Abstract
Voltage-gated calcium channels (CaVs) are large (~0.5 MDa), multisubunit, macromolecular machines that control calcium entry into cells in response to membrane potential changes. These molecular switches play pivotal roles in cardiac action potentials, neurotransmitter release, muscle contraction, calcium-dependent gene transcription and synaptic transmission. CaVs possess self-regulatory mechanisms that permit them to change their behaviour in response to activity, including voltage-dependent inactivation, calcium-dependent inactivation and calcium-dependent facilitation. These processes arise from the concerted action of different channel domains with CaV β-subunits and the soluble calcium sensor calmodulin. Until recently, nothing was known about the CaV structure at high resolution. Recent crystallographic work has revealed the first glimpses at the CaV molecular framework and set a new direction towards a detailed mechanistic understanding of CaV function.
Keywords: calcium-dependent facilitation (CDF), calcium-dependent inactivation (CDI), calmodulin (CaM), IQ domain, structural biology, voltage-gated calcium channel (CaV
Introduction
Voltage-gated calcium channels (CaVs) play a unique role in the interplay between electrical and chemical signalling in biological systems [1,2]. CaVs respond to membrane depolarization by opening calcium-selective pores. Calcium ion entry causes further membrane potential changes and, because calcium ions act as chemical messengers, may also ignite intracellular signalling cascades. As one of the principal sources of calcium in excitable cells, CaVs are under intense pressure to control calcium influx and to detect and respond to changes in intracellular calcium concentrations.
Three intrinsic well-characterized processes regulate CaVs: VDI (voltage-dependent inactivation), CDI (calcium-dependent inactivation) and CDF (calcium-dependent facilitation). VDI and CDI aid in calcium influx termination. CDI and CDF both act in a manner that can be thought of as ‘molecular short-term memory’, as channels that have already been opened ‘remember’ this fact and change their inactivation properties or are more readily opened by subsequent stimulation. Despite extensive functional characterization, it is only in the last 3 years that the molecular architecture that underpins VDI, CDI and CDF has begun to be revealed. As CaVs are major targets for drugs to treat hypertension, arrhythmias and pain [3], understanding channel architecture in atomic detail has implications that reach far beyond basic science.
CaV global architecture
Reconstitution of high-voltage-activated CaVs (CaV1s and CaV2s) requires four basic components: CaVα1 (CaV α1-subunit), CaVβ, CaM (calmodulin) and CaVα2δ (Figure 1A) [1,4]. CaVα1 is the transmembrane subunit (~1800–2300 amino acids long) that makes the voltage-sensitive, calcium-selective pore [1]. Primary structure analysis indicates that CaVα1 encodes four homologous repeats that each bears six transmembrane segments. These individual repeats share a common architecture with other members of the voltage-gated ion channel superfamily [2]. The first four transmembrane segments of each repeat (segments S1–S4) form the voltage-sensing domain, while the remaining two segments (S5 and S6) form the calcium-selective pore. Approximately half of the CaVα1 polypeptide forms the intracellular mass of the channel comprising the N- and C-terminal cytoplasmic domains and three interdomain regions (called the I-II, II-III and III-IV loops). These domains are the sites of action for a range of intracellular regulatory and signalling factors.
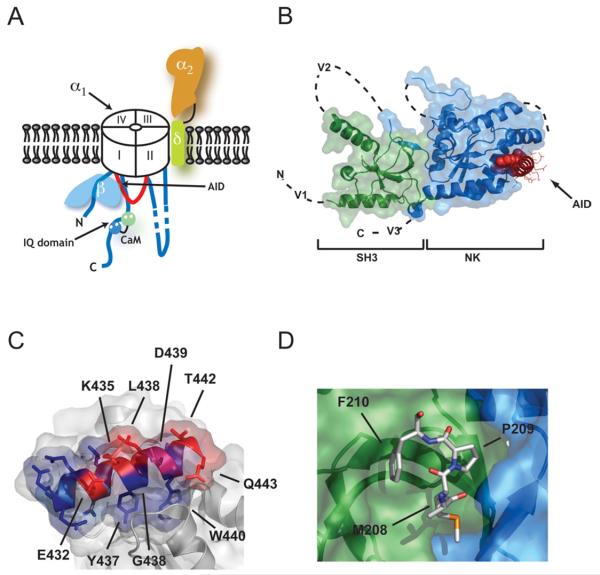
Figure 1. Voltage-gated calcium channel: CaVβ interactions.
