Abstract
RNA polymerase III (RNA pol III) transcribes many small structural RNA molecules involved in RNA processing and translation, and thus regulates the growth rate of a cell. Accurate initiation by RNA pol III requires the initiation factor TFIIIB. TFIIIB has been demonstrated to be regulated by tumor suppressors, including ARF, p53, RB, and the RB-related pocket proteins, and is a target of the oncogene c-myc and the mitogen-activated protein kinase ERK. EGCG has been demonstrated to inhibit the growth of a variety of cancer cells, induce apoptosis and regulate the expression of p53, myc, and ERK. Thus, we hypothesized that EGCG may regulate RNA pol III transcription in cells. Here, we report that EGCG (1) inhibits RNA pol III transcription from gene internal and gene external promoters (2) EGCG inhibits protein expression of the TFIIIB subunits Brf1 and Brf2, and (3) EGCG inhibits Brf2 promoter activity in cervical carcinoma cells.
Keywords: RNA polymerase III, TFIIIB, Brf1, Brf2, EGCG
Catechins are flavonoids found in many plant-derived food products including fruits, berries, chocolate, wine, and green tea (Camellia sinensis)[1]. (−)-Epigallocatechin gallate (EGCG) (Figure 1) constitutes the largest percentage of catechins found in green tea, accounting for 65% of the total catechin content[2]. Epidemiological and animal studies have demonstrated that EGCG provides protection against a variety of cancers including those of the skin, lung, prostate and breast [3–6]. EGCG also possess anti-proteolytic [7, 8], anti-mutagenic [9, 10], and anti-proliferative [5, 11–13] activity, thereby regulating the growth of a cell.
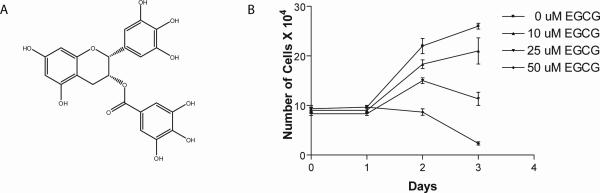
Figure 1. EGCG inhibits HeLa cell proliferation.
(A) Structure of (−) - epigallocatechin gallate (EGCG). (B) Hela cells treated with increasing concentrations of EGCG as indicated and counted using a hemocytometer at the time points indicated. Each dose and time point was performed in triplicate.
RNA polymerase III (RNA pol III) is the largest of the eukaryotic DNA dependent RNA polymerases, with 17 subunits, and transcribes many of the genes involved in processing (U6 snRNA) and translation (tRNA), thereby dictating the growth rate of a cell [14]. RNA pol III activity has been shown to be deregulated in a variety of cancers, reviewed in [15–17]. Like all other eukaryotic RNA polymerases, RNA pol III cannot recognize its target promoters directly. Proper initiation by RNA pol III is dependent on the transcription factor TFIIIB [14, 18]. In higher eukaryotes, at least two forms of TFIIIB have been identified [19–21]. TFIIIB required for initiation from gene internal RNA pol III promoters is comprised of TBP, Bdp1, and Brf1 [22, 23]. Initiation from gene external RNA pol III promoters requires TBP, Bdp1, and Brf2 [19–21].
TFIIIB is a molecular target of regulation by tumor suppressors, including ARF [24], p53 [25–29], RB [30, 31], and the RB-related pocket proteins [32], the oncogene c-myc [30, 33] and the mitogen-activated protein kinase ERK [34]. RNA pol III and TFIIIB activity have been demonstrated to be negatively regulated by other proteins, such as Maf1 [35–37]. Deregulation of TFIIIB-mediated transcription could be an important step in tumor development. Hence, we speculate that TFIIIB-mediated transcription may be a novel target of regulation by chemopreventive agents.
EGCG and RNA polymerase III have both been demonstrated to regulate cell growth and proliferation. We hypothesized that EGCG may regulate cell growth through modulation of RNA pol III transcription in the cervical carcinoma cell line HeLa. Here, we report that: (1) EGCG treatment inhibits transcription by RNA pol III, from both gene internal (tRNA) and gene external (U6 snRNA) promoters, (2) EGCG inhibits expression of the TFIIIB subunits Brf1 and Brf2, and (3) activity from the human Brf2 promoter is inhibited by EGCG.
