Abstract
Nuclear receptors (NRs) are a family of highly conserved transcription factors that regulate transcription in response to small lipophilic compounds. They play a role in every aspect of development, physiology and disease in humans. They are also ubiquitous in and unique to the animal kingdom suggesting that they may have played an important role in their evolution. In contrast to the classical endocrine receptors that originally defined the family, recent studies suggest that the first NRs might have been sensors of their environment, binding ligands that were external to the host organism. The purpose of this review is to provide a broad perspective on NR ligands and address the issue of exactly what constitutes a NR ligand from historical, biological and evolutionary perspectives. This discussion will lay the foundation for subsequent reviews in this issue as well as pose new questions for future investigation.
Introduction
Nuclear receptors (NRs) are proteins that share considerable amino acid sequence similarity in two highly conserved domains – the DNA binding (DBD) and the ligand binding domains (LBD) (Renaud and Moras, 2000). As their names indicate, these domains are responsible for binding specific DNA sequences and small lipophilic ligands, respectively. The DBD in the N-terminal half of the protein is comprised of two zinc-binding motifs that each coordinate a zinc ion through four precisely positioned cysteines. The LBD in the C-terminal half consists of a series of ~12 alpha helices that create a hydrophobic pocket that can bind a hydrophobic ligand (Fig. 1A). NRs bind specific DNA response elements in the regulatory regions of genes and regulate transcription in response to ligand binding by recruiting co-regulatory molecules that subsequently modify the chromatin and contact the basal transcription machinery (Fig. 1B) (Gronemeyer et al., 2004; Rosenfeld et al., 2006). Examples of classical NRs include estrogen (ER, NR3A), progesterone (PR, NR3C3), androgen (AR, NR3C4), glucocorticoid (NR3C1), Vitamin A (RAR, NR1B), Vitamin D (VDR, NR1I1) and thyroid hormone (TR, NR1A) receptors. While there are an increasing number of examples of NRs playing non genomic roles at the plasma and other membranes, this review will concentrate on the role and definition of the ligands only in their nuclear capacity; it is in that capacity that we have the greatest understanding of NR action, and yet even that understanding is far from complete.
Figure 1. Structure and mode of action of nuclear receptors.

A. Domain structure of nuclear receptors (A–F). The DNA binding domain (DBD) consists of two zinc-binding motifs (Zn++) and often includes the hinge region. Shown is the crystallographic structure of the estrogen receptor ligand binding domain (LBD) bound to diethylstilbestrol (DES), a synthetic nonsteroidal estrogen that functions as a pure agonist, and the antagonist 4-hydroxy tamoxifen (4OHT) (dark blue), reprinted from Shaiu et al. (Shiau et al., 1998) with permission from Elsevier. AF1, AF2, activation function 1 and 2 (AF2, AF-2 helix); both contact co-regulatory molecules but AF-1 is typically ligand-independent while the AF-2 is ligand-dependent (colored magenta as helix 12). NRs have variable AF1 and hinge regions and F domains and range in size from ~450 to ~933 amino acids, although the majority are ~50 kDa. Many NR genes have multiple isoforms generated by alternative promoter usage and splice variants; some NR genes lack a DBD (not shown) (for a review of NR structures, see (Renaud and Moras, 2000)).
B. Simplified diagram of classical NR action. In the absence of ligand, NRs recruit multi-subunit co-repressor complexes, which contain histone deacetylase activity (HDAC), to promoter regions of genes. Transcription of target genes is activated when ligand (hexagon) binds the NR and induces a conformation change in the LBD that recruits co-activator complexes that contain histone acetylase transferase activity (HAT). Not shown are other co-regulatory complexes needed for transcription such as chromatin remodeling and mediator complexes, and the general transcription machinery (see (Rosenfeld et al., 2006)).
Evolutionary origins of NRs
NR genes are present and expressed in some of the simplest animal organisms. They are absent, however, in fungi, plants and choanoflagellates, the closest known relatives of metazoans (King et al., 2008). Therefore, they appear to have arrived on the evolutionary scene ~635 million years (Myr) ago when animals first appeared in the fossil record (Fig. 2). They have also been associated with the Cambrian explosion -- the massive diversification from relatively simple, primarily unicellular organisms to morphologically more complex multicellular organisms consisting of different cell types, tissues and eventually organs and organ systems. Assuming that NRs were also present in the ancestors of those primordial animals that evolved in the oceans more than 270 Myr before the first animals appeared on land (~365 Myr ago), this would indicate that NRs predate walking, breathing and even nervous and circulatory systems. They certainly predate endocrine systems. And yet our current notion of NRs and their ligands has been formed largely by studies in the context of endocrine signaling in mammals that evolved ~440 Myr after the appearance of the first NR gene. A reconsideration of NRs and their ligands in the appropriate evolutionary light should therefore greatly enhance our understanding of this important transcription factor family.