(A) Schematic cartoon of a voltage-gated calcium channel. CaVs are composed of five polypeptides. The pore-forming CaVα1 subunit (white) is approx. 1800–2300 amino acids long, depending on the subtype, and is composed of four homologous membrane-spanning domains (I–IV) that are bridged intracellular loops. The transmembrane domains are indicated. For clarity, the shortest intracellular loop, the III–IV loop, is not depicted. Although the cartoon shows an anticlockwise transmembrane domain arrangement, the pore-forming subunit handedness is not known. CaVα2δ (orange, α2, and light green, δ) is a single pass transmembrane subunit formed from two disulfide-linked, glycosylated polypeptides: the extracellular α2 subunit (~930 amino acids) and the transmembrane δ subunit (~150 amino acids). Two intracellular proteins complete the complex. CaVβ (light blue) is an approx. 480–600-amino-acid subunit that binds with high affinity to the I-II intracellular loop (shown in red) at a binding domain known as the AID. CaVs also bind CaM. One of the sites of action of CaM is a conserved domain, known as an IQ domain, that is located on the C-terminal cytoplasmic segment of the α1 subunit. The N- and C-terminal lobes of CaM are shown in green and blue respectively. (B) The CaVβ–AID complex structure. Co-ordinates are for the CaVβ2a–CaV1.2 AID complex [22]. SH3 (green) and NK (blue) domains are indicated. V1, V2 and V3 show the locations of the three variable domains that are absent from the structure. The AID (red) binds to a deep groove (the ABP) in the NK domain. AID residues tyrosine, tryptophan and isoleucine are shown as CPK. The remaining residues are shown as lines. (C) The CaVβ–AID interaction uses conserved interactions. The degree of conservation among the CaVα1 subunits is shown. Blue, invariant; magenta, conserved; red, variable. Selective CaV1.2 AID residues are indicated. (D) Protein–protein interaction observed in the CaVβ2a structure between a portion of the V2 loop (residues 208–210) and a hydrophobic pocket on the SH3 surface. The binding site is on the opposite side of the SH3 domain from that shown in (B).
The two CaV intracellular subunits, CaVβ and CaM, bind to the I-II loop and the C-terminal cytoplasmic domain respectively and have prominent roles in VDI, CDI and CDF. Despite the fact that CaM is a highly expressed, ubiquitous protein, both forms, apo-CaM [5-7] and Ca2+/CaM [6-13], bind CaVα1. Thus, by virtue of its calcium-independent tethering to CaVα1, CaM can be considered an essential CaV subunit [14].
CaVs have one other essential protein, CaVα2δ [15]. CaVα2δ is made from a single gene product that is proteolytically processed into extracellular (α2) and membrane-spanning (δ) components that remain covalently associated by a disulfide bond. CaVα2δ is required for proper cell-surface expression of CaV complexes, but whether it also affects the kinetic properties of the channel remains unclear [15]. Perhaps the most interesting CaVα2δ property is that it is the target for gabapentin, an anti-epileptic and analgesic drug that acts via a yet unknown mechanism [16]. A final subunit, CaVγ , associates with skeletal-muscle calcium channels, but its general importance for other channel types remains uncertain [17].
The quaternary arrangement of CaV components remains undefined. Three low-resolution electron microscopy studies of skeletal-muscle CaVs have yielded very different images [18-20]. The limited resolution, together with the lack of a consensus, hinders the mechanistic utility of these studies. The present limitations in the use of native CaVs for high-resolution structural studies has inspired a different approach to obtain insight into the structural basis of CaV function. Division of the channel into functional parts has yielded the first two high-resolution structures of intracellular CaV components: CaVβ, alone and complexed to a part of the I–II loop [21-23], and the Ca2+/CaM-CaV1.2 IQ (Ile-Gln) domain complex [24,25]. Like many structural studies, these advances have settled some questions and raised others.