MATERIALS AND METHODS
Cell culture and luciferase assays
HeLa cells were maintained in 5% FBS DMEM (Biowhittaker). RNA pol III luciferase vectors and assays have been previously described[37]. For EGCG experiments, cells were treated with increasing concentrations of EGCG (Alexis Corp.), in 1% DMSO, 24 hours post transfection, and harvested at times indicated in the figure legends. Experiments were performed in triplicate, repeated three independent times, and the data presented are representative. Statistical analysis was performed using one-way ANOVA with a Tukey post-test with a 95% confidence interval (GraphpadPrism3.03, San Diego California USA).
Western blot analysis
Untransfected and HeLa cells transiently transfected with an empty Flag vector or 3XFlagMaf1[37] were treated with 0 uM or 25 uM EGCG, harvested, and nuclear extracts were prepared [20], and western blot were performed as previously described [20, 37].
Cloning of the Brf2 promoter
Gene2Promoter analysis software program of Genomatix Suite (www.genomatixsuite.de) was utilized to identify a putative promoter for human Brf2. Putative transcription factor sites within the human Brf2 promoter were identified using MatInspector [38] of the Genomatix Suite. PCR primers flanking the Brf2 promoter sequence were designed with Kpn I and Bgl II restriction sites and used to PCR amplify the promoter sequence from human genomic DNA and cloned into the Kpn I and Bgl II sites of the promoterless pGL3 Basic vector.
RESULTS AND DISCUSSION
EGCG inhibits RNA polymerase III transcription in HeLa cells
The human cervical carcinoma cell line HeLa was treated with 10uM, 25uM, and 50uM EGCG (Figure1B) for 24, 48 and 72 hours. As demonstrated in Figure 1B, treatment with increasing concentrations of EGCG inhibited HeLa cell proliferation in a dose dependent manner, consistent with previously published reports [12, 39]. We then determined if EGCG could modulate RNA pol III transcription in these cells. Asynchronous HeLa cells were transiently transfected with either pGL3-U6 (gene external RNA pol III U6 promoter), pGL3-VAI (prototypical gene internal RNA pol III tRNA-like promoter), or a promoterless pGL3 vector and treated with increasing concentrations of EGCG (5uM, 10uM, 25uM, 50uM) and analyzed using our recently characterized RNA pol III luciferase assay [37]. Treatment of HeLa cells with increasing concentrations of EGCG inhibits transcription from the U6 (Figure 2A) promoter in a dose-dependent manner. At the lowest dose tested, 5uM, the data was highly statistically significant, p<0.001. Interestingly, U6 transcription appears to be more sensitive to treatment with EGCG at lower concentrations than VAI transcription, compare Figure 2A to 2B. Inhibition of VAI transcription was statistically significant only at higher EGCG doses of 25uM and 50uM. The observed inhibition of RNA pol III transcription in Figure 2 is a direct consequence of EGCG treatment, as the total cellular protein concentrations remained unchanged despite treatment with increasing concentrations of EGCG (Figures 2 and 2B, lower panels).
Figure 2. EGCG inhibits RNA pol III transcription in HeLa cells.
(A) HeLa cells transiently transfected with pGL3-U6 (100ng), or empty pGL3 vector (100ng), treated with increasing concentrations of EGCG (0uM, 5uM, 10uM, 25uM, 50uM). Lower panel corresponds to total cellular protein concentrations used in luciferase assay as determined by Lowry assay. (B) HeLa cells transiently transfected with pGL3-VAI (100ng), or empty pGL3 vector (100ng), treated with increasing concentrations of EGCG. Lower panel corresponds to total cellular protein concentrations used in luciferase assay as determined by Lowry assay. All luciferase assay results are expressed as relative light units (RLU): the average of the Photinus pyralis firefly activity observed divided by the average of the activity recorded from the Renilla luciferase vector. Experiments were done in triplicate, repeated three times, and representative experiments are depicted. Statistical analysis was performed using one-way ANOVA with a Tukey post-test with a 95% confidence interval (Graphpad Prism 3.03); * = p <0.05; ** = p < 0.01; *** = p < 0.001.
