] [PubMed] [Google Scholar]
- Parisi M, Nuttall R, Edwards P, Minor J, Naiman D, Lu JN, Doctolero M, Vainer M, Chan C, Malley J, Eastman S, Oliver B. A survey of ovary-, testis-, and soma-biased gene expression in Drosophila melanogaster adults. Genome Biology. 2004;5:18. doi: 10.1186/gb-2004-5-6-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paspalas C, Consoulas C, Theophilidis G. The fate of specific motoneurons and sensory neurons of the pregenital abdominal segments in Tenebrio molitor (Insecta: Coleoptera) during metamorphosis. Rouxs Archives of Developmental Biology. 1993;202:204–213. doi: 10.1007/BF02427881. [DOI] [PubMed] [Google Scholar]
- Pereboom JJM, Jordan WC, Sumner S, Hammond RL, Bourke AFG. Differential gene expression in queen-worker caste determination in bumble-bees. Proceedings of the Royal Society B-Biological Sciences. 2005;272:1145–1152. doi: 10.1098/rspb.2005.3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfennig DW, Rice AM, Martin RA. Field and experimental evidence for competition’s role in phenotypic divergence. Evolution. 2007;61:257–271. doi: 10.1111/j.1558-5646.2007.00034.x. [DOI] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J. TM4: A free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Salathe M, Ackermann M, Bonhoeffer S. The effect of multifunctionality on the rate of evolution in yeast. Molecular Biology and Evolution. 2006;23:721–722. doi: 10.1093/molbev/msj086. [DOI] [PubMed] [Google Scholar]
- Sanchez L. Sex-determining mechanisms in insects. International Journal of Developmental Biology. 2008;52:837–856. doi: 10.1387/ijdb.072396ls. [DOI] [PubMed] [Google Scholar]
- Scharf ME, Wu-Scharf D, Pittendrigh BR, Bennett GW. Caste- and development-associated gene expression in a lower termite. Genome Biology. 2003;4 doi: 10.1186/gb-2003-4-10-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafiei M, Moczek AP, Nijhout HF. Food availability controls the onset of metamorphosis in the dung beetle Onthophagus taurus (Coleoptera: Scarabaeidae) Physiological Entomology. 2001;26:173–180. [Google Scholar]
- Simmons LW, Emlen DJ. Evolutionary trade-off between weapons and testes. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:16346–16351. doi: 10.1073/pnas.0603474103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons LW, Tomkins JL, Hunt J. Sperm competition games played by dimorphic male beetles. Proceedings of the Royal Society of London Series B-Biological Sciences. 1999;266:145–150. [Google Scholar]
- Simmons LW, Emlen DJ, Tomkins JL. Sperm competition games between sneaks and guards: A comparative analysis using dimorphic male beetles. Evolution. 2007;61:2684–2692. doi: 10.1111/j.1558-5646.2007.00243.x. [DOI] [PubMed] [Google Scholar]
- Smyth G. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Statistical Applications in Genetics and Molecular Biology. 2004;3:3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Smyth G. Limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and Compuatational Biology Solutions Using R and Bioconductor. Springer; New York: 2005. pp. 397–420. [Google Scholar]
- Snell-Rood E, JDVD, Cruickshank T, Wade M, Moczek A. Toward a population genetic framework of developmental evolution: costs, limits, and consequences of phenotypic plasticity. BioEssays. 2010;32:71–81. doi: 10.1002/bies.200900132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner CC, Weber JN, Hoekstra HE. Adaptive variation in beach mice produced by two interacting pigmentation genes. Plos Biology. 2007;5:1880–1889. doi: 10.1371/journal.pbio.0050219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern DL, Orgogozo V. The loci of evolution: How predictable is genetic evolution? Evolution. 2008;62:2155–2177. doi: 10.1111/j.1558-5646.2008.00450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumner S, Pereboom JJM, Jordan WC. Differential gene expression and phenotypic plasticity in behavioural castes of the primitively eusocial wasp, Polistes canadensis. Proceedings of the Royal Society B-Biological Sciences. 2006;273:19–26. doi: 10.1098/rspb.2005.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang ZJ, Choi JH, Hemmerich C, Sarangi A, Colbourne JK, Dong QF. ESTPiper - a web-based analysis pipeline for expressed sequence tags. BMC Genomics. 2009;10:8. doi: 10.1186/1471-2164-10-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomkins JL, Kotiaho JS, LeBas NR. Matters of scale: Positive allometry and the evolution of male dimorphisms. American Naturalist. 2005;165:389–402. doi: 10.1086/427732. [DOI] [PubMed] [Google Scholar]