CaVβs
CaVβs are approx. 500-amino-acid cytoplasmic proteins that bind to the CaV I–II intracellular loop [26] and affect channel gating properties [27,28], trafficking [29,30], regulation by neurotransmitter receptors through G-protein βγ subunit activation [31] and sensitivity to drugs [32]. There are four mammalian isoforms and each has separate effects on CaV VDI properties [33]. The CaVβ primary sequence encodes five domains, arranged V1-C1-V2-C2-V3. V1, V2 and V3 are variable domains, whereas C1 and C2 are conserved [34]. C1-V2-C2 makes the CaVβ functional core [23,35,36]. The principal CaVβ–CaVα1 interaction site is a conserved 18-residue sequence in the I–II loop called the AID (α-interaction domain) [37,38].
Structural studies reveal that C1 and C2 form an SH3 domain (Src homology 3 domain) and an NK (nucleotide kinase) domain respectively (Figure 1B) [21-23]. The five-stranded SH3 domain has a split architecture in which the last β-strand that completes the fold (β5) occurs in the primary sequence following the V2 domain (Figure 1B). CaVβs share structural features with a family of scaffolding molecules, the MAGUKs (membrane-associated guanylate kinases) [39], but have some notable differences that include different relative orientation of the SH3 and NK domains as well as the absence of a number of essential MAGUK domains [21-23].
The structural work resolved how CaVβ binds the AID. A contiguous 41-amino-acid region, known as the BID (β-interaction domain) [34], which was thought to be the interaction site, was completely buried in the NK domain core and thus could not be the AID interaction site. Co-crystal structures of CaVβ–AID complexes [21-23] showed that the AID binds in a conserved, deep groove at the distal end of the CaVβ NK domain [named the ABP (α-binding pocket) [22]]. The AID interacts with CaVβ through a set of conserved residues that include two aromatic residues that are deeply buried in the ABP (Figure 1C).
While the AID–ABP complex forms the high-affinity CaVβ–CaVα1 site, there is evidence for lower affinity CaVβ–CaVα1 interactions [36,40], although recent studies challenge this idea for the I-II loop [41]. By virtue of size (CaVβ ~55 kDa), the varied effects of CaVβ subtypes on channel gating and the sheer amount of CaVα1 cytoplasmic mass (~1300 amino acids), it is inescapable that there must be other points of CaVβ–CaVα1 interaction. Although the exact sites are unknown, the CaVβ2a crystal structure provides clues to candidate sites. One is the fairly deep groove between the SH3 and NK domains on the CaVβ side that is opposite to the ABP [22]. A second is defined by a site where a small portion of the V2 loop of one subunit in the crystal lattice binds to a hydrophobic pocket on the SH3 domain surface of the adjacent CaVβ (Figure 1D) [22]. Such crystallographically defined protein–protein contacts often indicate sites that are in want of a binding partner, even if the observed partner is not the natural one [42]. Finally, the V1, V2 and V3 regions seem likely candidates for elements that alter channel gating through contacts with CaVα1.
Despite extensive functional study, the molecular mechanisms by which CaVβs act remain imperfectly understood. In addition to the biophysical changes, CaVα1–CaVβ interactions markedly influence cell-surface expression [29,30]. Control of CaV trafficking by cellular signalling proteins that regulate CaVα1–CaVβ interactions may provide an important means to modulate cellular excitability [29]. There also remains a question surrounding the role of the CaVβ–AID interactions in CaV inhibition by G-protein βγ subunits. Biochemical experiments show that Gβγ binds the AID peptide, albeit with weaker affinity than CaVβ [43]. The relevance of the Gβγ –AID interaction for physiological regulation is unclear and is challenged by the demonstration that key positions required for Gβγ –AID interactions are buried deep in the ABP [22]. Whether the mechanism of Gβγ regulation of CaV channels occurs by antagonizing CaVβ binding to the AID or through other mechanisms remains to be determined [31]. Finally, recent studies of CaVβ3 knockout mice have shown in pancreatic β-cells that CaVβs have a role in Ca2+ oscillation modulation that is independent of CaVα1 [44]. Defining such ‘non-channel’ CaVβ functions represents an exciting new direction. Because CaV channel subtypes are major targets for drugs for cardiovascular disease, migraine and pain [3], the development of compounds that could interfere with the CaVα1–CaVβ interactions based on structural information may provide new ways to modulate CaV function in pathological states.