Inhibition of RNA pol III transcription does not appear to be due to the direct inhibition of the enzymatic activity of RNA pol III by EGCG, as the observed inhibition from gene internal and gene external promoters was not equivalent. We therefore investigated the effect of EGCG on the RNA pol III initiation factor TFIIIB.
EGCG inhibits expression of the TFIIIB subunits Brf1 and Brf2
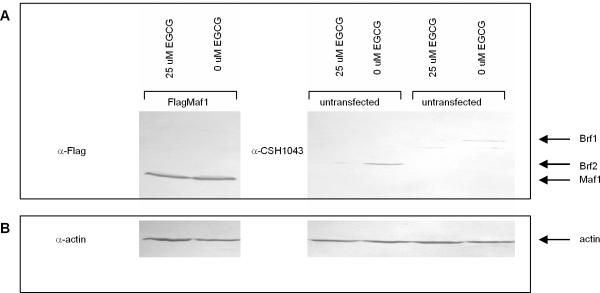
To elucidate the mechanism by which EGCG inhibits RNA pol III transcription, we determined if EGCG affects expression of TFIIIB. We examined the expression of the TFIIIB subunits Brf1, specific for gene internal RNA pol III promoters, and Brf2, specific for expression from gene external RNA pol III promoters in response to EGCG treatment. Nuclear extract was prepared from untransfected HeLa cells and HeLa cells transiently transfected with FlagMaf1 treated with 0 or 25uM EGCG for 72 hours. A 25uM dose of EGCG was used since this dose inhibited both U6 and VAI transcription and was statistically significant (Figure 2). The nuclear extracts were resolved by SDS-PAGE electrophoresis, transferred to nitrocellulose, immunoblotted with anti-Flag, or a previously characterized CSH1043 antibody known to cross react with both Brf1 and Brf2 [20]. Expression of the TFIIIB subunits Brf1 and Brf2 was severely diminished with 25uM EGCG treatment (Figure 3A). Interestingly, expression of the Maf1 protein, a negative regulator RNA pol III transcription in response to stress in vitro [35, 36] and in vivo [37], was unaffected by EGCG (Figure 3A), suggesting the mechanism by which EGCG inhibits RNA pol III transcription may occur via a pathway independent of Maf1. Maf1 is a common component of at least three signaling pathways leading to RNA pol III transcription repression: the secretory defect signaling pathway, the target of rapamycin (TOR) signaling pathway, and the DNA damage signaling pathway ([40], reviewed in [41, 42]). Although the protein levels of exogenously Flag-tagged Maf1 remain constant in response to EGCG, we cannot speculate if this is true for endogenous Maf1 expression.
Figure 3. EGCG inhibits expression of the TFIIIB subunits Brf1 and Brf2.
(A) Nuclear extract prepared from HeLa cells transiently transfected with FlagMaf1 and untransfected HeLa cells treated with 0uM or 25uM EGCG and immunoblotted with an anti-flag and CSH1043 antibodies. Arrows depict migration of Brf1, Brf2 and Maf1. (B) Same immunoblot from 2A reprobed using anti-actin antibody as loading control.
The human Brf2 promoter activity is inhibited by EGCG
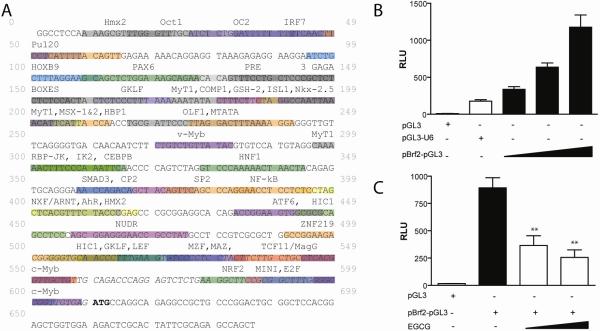
The observed decrease in Brf1 and Brf2 protein expression in Figure 3 raises the possibilities that EGCG may inhibit RNA pol III transcription by: (1) increasing the rate of Brf1 and Brf2 protein turnover, (2) decreasing the stability of Brf1 and Brf2 mRNA, or (3) decreasing expression of Brf1 and Brf2 genes. Previous results have demonstrated that EGCG inhibits proteasome activity, reviewed in [43]. Therefore, to determine the mechanism(s) by which Brf1 and Brf2 expression is inhibited by EGCG we cloned the human Brf2 promoter. Using Gene2Promoter software of the Genomatix Suite, we identified a putative promoter for Brf2, Figure 4A. We amplified this putative Brf2 promoter by PCR and sub-cloned it into the promoterless pGL3-Basic vector, generating Brf2-pGL3. As expected, Brf2-pGL3 was able to drive luciferase expression in a DNA concentration-dependent manner in HeLa cells (Figure 4B). Treatment of Brf2-pGL3 with increasing concentrations of EGCG showed a statistically significant (p<0.01) decrease in luciferase expression (Figure 4C). These data suggest that the observed decrease in Brf2 expression (Figure 3) may be a direct result of inhibition of Brf2 promoter activity by EGCG.