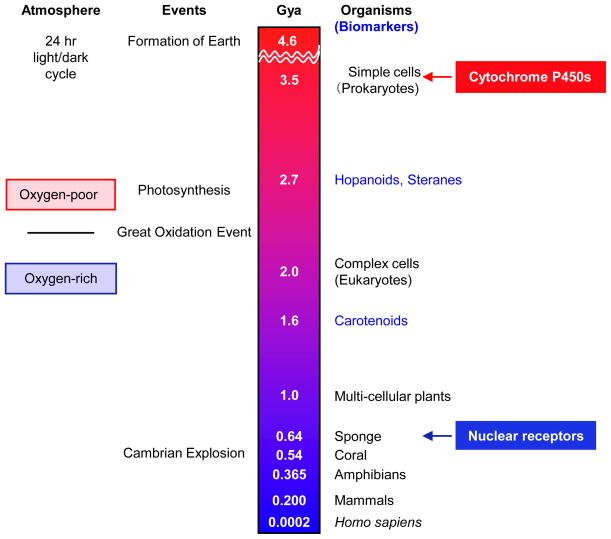
Fig. 2. Timeline of life on Earth.
Shown is a highly simplified depiction of some of the major biological events and first appearance of various life forms, as evidenced in the fossil record, during the history of the Earth in billions of years (Gya) before present day. NRs are found in living relatives of the simplest animals (sponges, in the phylum Porifera), the first evidence of which appears ~635 Myr ago in the form of molecular fossils of the biomarker 24-isopropylcholestanes (Love et al., 2009). Cytochrome P450s, mono-oxygenases that bind heme, are in every living organism, including prokaryotes (Nelson, 2009). Biomarkers for bacteria (hopanoids, 2700 Myr ago), eukaryotes (steranes such as stigmastane and cholestane, 2700 Myr ago) and purple sulfur bacteria (carotenoids such okenone, 1600 Myr) are indicated in blue (structures are shown in Fig. 5); molecular fossils of steranes are often used as evidence that eukaryotes might have evolved much earlier than 2000 Myr ago (Brocks and Banfield, 2009; Brocks et al., 1999). The Earth’s atmosphere and oceans changed from an oxygen-poor to an oxygen-rich environment during the Great Oxygenation Event (~2300 Myr ago), ~300 million years after the advent of oxygenic photosynthesis by cyano- and other bacteria (~2600 Myr ago) (Reinhard et al., 2009). Cambrian explosion (520–550 Myr ago), the rapid appearance of most major groups of complex animals. Not indicated is the whole genome duplication of teleosteans (bony fishes) that occurred at 0.250 Gya and facilitated the expansion of the NR gene family.
All told more than >900 NR genes have been identified in every animal examined thus far, from the simplest to the most morphologically complex (NureXbase, http://nurexbase.prabi.fr/). The simplest animal organisms, such as sponges and trichoplax that have no distinct tissues or organs, tend to have just a few NR genes (Baker, 2008; Larroux et al., 2006; Tarrant et al., 2008; Wiens et al., 2003) (Fig. 3). Somewhat more morphologically complex organisms, such as coral and sea anemones (Cnidarians), have more NR genes (~10–17), while early bilaterian animals (i.e., animals with a front and back end, as well as an upside and downside), such insects and tetrapod vertebrates have even more NRs (~21 to 25, respectively). The proposed last common ancestor of all bilaterians (Urbilateria) is predicted to have ~25 NR genes (Bertrand et al., 2004). Mammals, including rat, mouse and humans, have ~48 genes. Hence, there appears to be a rough relationship between the number and complexity of cell types in an organism and the number of NR genes, supporting the notion that NRs may have played a role in the rapid diversification of metazoans (Escriva et al., 2004; Escriva et al., 2000).
Fig. 3. Evolution of nuclear receptors: Relationship with morphological complexity of organisms?