The CaV Ca2+ /CaM–IQ domain interaction
Association with CaM endows CaVs with CDI and CDF properties [14]. The Ca2+/CaM target site is part of the CaV C-terminal cytoplasmic tail known as the IQ domain, a class of domains found in many Ca2+/CaM modulated proteins [45] (Figure 2A). Crystallographic studies have provided the first high-resolution structures of a Ca2+/CaM-IQ domain complex [24,25] and reveal features of the Ca2+/CaM-IQ domain interaction that have important consequences for understanding how CDI and CDF work. CaM has two lobes (N-lobe and C-lobe). Both wrap around a helix formed by the CaV1.2 IQ domain and each engages a set of three aromatic anchor residues from the IQ domain (Figures 2B and 2C). The complex is unusual in that the relative orientation of Ca2+/CaM and the IQ helix is the opposite to that in most other Ca2+/CaM–peptide complexes. Ca2+/CaM binds to the IQ domain helix in a parallel manner in which the N-lobe binds the N-terminal end of the IQ helix, while the C-lobe binds to the C-terminal end rather than in the usual antiparallel mode.
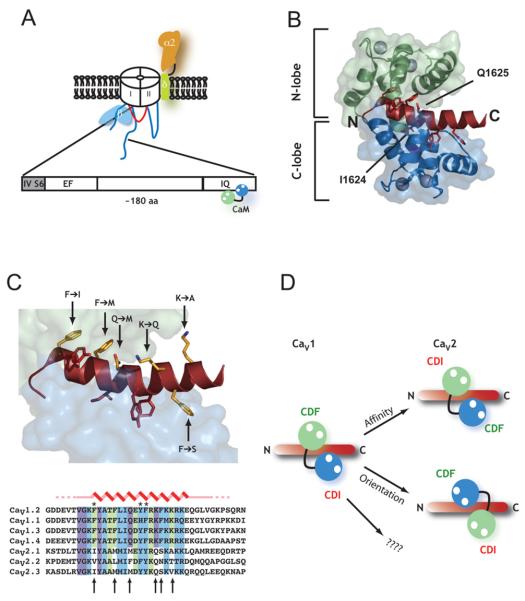
Figure 2. Structural insights into Ca2+-CaM regulation of CaVs.
(A) Schematic diagram of the conserved CaV C-terminal domain involved in calcium-dependent CaV regulation. The domain begins immediately after the last transmembrane segment of transmembrane domain IV (IVS6) and contains an EF-hand motif, a conserved intervening region and the IQ domain. Ca2+/CaM binds tightly (Kd in nM) to the IQ domain. (B) High-resolution structure of the Ca2+/CaM–CaV1.2 IQ domain complex [24]. Ca2+/CaM N-lobe (green) and C-lobe (blue) and IQ domain (dark red) are shown. Aromatic anchor positions on the IQ domain are shown in stick representation. The positions of Ile1624 and Gln1625 that define the IQ domain are also shown. Ca2+ ions are shown as spheres. (C) Six key Ca2+/N-lobe–IQ domain and Ca2+/C-lobe–IQ domain contact positions are conserved within CaV1s and CaV2s, but differ between the two subfamilies. Top: structure from (B) in which CaV1.2 residues are shown as sticks with the amino acid differences between CaV1.2 and CaV2.1 indicated. Bottom: sequence alignment of CaV1 and CaV2 IQ domains. Colours indicate residues that contact the Ca2+/N-lobe (green), Ca2+/C-lobe (blue) and both lobes (purple). Arrows indicate positions of the amino acid differences between CaV1s and CaV2s. (D) Hypotheses for the swapping of lobe-specific roles for Ca2+/CaM in CDF and CDI of CaV1s and CaV2s. The CaV1 schematic diagram shows the parallel orientation of the Ca2+/CaM-CaV1.2 IQ domain complex. The changes in key positions indicated in (C) may cause the swapping of CDF and CDI roles by causing affinity changes (right-hand side top), a reversal of Ca2+/N-lobe and Ca2+/C-lobe binding positions to an antiparallel orientation (bottom), or interactions with yet to be characterized structural elements.