Figure 4. EGCG inhibits activity from the human Brf2 promoter.
(A) Schematic representation of the human Brf2 promoter as determined by Gene2Promoter software of Genomatix Suite. Putative transcription factor binding sites are depicted as identified by MapInspector [38]. Bolded ATG indicates start of translation, and italicized sequence denotes 5' untranslated sequence. (B) HeLa cells transiently transfected with pGL3 (200ng), pGL3-U6 (200ng), Brf2-pGL3 (50ng, 100ng, 200ng). (3) HeLa cells transiently transfected with pGL3 (100ng), Brf2-pGL3 (100ng) and treated with increasing concentrations of EGCG. Statistical analysis was performed using one-way ANOVA with a Tukey post-test with a 95% confidence interval (GraphpadPrism3.03);**=p<0.01.
Analysis of the Brf2 promoter (Figure 4A) reveals a single binding site for nuclear factor kappa B (NF-κB). Hence, we speculate that regulation of the human Brf2 promoter may involve NF-κB which has been previously demonstrated to be regulated by EGCG [11, 44, 45]. NF-κB is an oxidative stress-sensitive transcription factor regulating genes involved in inflammation, immunity, growth and cell death [43]. The ROS scavenging property of EGCG inhibits NF-κB activation, leading to decreased expression of pro-inflammatory and survival genes [46]. The Brf2 promoter also has a putative binding site for the HMG-Box containing protein 1 (HBP1) (Figure 4A) a known transcription repressor which has been demonstrated to be induced upon EGCG treatment [12]. Also, activity from the Brf2 promoter could be inhibited by the activity of HBP1 in response to EGCG, leading to the inhibition of RNA pol III transcription. Additionally, it has been demonstrated that EGCG can inhibit DNA methyltransferases (DNMT) in vitro [47]. We speculate that demethylation of the hypermethylated in cancer (HIC1) promoter allows for expression of this tumor suppressor, and ultimately leading to inhibition of Brf2 expression (Figure 4A). Future mutation analysis of the Brf2 promoter will be paramount in determining the mechanism of EGCG regulation.
To the best of our knowledge, this is the first report of the modulation of the activity of RNA polymerase III by a natural product with known anticancer activity. Our data demonstrates that EGCG negatively regulates RNA pol III transcription in HeLa cells. This observed RNA pol III inhibition may be a direct result of the inhibition of Brf1 and Brf2 expression by EGCG. We further demonstrate, in the case of U6 RNA pol III transcription, Brf2 expression is regulated at the transcriptional level by EGCG. Future studies to determine the effect of EGCG on the expression of other TFIIIB subunits Bdp1 and TBP will provide more detailed insight into the mechanism(s) by which EGCG regulates RNA pol III transcription from both gene internal and gene external promoters.
RNA pol III regulates cell proliferation and has been shown to be de-regulated by oncogenic signals [15–17]. Polyphenolic antioxidants such as EGCG have been shown to possess cancer chemopreventative activity, mediated by several mechanisms that are postulated to stem either from its antioxidant effects as a ROS (reactive oxygen species) scavenger [6] or by direct inhibition of molecular targets [47–49]. The antioxidant activity of EGCG has been shown to regulate multiple signal transduction pathways [43, 50] including the activities of several oxidative stress-responsive transcription factors such as NF-κB, AP-1 and Nrf2. However, low concentrations of EGCG has also been shown to be pro-oxidative and to increase the concentrations of H2O2 in the culture medium, and postulated to be responsible for the pro-apoptotic and antiproliferative effects of EGCG [51, 52]. In our studies, we tested concentrations of EGCG (up to 25uM), previously demonstrated to inhibit cell proliferation and intracellular oncogenic signaling networks [53]. Whether the inhibition of Brf2 expression and RNA pol III transcription is due to the direct antioxidant effect of EGCG, or via the generation of H2O2 can be determined by repeating the experiments in Figure 2 in the presence of catalase, previously shown to decompose hydrogen peroxide in culture systems [51, 52]. Further investigation of the transcriptional control of Brf2 expression via kinase-dependent signaling networks will also shed light into the mechanism of inhibition of RNA polymerase III by EGCG. These studies may reveal novel mechanisms of the anti-tumor properties of this natural product.