Shown are the number of NR genes and their proposed role in various organisms representing different taxa, along with key features that distinguish each group from the one above it (complexity increases from top to bottom). Dashed line, there is some controversy over which organism – Trichoplax or Porifera (sponge) -- is at the base of the Metazoan Tree (Schierwater et al., 2009; Sperling et al., 2007). (*) Many nematode genes are expansions of a single NR gene (HNF4-like, NR2A). (**) Fish (zebrafish and fugu) NR numbers are higher than anticipated due to a recent genome-wide duplication of that genome. The first evidence of NRs acting in an endocrine fashion occurs in nematodes (Motola et al., 2006). References for NR gene numbers: Porifera (Larroux et al., 2006; Wiens et al., 2003); Trichoplax (Srivastava et al., 2008); Coral Acropora (Grasso et al., 2001); Sea Anemone (N. vectensis) (Reitzel and Tarrant, 2009); Nematodes (C. elegans, M. incognita, B. malayi) ((Abad et al., 2008) and references therein); other (Bertrand et al., 2007; Markov et al., 2010). See text for additional details.
There are, of course, as with everything in biology, some salient exceptions. The puffer fish genome (Fugu rubripes), for example, contains 68 NR genes due to a genome-wide duplication event common to all teleostean fishes (Bertrand et al., 2007; Jaillon et al., 2004; Maglich et al., 2003). Several nematodes also have an unusually high number of NR genes due to an expansion of a single NR gene. For example, the free-living nematode that is a model for developmental biology, Caenorhabditis elegans, has >270 NR genes, although all but ~15 of those genes are variations of a single NR gene (HNF4, NR2A) (Robinson-Rechavi et al., 2005; Sluder and Maina, 2001). These additional genes are sometimes referred to as supplementary nuclear receptors (SupNRs). The plant-parasitic nematode Meloidogyne incognita has 92 predicted NR genes which includes a 41-member lineage-specific expansion of a SupNR (Abad et al., 2008). In contrast, Brugia malayi, which causes lymphatic filariasis (elephantiasis) in humans, has only 27 NR genes, 14 of which are SupNRs (Ghedin et al., 2007). Hence, while the notion that NRs play a role in metazoan patterning and development seems to be well founded, additional investigation is required to determine the precise relationship between NRs and morphological complexity. What is unambiguous is that NRs were present long before endocrine systems developed. This then raises the issue of the role of the early NR ligands. While it has been previously proposed that the original NRs did not respond to ligands because they fell into the class of orphan receptors (Laudet, 1997), that view is rapidly changing.
Roughly half the 48 receptors found in man have ligands associated with them; the other half are orphans in that no ligand has yet been identified for them (Benoit et al., 2004). When a ligand is identified for a NR it is said to be de-orphanized. While there are a few mammalian NRs that are known to have no ligand binding pocket, due to amino acid side chains that fill the cavity formed by the 12 helices (e.g., Nurr1 (NR4A2)), the current view is that most if not all NRs with a hydrophobic pocket will eventually have ligands associated with them (Benoit et al., 2004; Ingraham and Redinbo, 2005). The number of NRs with associated ligands is much fewer from non mammalian organisms but that will likely change as investigation continues on those receptors.
Historical perspective of NR Ligands
Our current concept of what NR ligands are and what they do comes primarily from studies on a relatively few receptor·ligand pairs from man and related mammals. The reason for the anthropocentric bias of NR ligands is an obvious, and practical, one. Most of the first full length NRs cDNAs were cloned from human within a three-year time frame between 1985 and 1987 (GR, PR, ER, VDR, TR, RAR, and mineralcorticoid (MR, NR3C2)); these receptors subsequently defined the steroid and thyroid hormone receptor superfamily, as the NR family was originally called (Evans, 1988). NRs are also among the most popular of drug targets for human diseases, for everything ranging from diabetes (thiazolenediones for PPARγ, NR1C3) to cancer (tamoxifen for ER) to heart disease (fenofibrates for PPARα, NR1C1), as well as for lifestyle (birth control pills for ER and PR) and behavioral conditions (performance enhancing steroids for AR) (Ottow and Weinmann, 2008; Overington et al., 2006). The popularity of NRs as drug targets is due to the fact that, in addition to playing a key role in these physiological processes, the receptors naturally contain a hydrophobic pocket that binds small hydrophobic molecules, and the most effective drugs are typically small hydrophobic compounds that can cross the plasma membrane (e.g., (Chen, 2008; Schulman and Heyman, 2004)). Hence, for medical as well as financial reasons (drugs such as Advair and Avandia that target GR and PPARγ, respectively, have grossed $3–6 billion dollars annually at their peak (www.gsk.com)), there has been intense interest in identifying ligands for human NRs, synthetic as well as natural. There has also been a tremendous effort to determine the function of human NR ligands, down to the atomic level, so that ligands/drugs can be tweaked to yield the optimal desired effect (e.g., (Cornell and Nam, 2009; Miyachi, 2007)). It is from these investigations, performed in academia and industry alike, that we have developed our current understanding of NR ligands. In this issue, however, we are concerned with more basic evolutionary issues regarding NRs and their ligands, and therefore must broaden our perspective.