One puzzle in understanding CDI and CDF has been that the action of opposite CaM lobes on the conserved IQ domain has different roles in the CaV1s and CaV2s [14]. In CaV1s, the Ca2+/C-lobe controls CDI, but CDI is governed by the Ca2+/N-lobe in CaV2s. There is a clear role for the Ca2+/C-lobe in CaV2 channel CDF but no previous characterization of the origin of CaV1 CDF. Titration calorimetry experiments establish two Ca2+/N-lobe binding sites (Kd ≈ 50 nM and Kd ≈ 20 μM) on the CaV1.2 IQ domain and a single high-affinity Ca2+/C-lobe site (Kd ≈ 2 nM) [24]. The Ca2+/C-lobe site matches well what is known regarding the prominence of the Ca2+/C-lobe in CaV1 CDI [9,46]. Disruption of the CaV1Ca2+/N-lobe binding site by simultaneous mutation of the three N-lobe aromatic anchors to alanine eliminates CDF and indicates a previously unknown role for the Ca2+/N-lobe interaction site in CaV1 CDF [24]. Together with previous work, this observation suggests that there is an exchange of roles for the two CaM lobes between the two channel types.
How might such a role exchange happen? In light of the structural data, comparison of the IQ domains from CaV1s and CaV2s reveals six amino acids that are conserved within the subtypes but not between the subtypes (Figure 2C). Notably, three positions are aromatic anchor sites. How might these changes cause exchange of N-lobe and C-lobe roles (Figure 2D)? One possibility is that the differences alter the relative affinities of the lobes for their respective IQ domain binding sites (and by linkage the calcium affinities of the individual CaM lobes) and that these changes tip the balance so that N-lobe binds more tightly in CaV2 channels and thereby leads to CDI. Another possibility is that the IQ domain residue differences result in an exchange of binding sites so that in CaV2 channels the Ca2+/CaM complex binds the IQ domain in the more common antiparallel arrangement (Figure 2D). In this way, the CDF site is in the same relative place (at the N-terminal end of the IQ peptide) but occupied by the other CaM lobe. While such a binding site exchange would provide a simple and elegant explanation for the exchange of lobe-specific roles in CDI and CDF, the situation may be more complicated. There are potential roles for apo-CaM [13,47] and CaMKII [48] in binding to the IQ peptide and in CDI and CDF. There may also be other CaM binding sites on CaVα1 that affect these processes. Clearly, a Ca2+/CaM–CaV2 IQ domain structure and structures of larger portions of the C-terminal domain will help clarify the issue.
Intracellular domains and gating
Given the structures of CaV intracellular components, a key question is how these domains influence the opening and closing of the CaV pore. The simplest model for understanding CaVβ regulation is that the AID helix is part of a longer helix that continues into the IS6 transmembrane segment (Figure 3A). As IS6 is implicated in channel closing, restriction of the movement of IS6 by CaVβ2a, which is anchored to the membrane by a palmitoylation site at the N-terminus of the V1 segment [26], may be an important mechanism for slowing channel inactivation. It is less obvious how the other CaVβs work. CaVβ1b and CaVβ3 speed inactivation and CaVβ4 causes moderate slowing [33]. None of these isoforms has an N-terminal lipid anchor. Because the core SH3 and NK domains are identical, it seems likely that variability in the V1, V2 and V3 segments will be responsible for the other properties via interaction with parts of CaVα1.
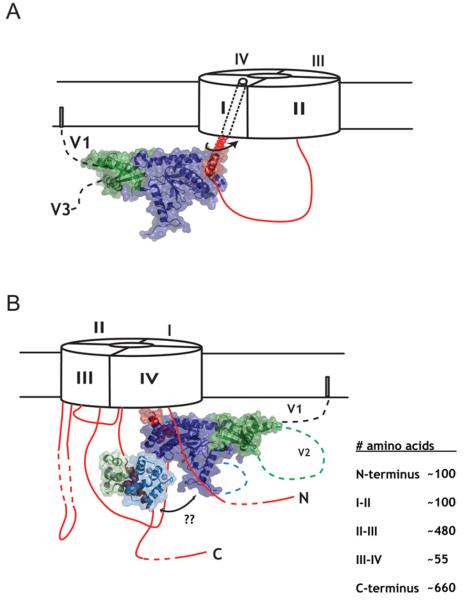
Figure 3. Structural insights into CaV gating.
(A) Cartoon model of how CaVβ affects CaVα1 gating. For simplicity, only the I–II loop is shown. CaVβ influences the movement of IS6. (B) Potential interplay between Ca2+/CaM regulation and CaVβ regulation of CaV channel gating. How such interactions occur remains to be revealed. The lengths of the intracellular domains are indicated.