ACKNOWLEDGEMENTS
This work was supported in part by the Henry Luce foundation (L. Schramm), a St. John's University faculty growth grant (L. Schramm) and by the Department of Education's Graduate Assistance in Areas of National Need (GAANN) Grant P200A010130 (I. Veras and S. Cabarcas).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Zaveri NT. Green tea and its polyphenolic catechins: medicinal uses in cancer and noncancer applications. Life Sci. 2006;78(18):2073–80. doi: 10.1016/j.lfs.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 2.Horie N, Hirabayashi N, Takahashi Y, Miyauchi Y, Taguchi H, Takeishi K. Synergistic effect of green tea catechins on cell growth and apoptosis induction in gastric carcinoma cells. Biol Pharm Bull. 2005;28(4):574–9. doi: 10.1248/bpb.28.574. [DOI] [PubMed] [Google Scholar]
- 3.Mukhtar H, Ahmad N. Tea polyphenols: prevention of cancer and optimizing health. Am J Clin Nutr. 2000;71(6 Suppl):1698S–702S. doi: 10.1093/ajcn/71.6.1698S. discussion 1703S–4S. [DOI] [PubMed] [Google Scholar]
- 4.Yang CS, Lambert JD, Hou Z, Ju J, Lu G, Hao X. Molecular targets for the cancer preventive activity of tea polyphenols. Mol Carcinog. 2006;45(6):431–5. doi: 10.1002/mc.20228. [DOI] [PubMed] [Google Scholar]
- 5.Yang CS, Maliakal P, Meng X. Inhibition of carcinogenesis by tea. Annu Rev Pharmacol Toxicol. 2002;42:25–54. doi: 10.1146/annurev.pharmtox.42.082101.154309. [DOI] [PubMed] [Google Scholar]
- 6.Yang CS, Sang S, Lambert JD, Hou Z, Ju J, Lu G. Possible mechanisms of the cancer-preventive activities of green tea. Mol Nutr Food Res. 2006;50(2):170–5. doi: 10.1002/mnfr.200500105. [DOI] [PubMed] [Google Scholar]
- 7.Annabi B, Lachambre MP, Bousquet-Gagnon N, Page M, Gingras D, Beliveau R. Green tea polyphenol (−)-epigallocatechin 3-gallate inhibits MMP-2 secretion and MT1-MMP-driven migration in glioblastoma cells. Biochim Biophys Acta. 2002;1542(1–3):209–20. doi: 10.1016/s0167-4889(01)00187-2. [DOI] [PubMed] [Google Scholar]
- 8.Nam S, Smith DM, Dou QP. Ester bond-containing tea polyphenols potently inhibit proteasome activity in vitro and in vivo. J Biol Chem. 2001;276(16):13322–30. doi: 10.1074/jbc.M004209200. [DOI] [PubMed] [Google Scholar]
- 9.Hernaez JF, Xu M, Dashwood RH. Antimutagenic activity of tea towards 2-hydroxyamino-3-methylimidazo[4,5-f]quinoline: effect of tea concentration and brew time on electrophile scavenging. Mutat Res. 1998;402(1–2):299–306. doi: 10.1016/s0027-5107(97)00309-6. [DOI] [PubMed] [Google Scholar]
- 10.Kuroda Y, Hara Y. Antimutagenic and anticarcinogenic activity of tea polyphenols. Mutat Res. 1999;436(1):69–97. doi: 10.1016/s1383-5742(98)00019-2. [DOI] [PubMed] [Google Scholar]
- 11.Lambert JD, Yang CS. Mechanisms of cancer prevention by tea constituents. J Nutr. 2003;133(10):3262S–3267S. doi: 10.1093/jn/133.10.3262S. [DOI] [PubMed] [Google Scholar]
- 12.Kim J, Zhang X, Rieger-Christ KM, Summerhayes IC, Wazer DE, Paulson KE, Yee AS. Suppression of Wnt signaling by the green tea compound (−)-epigallocatechin 3-gallate (EGCG) in invasive breast cancer cells. Requirement of the transcriptional repressor HBP1. J Biol Chem. 2006;281(16):10865–75. doi: 10.1074/jbc.M513378200. [DOI] [PubMed] [Google Scholar]