NR ligands are hydrophobic molecules that are derivatives of retinoids, fatty acids, cholesterol, lipophilic hormones and vitamins, as well as antibiotics, xenobiotics and synthetic drugs (Fig. 4). The ligands, which bind in the hydrophobic pocket that ranges in size from ~220 to 1600 angstroms (Benoit et al., 2004; Ingraham and Redinbo, 2005), induce a conformational change in the receptor resulting in the 12th alpha helix (also called the AF-2 helix) making contact with the ligand. For agonist ligands, this results in a disassociation of co-repressor molecules and recruitment of co-activator molecules (Fig. 1B) (Gronemeyer et al., 2004; Rosenfeld et al., 2006). Hence, slight changes in the relative position of the AF-2 helix, induced by ligand binding, initiates a cascade of molecular events on the promoters of genes that ultimately results in changes in patterns of gene expression.
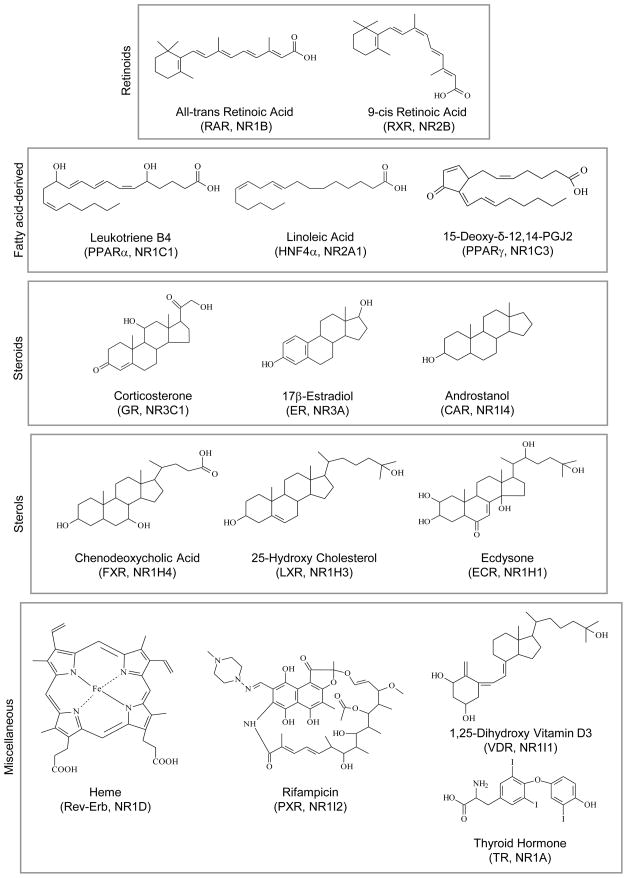
Figure 4. Structure of nuclear receptor ligands.
Shown are the structures of some of the >250 ligands in the Nuclear Receptor Signaling Atlas (http://www.nursa.org/) grouped according to their structures.
This is the current view of NR ligands, a very precise and yet sophisticated concept based on the actions of some of the first NRs identified. And yet, we have learned bit by bit over the years that those first NRs are not necessarily typical of all NRs. For example, GR, the first NR cloned (Evans, 1988), is located primarily in the cytoplasm complexed with heat shock protein (HSP) until it binds its ligand, whereupon it dissociates from HSP and translocates to the nucleus. And yet, we now know that the majority of NRs do not typically reside in the cytoplasm as does GR. Similarly, it is becoming increasingly clear that the actions identified for the ligands of the first NRs identified will not be completely applicable to all the other NRs, especially those found in organisms that are more distantly related to man.
Biological perspective of NR ligands
The word “ligand” comes from the Latin ligare which means “to bind.” Whereas in inorganic chemistry a ligand is an atom or molecule that binds a metal, in biochemistry and pharmacology a ligand is defined as a substance that is able to bind to and form a complex with a biomolecule to serve a biological purpose, such as heme binding to myoglobin. This definition is much simpler than the classical notion of a NR ligand as one that induces a conformation change that triggers a cascade of events. There are also different classes of ligands, such as high affinity agonists that require only low concentrations of ligands to bind with maximal occupancy and elicit a maximal physiological response (e.g., glucocorticoids that bind in the nanomolar range and are present in nanomolar concentrations). There are antagonist ligands that bind but do not elicit the expected physiological response (e.g., tamoxifen for ER in breast tissue), as well as inverse agonists that bind and elicit the opposite effect of the agonists (androstanol for CAR, NR1I3). Finally, there are low affinity ligands that must be present at relatively high concentrations before the binding site is maximally occupied and the maximum physiological response to the ligand is achieved, as well as partial agonists and antagonists, etc. (Gilman et al., 1980). In short, there are many different types of biochemical ligands and the only thing that they all have in common is that they bind a biomolecule.