The IQ domain is too far in primary sequence from IVS6 to postulate a simple model for how Ca2+/CaM effects gating. One interesting question is whether CaVβ and the CaM domain collaborate in some way to affect the movement of a common element in the pore (Figure 3B). The possibility that such a linkage happens via the isoleucine residue of the IQ domain [49] is unlikely as this residue is completely buried by the Ca2+/C-lobe [24].
There are other large CaV cytoplasmic regions. Defining interactions between these domains, their structures, their relationships to CaVβ and the CaM/IQ domain and their conformational changes are at the heart of determining how CaVs work. Eventually, we will need to see the architecture of the entire approx. 0.5 MDa complex and how CaVsintegrate into signalling networks [50]. Until then, pairing the piece-by-piece dissection of CaV structure with biochemical and functional studies is likely to be fruitful for understanding CaV design, function and regulation.
Acknowledgments
F.V.P. is an American Heart Association Postdoctoral Fellow. D.L.M. acknowledges support from the McKnight Foundation for Neuroscience, Rita Allen Foundation, Searle Scholars Award, Beckman Young Investigator Award and the National Institutes of Health.
Abbreviations used
- ABP
α-binding pocket
- AID
α-interaction domain
- Ca
voltage-gated calcium channel
- CDF
calcium-dependent facilitation
- CDI
calcium-dependent inactivation
- CaM
calmodulin
- MAGUK
membrane-associated guanylate kinase
- NK
nucleotide kinase
- SH3 domain
Src homology 3 domain
- VDI
voltage-dependent inactivation
References
- 1.Catterall WA. Annu. Rev. Cell Dev. Biol. 2000;16:521–555. doi: 10.1146/annurev.cellbio.16.1.521. [DOI] [PubMed] [Google Scholar]
- 2.Hille B. Ion Channels of Excitable Membranes. Sinauer Associates; Sunderland, MA: 2001. [Google Scholar]
- 3.Kochegarov AA. Cell Calcium. 2003;33:145–162. doi: 10.1016/s0143-4160(02)00239-7. [DOI] [PubMed] [Google Scholar]
- 4.Arikkath J, Campbell KP. Curr. Opin. Neurobiol. 2003;13:298–307. doi: 10.1016/s0959-4388(03)00066-7. [DOI] [PubMed] [Google Scholar]
- 5.Erickson MG, Alseikhan BA, Peterson BZ, Yue DT. Neuron. 2001;31:973–985. doi: 10.1016/s0896-6273(01)00438-x. [DOI] [PubMed] [Google Scholar]
- 6.Peterson BZ, DeMaria CD, Adelman JP, Yue DT. Neuron. 1999;22:549–558. doi: 10.1016/s0896-6273(00)80709-6. [DOI] [PubMed] [Google Scholar]
- 7.Pitt GS, Zühlke RD, Hudmon A, Schulman H, Reuter H, Tsien RW. J. Biol. Chem. 2001;276:30794–30802. doi: 10.1074/jbc.M104959200. [DOI] [PubMed] [Google Scholar]
- 8.Qin N, Olcese R, Bransby M, Lin T, Birnbaumer L. Proc. Natl. Acad. Sci. U.S.A. 1999;96:2435–2438. doi: 10.1073/pnas.96.5.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zühlke RD, Pitt GS, Deisseroth K, Tsien RW, Reuter H. Nature. 1999;399:159–162. doi: 10.1038/20200. [DOI] [PubMed] [Google Scholar]
- 10.Pate P, Mochca-Morales J, Wu Y, Zhang JZ, Rodney GG, Serysheva II, Williams BY, Anderson ME, Hamilton SL. J. Biol. Chem. 2000;275:39786–39792. doi: 10.1074/jbc.M007158200. [DOI] [PubMed] [Google Scholar]