- 13.Kumar N, Shibata D, Helm J, Coppola D, Malafa M. Green tea polyphenols in the prevention of colon cancer. Front Biosci. 2007;12:2309–15. doi: 10.2741/2233. [DOI] [PubMed] [Google Scholar]
- 14.Schramm L, Hernandez N. Recruitment of RNA polymerase III to its target promoters. Genes Dev. 2002;16(20):2593–620. doi: 10.1101/gad.1018902. [DOI] [PubMed] [Google Scholar]
- 15.White RJ. RNA polymerase III transcription and cancer. Oncogene. 2004;23(18):3208–16. doi: 10.1038/sj.onc.1207547. [DOI] [PubMed] [Google Scholar]
- 16.White RJ. RNA polymerase III transcription--a battleground for tumour suppressors and oncogenes. Eur J Cancer. 2004;40(1):21–7. doi: 10.1016/j.ejca.2003.09.027. [DOI] [PubMed] [Google Scholar]
- 17.White RJ. RNA polymerases I and III, growth control and cancer. Nat Rev Mol Cell Biol. 2005;6(1):69–78. doi: 10.1038/nrm1551. [DOI] [PubMed] [Google Scholar]
- 18.Kassavetis GA, Geiduschek EP. Transcription factor TFIIIB and transcription by RNA polymerase III. Biochem Soc Trans. 2006;34(Pt 6):1082–7. doi: 10.1042/BST0341082. [DOI] [PubMed] [Google Scholar]
- 19.McCulloch V, Hardin P, Peng W, Ruppert JM, Lobo-Ruppert SM. Alternatively spliced hBRF variants function at different RNA polymerase III promoters. Embo J. 2000;19(15):4134–43. doi: 10.1093/emboj/19.15.4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schramm L, Pendergrast PS, Sun Y, Hernandez N. Different human TFIIIB activities direct RNA polymerase III transcription from TATA-containing and TATA-less promoters. Genes Dev. 2000;14(20):2650–63. doi: 10.1101/gad.836400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Teichmann M, Wang Z, Roeder RG. A stable complex of a novel transcription factor IIB- related factor, human TFIIIB50, and associated proteins mediate selective transcription by RNA polymerase III of genes with upstream promoter elements. Proc Natl Acad Sci U S A. 2000;97(26):14200–5. doi: 10.1073/pnas.97.26.14200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mital R, Kobayashi R, Hernandez N. RNA polymerase III transcription from the human U6 and adenovirus type 2 VAI promoters has different requirements for human BRF, a subunit of human TFIIIB. Mol Cell Biol. 1996;16(12):7031–42. doi: 10.1128/mcb.16.12.7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Z, Roeder RG. Structure and function of a human transcription factor TFIIIB subunit that is evolutionarily conserved and contains both TFIIB- and high-mobility-group protein 2-related domains. Proc Natl Acad Sci U S A. 1995;92(15):7026–30. doi: 10.1073/pnas.92.15.7026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morton JP, Kantidakis T, White RJ. RNA polymerase III transcription is repressed in response to the tumour suppressor ARF. Nucleic Acids Res. 2007;35(9):3046–52. doi: 10.1093/nar/gkm208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stein T, Crighton D, Warnock LJ, Milner J, White RJ. Several regions of p53 are involved in repression of RNA polymerase III transcription. Oncogene. 2002;21(36):5540–7. doi: 10.1038/sj.onc.1205739. [DOI] [PubMed] [Google Scholar]
- 26.Stein T, Crighton D, Boyle JM, Varley JM, White RJ. RNA polymerase III transcription can be derepressed by oncogenes or mutations that compromise p53 function in tumours and Li-Fraumeni syndrome. Oncogene. 2002;21(19):2961–70. doi: 10.1038/sj.onc.1205372. [DOI] [PubMed] [Google Scholar]