Just as our notion of NRs has evolved, so too has our notion of NR ligands. For example, in 1992 when fatty acids were first described as ligands for PPARs (Gottlicher et al., 1992), there was considerable resistance in the NR field to accept these newcomers as bona fide ligands. Concentrations of fatty acids that were ~100,000 times greater than what were needed for the steroids to activate their cognate receptors were required to activate PPARs (>100 μM vs. 0.5 nM amounts for 17β-estradiol to bind ER) (Gottlicher et al., 1992; Green et al., 1986). Only once it was realized that fatty acids were of course present at much higher concentrations than steroids in the body, did fatty acids gain full acceptance as ligands (conditions were also optimized to a more physiological range of ~30 μM (Forman et al., 1997)). But even then it is important to note that the typical 100-fold activation in transcriptional activity that steroids elicited (e.g., (Webster et al., 1988)) was never observed for fatty acids on PPARs. Rather, the effects were typically a 3- to10-fold effect (e.g., (Forman et al., 1997)). Nonetheless, fatty acids as ligands for PPARs eventually became part of the pantheon of accepted NR ligands (e.g., (Michalik et al., 2006)) and ushered in the notion of NR ligands being metabolic sensors as well as high affinity endocrine receptors (Benoit et al., 2004; Gronemeyer et al., 2004).
Evolutionary perspective of NR ligands
From an evolutionary perspective, the notion of NR ligands as signaling molecules that help recruit co-regulatory molecules requires that at least some of those co-regulatory molecules (e.g., p300/CBP, GRIP1/SRC, PGC1, SMRT, N-CoR, SWI/SNF, Mediator, etc.) be present in the organisms in which the first ligand-dependent NR arose. This appears to be the case. Whereas several co-regulatory molecules were first identified as interacting with NRs (e.g., GRIP1/SRC, PGC1, SMRT, N-CoR), they are now known to interact with and regulate other transcription factor families as well, some of which were also present in early metazoans (Degnan et al., 2009; Rosenfeld et al., 2006). Furthermore, the first HAT-containing co-regulator identified, GCN5, was identified in yeast (Grant et al., 1997) as part of a complex (SAGA/STAGA) that is conserved in humans (Martinez, 2002). Likewise, the Mediator complex, which was identified as binding VDR (DRIPs) and TR (TRAPs) in a ligand-specific fashion, and the chromatin remodeling complex SWI/SNF, are known to interact with non NR transcription factors and were first identified in yeast (Martinez, 2002). However, what is not known is whether the ancient NR ligands induced a conformational switch that facilitated the recruitment of these co-regulators. This will require additional investigation, including solving the three dimensional structures of NRs from primitive organisms.
NR co-regulatory molecules were not the only components of the NR system that were present before the first animal organisms evolved; potential ligands for NRs were also abundant. Some of the most common biomarkers for early microorganisms share a great deal of structural similarity to modern day NR ligands (Fig.s 4, 5). For example, the molecular fossil okenane, which has been extracted from sedimentary rocks more than 1600 Myr old, is considered a biomarker for Chromatiaceae (planktonic purple sulfur bacteria). Okenane is a derivative of okenone, which is a carotenoid; β-carotene from land plants such as carrots is the precursor to retinoic acid used by mammals (Brocks and Banfield, 2009). Likewise, a variety of steranes (C26 to C30 compounds that are the basis of all sterols and steroids) are considered biomarkers for early eukaryotes such as diatoms and various types of algae, and have been found in 2700 Myr old rock, which suggests that eukaryotes might have evolved earlier than previously thought (Brocks et al., 1999) (Fig.s 2, 5). Another biomarker is 24-isopropylcholestane (a C30 steroid) that is produced by demosponges and is considered to be some of the oldest (>635 Myr) credible evidence in the geological record for animal life (Brocks and Banfield, 2009). Therefore, the modern day retinoids, steroids and cholesterol-derived compounds that are known NR ligands have very ancient origins that pre-date the evolution of animals, and hence NRs, by billions of years.