- 11.DeMaria CD, Soong TW, Alseikhan BA, Alvania RS, Yue DT. Nature. 2001;411:484–489. doi: 10.1038/35078091. [DOI] [PubMed] [Google Scholar]
- 12.Lee A, Zhou H, Scheuer T, Catterall WA. Proc. Natl. Acad. Sci. U.S.A. 2003;100:16059–16064. doi: 10.1073/pnas.2237000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang W, Halling DB, Black DJ, Pate P, Zhang JZ, Pedersen S, Altschuld RA, Hamilton SL. Biophys. J. 2003;85:1538–1547. doi: 10.1016/s0006-3495(03)74586-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Halling DB, Aracena-Parks P, Hamilton SL. Science STKE. 20062006:er1. doi: 10.1126/stke.3182006er1. [DOI] [PubMed] [Google Scholar]
- 15.Cantí C, Davies A, Dolphin AC. Curr. Neuropharmacol. 2003;1:209–217. [Google Scholar]
- 16.Sills GJ. Curr. Opin. Pharmacol. 2006;6:108–113. doi: 10.1016/j.coph.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 17.Kang MG, Campbell KP. J. Biol. Chem. 2003;278:21315–21318. doi: 10.1074/jbc.R300004200. [DOI] [PubMed] [Google Scholar]
- 18.Wolf M, Eberhart A, Glossmann H, Striessnig J, Grigorieff N. J. Mol. Biol. 2003;332:171–182. doi: 10.1016/s0022-2836(03)00899-4. [DOI] [PubMed] [Google Scholar]
- 19.Serysheva II, Ludtke SJ, Baker MR, Chiu W, Hamilton SL. Proc. Natl. Acad. Sci. U.S.A. 2002;99:10370–10375. doi: 10.1073/pnas.162363499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang MC, Velarde G, Ford RC, Berrow NS, Dolphin AC, Kitmitto A. J. Mol. Biol. 2002;323:85–98. doi: 10.1016/s0022-2836(02)00890-2. [DOI] [PubMed] [Google Scholar]
- 21.Opatowsky Y, Chen CC, Campbell KP, Hirsch JA. Neuron. 2004;42:387–399. doi: 10.1016/s0896-6273(04)00250-8. [DOI] [PubMed] [Google Scholar]
- 22.Van Petegem F, Clark KA, Chatelain FC, Minor DL., Jr Nature. 2004;429:671–675. doi: 10.1038/nature02588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen YH, Li MH, Zhang Y, He LL, Yamada Y, Fitzmaurice A, Shen Y, Zhang H, Tong L, Yang J. Nature. 2004;429:675–680. doi: 10.1038/nature02641. [DOI] [PubMed] [Google Scholar]
- 24.Van Petegem F, Chatelain FC, Minor DL., Jr Nat. Struct. Mol. Biol. 2005;12:1108–1115. doi: 10.1038/nsmb1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fallon JL, Halling DB, Hamilton SL, Quiocho FA. Structure. 2005;13:1881–1886. doi: 10.1016/j.str.2005.09.021. [DOI] [PubMed] [Google Scholar]
- 26.Dolphin AC. J. Bioenerg. Biomembr. 2003;35:599–620. doi: 10.1023/b:jobb.0000008026.37790.5a. [DOI] [PubMed] [Google Scholar]
- 27.Colecraft HM, Alseikhan B, Takahashi SX, Chaudhuri D, Mittman S, Yegnasubramanian V, Alvania RS, Johns DC, Marban E, Yue DT. J. Physiol. 2002;541:435–452. doi: 10.1113/jphysiol.2002.018515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hullin R, Khan IF, Wirtz S, Mohacsi P, Varadi G, Schwartz A, Herzig S. J. Biol. Chem. 2003;278:21623–21630. doi: 10.1074/jbc.M211164200. [DOI] [PubMed] [Google Scholar]
- 29.Beguin P, Nagashima K, Gonoi T, Shibasaki T, Takahashi K, Kashima Y, Ozaki N, Geering K, Iwanaga T, Seino S. Nature. 2001;411:701–706. doi: 10.1038/35079621. [DOI] [PubMed] [Google Scholar]