- 27.Crighton D, Woiwode A, Zhang C, Mandavia N, Morton JP, Warnock LJ, Milner J, White RJ, Johnson DL. p53 represses RNA polymerase III transcription by targeting TBP and inhibiting promoter occupancy by TFIIIB. Embo J. 2003;22(11):2810–20. doi: 10.1093/emboj/cdg265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cairns CA, White RJ. p53 is a general repressor of RNA polymerase III transcription. Embo J. 1998;17(11):3112–23. doi: 10.1093/emboj/17.11.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chesnokov I, Chu WM, Botchan MR, Schmid CW. p53 inhibits RNA polymerase III-directed transcription in a promoter-dependent manner. Mol Cell Biol. 1996;16(12):7084–8. doi: 10.1128/mcb.16.12.7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Felton-Edkins ZA, Kenneth NS, Brown TR, Daly NL, Gomez-Roman N, Grandori C, Eisenman RN, White RJ. Direct regulation of RNA polymerase III transcription by RB, p53 and c-Myc. Cell Cycle. 2003;2(3):181–4. [PubMed] [Google Scholar]
- 31.Chu WM, Wang Z, Roeder RG, Schmid CW. RNA polymerase III transcription repressed by Rb through its interactions with TFIIIB and TFIIIC2. J Biol Chem. 1997;272(23):14755–61. doi: 10.1074/jbc.272.23.14755. [DOI] [PubMed] [Google Scholar]
- 32.Sutcliffe JE, Cairns CA, McLees A, Allison SJ, Tosh K, White RJ. RNA polymerase III transcription factor IIIB is a target for repression by pocket proteins p107 and p130. Mol Cell Biol. 1999;19(6):4255–61. doi: 10.1128/mcb.19.6.4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gomez-Roman N, Grandori C, Eisenman RN, White RJ. Direct activation of RNA polymerase III transcription by c-Myc. Nature. 2003;421(6920):290–4. doi: 10.1038/nature01327. [DOI] [PubMed] [Google Scholar]
- 34.Felton-Edkins ZA, Fairley JA, Graham EL, Johnston IM, White RJ, Scott PH. The mitogen-activated protein (MAP) kinase ERK induces tRNA synthesis by phosphorylating TFIIIB. Embo J. 2003;22(10):2422–32. doi: 10.1093/emboj/cdg240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reina JH, Azzouz TN, Hernandez N. Maf1, a New Player in the Regulation of Human RNA Polymerase III Transcription. PLoS ONE. 2006;1:e134. doi: 10.1371/journal.pone.0000134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson SS, Zhang C, Fromm J, Willis IM, Johnson DL. Mammalian maf1 is a negative regulator of transcription by all three nuclear RNA polymerases. Mol Cell. 2007;26(3):367–79. doi: 10.1016/j.molcel.2007.03.021. [DOI] [PubMed] [Google Scholar]
- 37.Rollins J, Veras I, Cabarcas S, Willis I, Schramm L. Human Maf1 negatively regulates RNA polymerase III transcription via the TFIIB family members Brf1 and Brf2. Int J Biol Sci. 2007;3(5):292–302. doi: 10.7150/ijbs.3.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21(13):2933–42. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- 39.Sartippour MR, Pietras R, Marquez-Garban DC, Chen HW, Heber D, Henning SM, Sartippour G, Zhang L, Lu M, Weinberg O, Rao JY, Brooks MN. The combination of green tea and tamoxifen is effective against breast cancer. Carcinogenesis. 2006;27(12):2424–33. doi: 10.1093/carcin/bgl066. [DOI] [PubMed] [Google Scholar]
- 40.Upadhya R, Lee J, Willis IM. Maf1 is an essential mediator of diverse signals that repress RNA polymerase III transcription. Mol Cell. 2002;10(6):1489–94. doi: 10.1016/s1097-2765(02)00787-6. [DOI] [PubMed] [Google Scholar]