Figure 5. Tree of life, nuclear receptors and potential ligand precursors.
The Tree of Life showing that the expression of NRs is limited to the animal kingdom while many potential NR ligand precursors are produced by organisms in all three domains of life – eukaryotes, archaea and bacteria (Brocks and Banfield, 2009). The Tree of Life diagram is reprinted with permission from S. Baldauf, Uppsala University, Sweden (Baldauf et al., 2004). See text for details.
Another indication that NR ligands predate NRs comes from some of the enzymes that generate and/or metabolize the ligands – the cytochrome P450 enzymes encoded by the Cyp genes. Cyp genes are found in every living organism, including eubacteria and archeabacteria, as well as mitochondria; more than 11,500 distinct enzymes have been identified thus far in every life form (Danielson, 2002; Nebert et al., 1989; Nelson, 2009) (Fig. 2). P450’s are primarily mono-oxygenases and it has been proposed that one of their initial purposes may have been to detoxify the oxygen produced by newly evolved photosynthetic organisms around 2.6 billion years ago (Wickramashighe and Villee, 1975). There is a close and well characterized connection between NR and Cyp genes, including a similar evolution from low specificity to high specificity of binding substrates/ligands (Markov et al., 2009). Furthermore, (Markov et al., 2009). NRs, including the most ancient ones such as HNF4, are some of the primary regulators of Cyp genes in mammals (Bolotin et al., 2009; Jover et al., 2009). It will be of interest to determine whether a similar regulation exists earlier in evolution.
There is one other intriguing clue as to the ancient origins of NRs and their ligands. All P450 enzymes, by definition, bind heme, an iron-binding biomolecule that absorbs light near 450 nm. For the first >2 billion years of the Earth’s history, the oceans had low amounts of oxygen but high amounts of dissolved iron (Fig. 2), a natural consequence of the extensive amount of iron in the Earth’s core. Recently, heme was identified as the ligand for the orphan receptors Rev-erbα/β (NR1D1/2) (Raghuram et al., 2007; Yin et al., 2007). What makes this finding so intriguing is the fact that the Rev-erb’s are key transcriptional regulators of the circadian clock (Burris, 2008), another biological phenomenon with ancient origins that pre-date animals and that has intricate links to metabolism (Kovac et al., 2009). Additionally, Rev-erb’s and ~50% of all mammalian NRs are regulated in a circadian fashion (Yang et al., 2006). Hence, even though NRs did not arrive on the evolutionary scene until 4 billion years after the formation of the Earth, via the 24 hour/light dark cycle, iron-binding heme and cytochrome P450 enzymes, they have links back to the very beginning of life on Earth.
Environmental Sensors vs. Endocrine Receptors
The first NRs identified (GR, ER, PR, VDR) were purified based on their ability to bind their respective ligands (Evans, 1988). Most subsequent NRs were cloned or identified via sequence similarity to these first receptors, and hence initially had no ligands associated with them. Indeed, even the retinoic acid receptor RARα (NR1B1) started out as an orphan; although by the end of the original paper describing its cloning retinoic acid had been identified as a ligand (Giguere et al., 1987). Progesterone, estrogen and glucocorticoids can be considered endogenous compounds, derived from cholesterol in the ovaries and the adrenal cortex, respectively. Likewise, thyroid hormone is produced in-house by the thyroid gland, while Vitamin D3 is made in the skin when ultraviolet light from sun activates 7-dehydrocholesterol. Therefore, the first receptors identified had ligands that were not only high affinity but were also made in and subsequently secreted from organs in an endocrine fashion. And yet we now know that NRs are present even in animals that do not have any endocrine system (Fig. 3). Indeed, sponges are often not even considered metazoans as they do not contain different tissues, let alone organs, circulatory or digestive systems, and yet they too have at least one NR.
Interestingly, the NRs in sponges have the greatest amino acid sequence similarity to “orphan” receptors in the NR2 subfamily (HNF4- and RXR-like) (Larroux et al., 2006; Wiens et al., 2003), as do most of the NRs that have been identified thus far in primitive metazoans such as cnidarians (e.g., coral and sea anemone) (HNF4, NR2A; COUP-TF, NR2F; TLL, NR2E; TR2/4, NR2C) (Grasso et al., 2001; Reitzel and Tarrant, 2009; Tarrant et al., 2008). Trichoplax, a morphologically very simple metazoan, likewise has NR2 family members (RXR, HNF4, COUP-TF) as well as an NR3B family member (ERR) (Baker, 2008). In short, the original NRs could not have been endocrine receptors, since there were no endocrine organs in the host organism. But they could have been environmental sensors, which also appears to be the case for many of the modern day “orphan” receptors.