- Tomkins JL, Moczek AP. Patterns of threshold evolution in polyphenic insects under different developmental models. Evolution. 2009;63:459–468. doi: 10.1111/j.1558-5646.2008.00563.x. [DOI] [PubMed] [Google Scholar]
- Toth AL, Varala K, Newman TC, Miguez FE, Hutchison SK, Willoughby DA, Simons JF, Egholm M, Hunt JH, Hudson ME, Robinson GE. Wasp gene expression supports an evolutionary link between maternal behavior and eusociality. Science. 2007;318:441–444. doi: 10.1126/science.1146647. [DOI] [PubMed] [Google Scholar]
- Van Dyken J, Wade M. On the evolutionary genetics of conditionally expressed genes. Genetics. 2010;184:557–570. doi: 10.1534/genetics.109.110163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vangelder RN, Vonzastrow ME, Yool A, Dement WC, Barchas JD, Eberwine JH. Amplified RNA synthesized from limited quantities of heterogeneous cDNA. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:1663–1667. doi: 10.1073/pnas.87.5.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson NL, Simmons LW. Mate choice in the dung beetle Onthophagus sagittarius: are female horns ornaments? Behavioral Ecology. 2010;21:424–430. [Google Scholar]
- Wegerhoff R. GABA and serotonin immunoreactivity during postembryonic brain development in the beetle Tenebrio molitor. Microscopy Research and Technique. 1999;45:154–164. doi: 10.1002/(SICI)1097-0029(19990501)45:3<154::AID-JEMT3>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- West-Eberhard MJ. Phenotypic plasticity and the origins of diversity. Annual Review of Ecology and Systematics. 1989;20:249–278. [Google Scholar]
- West-Eberhard MJ. Developmental plasticity and evolution. Oxford University Press; Oxford, UK: 2003. [Google Scholar]
- West-Eberhard MJ. Developmental plasticity and the origin of species differences. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:6543–6549. doi: 10.1073/pnas.0501844102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan S, Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Molecular Biology and Evolution. 2001;18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- Whitfield CW, Ben-Shahar Y, Brillet C, Leoncini I, Crauser D, LeConte Y, Rodriguez-Zas S, Robinson GE. Genomic dissection of behavioral maturation in the honey bee. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:16068–16075. doi: 10.1073/pnas.0606909103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams TM, Carroll SB. Genetic and molecular insights into the development and evolution of sexual dimorphism. Nature Reviews Genetics. 2009;10:797–804. doi: 10.1038/nrg2687. [DOI] [PubMed] [Google Scholar]
- Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Research. 2002;30 doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZH. PAML 4: Phylogenetic analysis by maximum likelihood. Molecular Biology and Evolution. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Zhang LQ, Li WH. Mammalian housekeeping genes evolve more slowly than tissue-specific genes. Molecular Biology and Evolution. 2004;21:236–239. doi: 10.1093/molbev/msh010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Each axis shows the M value for thoracic horn-abdominal epidermis comparisons, where positive values indicate greater expression in the thoracic horn relative to the abdomen. Shown is the correlation between such M values for microarrays conducted with O. taurus horned male tissue (X axis) and O. nigriventris horned male tissue (Y axis). Correlations are similarly significant for cross-species comparisons between sneaker male and female tissue. This figure illustrates only genes that were significantly expressed in at least one O. nigriventris array comparison (horned males, sneaker males, females).
Each axis shows a measure of morph-biased patterns of gene expression where positive values indicate greater expression in the head horn tissue of horned males relative to sneaker males. The X axis shows a direct measure of this value, where tissue from one male morph was directly hybridized from tissue with the other male morph. The Y axis shows an indirect measure of this value, the difference in M values between head-abdomen arrays of horned males and sneaker males. This figure illustrates only genes that were significantly differentially expressed in the head-abdomen arrays for either sneaker or horned male morphs.