- 41.Geiduschek EP, Kassavetis GA. Transcription: adjusting to adversity by regulating RNA polymerase. Curr Biol. 2006;16(19):R849–51. doi: 10.1016/j.cub.2006.08.071. [DOI] [PubMed] [Google Scholar]
- 42.Willis IM, Moir RD. Integration of nutritional and stress signaling pathways by Maf1. Trends Biochem Sci. 2007;32(2):51–53. doi: 10.1016/j.tibs.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 43.Khan N, Afaq F, Saleem M, Ahmad N, Mukhtar H. Targeting multiple signaling pathways by green tea polyphenol (−)-epigallocatechin-3-gallate. Cancer Res. 2006;66(5):2500–5. doi: 10.1158/0008-5472.CAN-05-3636. [DOI] [PubMed] [Google Scholar]
- 44.Lin JK. Cancer chemoprevention by tea polyphenols through modulating signal transduction pathways. Arch Pharm Res. 2002;25(5):561–71. doi: 10.1007/BF02976924. [DOI] [PubMed] [Google Scholar]
- 45.Ahmad N, Gupta S, Mukhtar H. Green tea polyphenol epigallocatechin-3-gallate differentially modulates nuclear factor kappaB in cancer cells versus normal cells. Arch Biochem Biophys. 2000;376(2):338–46. doi: 10.1006/abbi.2000.1742. [DOI] [PubMed] [Google Scholar]
- 46.Levites Y, Youdim MB, Maor G, Mandel S. Attenuation of 6-hydroxydopamine (6-OHDA)-induced nuclear factor-kappaB (NF-kappaB) activation and cell death by tea extracts in neuronal cultures. Biochem Pharmacol. 2002;63(1):21–9. doi: 10.1016/s0006-2952(01)00813-9. [DOI] [PubMed] [Google Scholar]
- 47.Fang M, Chen D, Yang CS. Dietary polyphenols may affect DNA methylation. J Nutr. 2007;137(1 Suppl):223S–228S. doi: 10.1093/jn/137.1.223S. [DOI] [PubMed] [Google Scholar]
- 48.Fujimura Y, Umeda D, Kiyohara Y, Sunada Y, Yamada K, Tachibana H. The involvement of the 67 kDa laminin receptor-mediated modulation of cytoskeleton in the degranulation inhibition induced by epigallocatechin-3-O-gallate. Biochem Biophys Res Commun. 2006;348(2):524–31. doi: 10.1016/j.bbrc.2006.07.086. [DOI] [PubMed] [Google Scholar]
- 49.Smith DM, Daniel KG, Wang Z, Guida WC, Chan TH, Dou QP. Docking studies and model development of tea polyphenol proteasome inhibitors: applications to rational drug design. Proteins. 2004;54(1):58–70. doi: 10.1002/prot.10504. [DOI] [PubMed] [Google Scholar]
- 50.Na HK, Surh YJ. Intracellular signaling network as a prime chemopreventive target of (−)-epigallocatechin gallate. Mol Nutr Food Res. 2006;50(2):152–9. doi: 10.1002/mnfr.200500154. [DOI] [PubMed] [Google Scholar]
- 51.Yamamoto T, Hsu S, Lewis J, Wataha J, Dickinson D, Singh B, Bollag WB, Lockwood P, Ueta E, Osaki T, Schuster G. Green tea polyphenol causes differential oxidative environments in tumor versus normal epithelial cells. J Pharmacol Exp Ther. 2003;307(1):230–6. doi: 10.1124/jpet.103.054676. [DOI] [PubMed] [Google Scholar]
- 52.Yamamoto T, Lewis J, Wataha J, Dickinson D, Singh B, Bollag WB, Ueta E, Osaki T, Athar M, Schuster G, Hsu S. Roles of catalase and hydrogen peroxide in green tea polyphenol-induced chemopreventive effects. J Pharmacol Exp Ther. 2004;308(1):317–23. doi: 10.1124/jpet.103.058891. [DOI] [PubMed] [Google Scholar]
- 53.Shimizu M, Deguchi A, Joe AK, McKoy JF, Moriwaki H, Weinstein IB. EGCG inhibits activation of HER3 and expression of cyclooxygenase-2 in human colon cancer cells. J Exp Ther Oncol. 2005;5(1):69–78. [PubMed] [Google Scholar]