- 30.Bichet D, Cornet V, Geib S, Carlier E, Volsen S, Hoshi T, Mori Y, De Waard M. Neuron. 2000;25:177–190. doi: 10.1016/s0896-6273(00)80881-8. [DOI] [PubMed] [Google Scholar]
- 31.Dolphin AC. Pharmacol. Rev. 2003;55:607–627. doi: 10.1124/pr.55.4.3. [DOI] [PubMed] [Google Scholar]
- 32.Hering S. Trends Pharmacol. Sci. 2002;23:509–513. doi: 10.1016/s0165-6147(02)02104-1. [DOI] [PubMed] [Google Scholar]
- 33.Stotz SC, Zamponi GW. Trends Neurosci. 2001;24:176–181. doi: 10.1016/s0166-2236(00)01738-0. [DOI] [PubMed] [Google Scholar]
- 34.De Waard M, Pragnell M, Campbell KP. Neuron. 1994;13:495–503. doi: 10.1016/0896-6273(94)90363-8. [DOI] [PubMed] [Google Scholar]
- 35.Opatowsky Y, Chomsky-Hecht O, Kang MG, Campbell KP, Hirsch JA. J. Biol. Chem. 2003;278:52323–52332. doi: 10.1074/jbc.M303564200. [DOI] [PubMed] [Google Scholar]
- 36.Maltez JM, Nunziato DA, Kim J, Pitt GS. Nat. Struct. Mol. Biol. 2005;12:372–377. doi: 10.1038/nsmb909. [DOI] [PubMed] [Google Scholar]
- 37.Pragnell M, De Waard M, Mori Y, Tanabe T, Snutch TP, Campbell KP. Nature. 1994;368:67–70. doi: 10.1038/368067a0. [DOI] [PubMed] [Google Scholar]
- 38.Witcher DR, De Waard M, Liu H, Pragnell M, Campbell KP. J. Biol. Chem. 1995;270:18088–18093. doi: 10.1074/jbc.270.30.18088. [DOI] [PubMed] [Google Scholar]
- 39.Funke L, Dakoji S, Bredt DS. Annu. Rev. Biochem. 2005;74:219–245. doi: 10.1146/annurev.biochem.74.082803.133339. [DOI] [PubMed] [Google Scholar]
- 40.Walker D, Bichet D, Campbell KP, De Waard M. J. Biol. Chem. 1998;273:2361–2367. doi: 10.1074/jbc.273.4.2361. [DOI] [PubMed] [Google Scholar]
- 41.Butcher AJ, Leroy J, Richards MW, Pratt WS, Dolphin AC. J. Physiol. 2006;574:387–398. doi: 10.1113/jphysiol.2006.109744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arkin MR, Wells JA. Nat. Rev. Drug Discov. 2004;3:301–317. doi: 10.1038/nrd1343. [DOI] [PubMed] [Google Scholar]
- 43.De Waard M, Liu H, Walker D, Scott VE, Gurnett CA, Campbell KP. Nature. 1997;385:446–450. doi: 10.1038/385446a0. [DOI] [PubMed] [Google Scholar]
- 44.Berggren PO, Yang SN, Murakami M, Efanov AM, Uhles S, Kohler M, Moede T, Fernstrom A, Appelskog IB, Aspinwall CA, et al. Cell. 2004;119:273–284. doi: 10.1016/j.cell.2004.09.033. [DOI] [PubMed] [Google Scholar]
- 45.Bahler M, Rhoads A. FEBS Lett. 2002;513:107–113. doi: 10.1016/s0014-5793(01)03239-2. [DOI] [PubMed] [Google Scholar]
- 46.Zühlke RD, Pitt GS, Tsien RW, Reuter H. J. Biol. Chem. 2000;275:21121–21129. doi: 10.1074/jbc.M002986200. [DOI] [PubMed] [Google Scholar]
- 47.Erickson MG, Liang H, Mori MX, Yue DT. Neuron. 2003;39:97–107. doi: 10.1016/s0896-6273(03)00395-7. [DOI] [PubMed] [Google Scholar]
- 48.Hudmon A, Schulman H, Kim J, Maltez JM, Tsien RW, Pitt GS. J. Cell Biol. 2005;171:537–547. doi: 10.1083/jcb.200505155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim J, Ghosh S, Nunziato DA, Pitt GS. Neuron. 2004;41:745–754. doi: 10.1016/s0896-6273(04)00081-9. [DOI] [PubMed] [Google Scholar]
- 50.Davare MA, Avdonin V, Hall DD, Peden EM, Burette A, Weinberg RJ, Horne MC, Hoshi T, Hell JW. Science. 2001;293:98–101. doi: 10.1126/science.293.5527.98. [DOI] [PubMed] [Google Scholar]