One of the primary distinctions between animals and plants and other eukaryotes, such as fungi, is that animals do not have cell walls. The lack of a cell wall could have made animal cells more accessible to potential lipophilic ligands. A less oft cited distinction between animals and non animal organisms is the fact that most non animal organisms are autotrophs which means that they directly use sources of energy, such as light, to produce organic carbon from inorganic carbon dioxide. In contrast, animals, the only organisms that have NRs, are heterotrophs. Hetero in Greek means “another” and trophe means “nutrition”. In other words, animals must ultimately get their energy from other organisms; they cannot derive it from light as do plants. So if animals are completely dependent on other organisms for their existence, perhaps the earliest NRs were also dependent on other organisms as a source of ligands. Indeed, the notion that some NRs act as sensors, either environmental or metabolic, has become increasingly popular as more and more ligands are identified for the “orphan” receptors (Benoit et al., 2004; Gronemeyer et al., 2004; Markov et al., 2008). This view of NRs as environmental sensors is also consistent with the abundance of potential ligand precursors in the ancient oceans where all life formed (Fig.s 4,5). (This discussion is not meant to imply that non animal organisms do not have ligand-dependent transcription factors. They of course do. However, while those transcription factors share functional, and occasionally structural, similarities with NRs, they do not share significant amino acid sequence similarity (see (Markov et al., 2010))).
Initially, it was proposed that orphan receptors would have as ligands endogenous compounds, possibly metabolic intermediates, which would explain some of their apparently constitutive transactivation activity and the difficulty encountered in trying to identify their ligands (O’Malley and Conneely, 1992). However, we now know that several receptors previously considered to be orphans bind compounds that originate outside of the organism and hence act as sensors of the environment. The traditional examples of NRs as environmental sensors are the pregnane X receptor (PXR; NR1I2) and the constitutive androstane receptor (CAR; NR1I4) that bind antibiotics and xenobiotics. However, even some of the more classical receptors can be considered environmental sensors. For example, the retinoid receptors (RARs, RXRs) bind Vitamin A and its derivatives that are formed from exogenously obtained β-carotene from plants and retinyl esters from animals. Even the bile acid receptors, Vitamin D receptor (VDR; NRNR1I1) and farnesoid X receptor (FXR; NR1H4), can be considered environmental sensors as they bind lithocholic acid (LCA) that is produced from chenodeoxycholic acid (CDCA) by intestinal bacteria (Makishima et al., 2002; Parks et al., 1999; Staudinger et al., 2001) (Fig.s 4 and 5).
Another example of a NR previously classified as an orphan receptor that senses its environment via ligand binding is hepatocyte nuclear factor 4α (HNF4α, NR2A1). HNF4α was originally purified based on its ability to bind DNA response elements in liver-specific genes and hence did not have a ligand associated with it (Sladek et al., 1990). Unlike many other orphan receptors, HNF4α exhibited constitutive transactivation activity in that no exogenous ligand was needed for HNF4α to activate transcription, even when expressed in yeast (Sladek et al., 1999). First, fatty acyl Co-enzyme A thioesters (Hertz et al., 1998) and then a mixture of fatty acids including palmitoleic (C16:1) and oleic acid (C18:1) were proposed as ligands for HNF4, based on work with bacterially expressed human HNF4α and HNF4γ (Dhe-Paganon et al., 2002; Wisely et al., 2002). However, there were problems with those ligands in that, among other things, the former were much too large for the proposed ligand binding pocket and the latter bound so tightly to the recombinant HNF4 that they were considered to be more like structural co-factors than reversible ligands (Bogan et al., 2000; Sladek, 2002). More recently, we have identified linoleic acid (LA) (cis, cis-9,12-octadecadienoic acid), a C18:2 ω-6 fatty acid, as the endogenous ligand for endogenous HNF4α expressed in mammalian tissues (Yuan et al., 2009). LA as a ligand is of particular interest in that it is an essential fatty acid that must be derived from the diet (Burr and Burr, 1929; Burr and Burr, 1930). No animal can synthesize LA as all animals lack an essential desaturase enzyme that converts monounsaturated fatty acids to polyunsaturated fatty acids (Gunstone, 1996). Also of note is the fact that HNF4 is considered to be one of the most ancient NRs in that it is found in all animals, including sponge, trichoplax and coral reef (Baker, 2008; Grasso et al., 2001; Larroux et al., 2006). Even coral, as animals, cannot make LA and must obtain it from zooplankton or phytoplankton (Al-Moghrabi et al., 1995; Latyshev et al., 1991). Therefore, assuming that the more primitive forms of HNF4 also bind LA, this would suggest that one of the first NRs was not only sensing its environment, but was sensing a compound that the host organism was completely dependent upon. Another example of an orphan NR with ancient origins that might be sensing its environment is COUP-TF. Human COUP-TF2 (NR2F2) was recently crystallized and its pocket was found to accommodate retinoic acid; the receptor also responded to high concentrations of retinoic acid in a transactivation assay (Kruse et al., 2008). This is of interest considering the ancient origins of carotenoids, the precursors to retinoids, mentioned above. It will be of great interest to identify the endogenous ligands for other NRs, especially those from organisms more distantly related to man, and see whether they too are derived from exogenous sources.
Criteria for NR ligand designation
Having broadened our notion of what constitutes a NR ligand, we must now decide on the precise criteria that should be used to classify a compound as a NR ligand. This will be an increasingly important issue as an increasing number of NRs are identified in newly sequenced genomes of all types of organisms. I propose that we use the simplest, least restrictive definition possible -- i.e., that the compound bind the NR. The next criterion would be that the compound bind in the ligand binding pocket. While it is possible that there might be small hydrophilic compounds that bind on the surface of a receptor rather than in the hydrophobic pocket created by the 12 helices, perhaps those could be considered as secondary ligands. Reversible binding might be considered desirable, especially for drug development, but there could be receptor·ligand pairs for which the affinity is so high that the binding is essentially irreversible. Requiring any one specific function as a result of the ligand binding, such as altered cellular location, a specific conformational change, or recruitment of co-regulators, would require assumptions that may not be valid for the receptor in question and may result in many ligands being overlooked. There are other ways to alter gene expression that do not require a highly specialized relocation of the AF-2 helix that we associate with classical NR ligands.
Again the identification of LA as the endogenous ligand for HNF4α will help illustrate the point. LA was identified based on the fact that it was the only compound found to bind HNF4α expressed in mammalian cells in a specific, and reversible, fashion using GC/MS. However, a direct effect of LA on the transactivation function of HNF4α could not be unequivocally demonstrated, even using the most inclusive genome-wide approaches (Yuan et al., 2009). Other classical functional assays developed for other NRs, such as conformation change by protease digestion, in vitro recruitment of co-regulatory proteins and intracellular localization, likewise failed to unveil a function for LA (Ta and Sladek, 2010). The only effect of LA on HNF4α that was observed was a reduction in HNF4α protein levels and a concomitant decrease in HNF4α target genes (Yuan et al., 2009). Hence, LA may decrease the ability of HNF4α to activate gene expression, but not via the classical method identified for other NR·ligand pairs of altering the position of the AF-2 helix and recruiting co-regulatory proteins. (The structural studies on bacterially expressed HNF4α also confirm that the AF-2 helix is constitutively in the active conformation, regardless of ligand binding (Duda et al., 2004).) Rather, the effect of LA may be a more direct and simpler one of affecting HNF4α protein levels. The notion of ligand binding affecting NR protein stability is not a novel one and has in fact been observed even for some of the classical NRs (ER, TR, RARs, PR, PPARs, VDR, AR) (Nawaz and O’Malley, 2004; Rosenfeld et al., 2006). Similar scenarios of ligand binding affecting novel functions may exist for other receptors which is why the simplest definition of a ligand – a compound that binds the receptor – is the least biased and most inclusive, and therefore perhaps the most useful one.
This then raises the question of whether a given compound must have some sort of functional effect on the receptor, whatever it may be, in order to be considered a true ligand. One can easily make the argument that if a given compound has no effect on function whatsoever, then it must not be a physiologically relevant ligand. However, that assumes that every function has in fact been examined thoroughly, and under the appropriate experimental conditions. That is a very tall order. Furthermore, it may be possible that a ligand previously elicited a function, but that function was lost over time; indeed studies in comparative endocrinology show that NRs and their ligands are both constantly evolving (Markov et al., 2008). For example, a given NR·ligand pair may represent an intermediate evolutionary state in which the function has been lost (or coopted by another receptor) but ligand binding has not yet been lost. Discreet evolutionary steps of NR·ligand pairs have been observed previously (Bridgham et al., 2006; Carroll et al., 2008; Ortlund et al., 2007; Thornton, 2001).
The next issue is the criteria that should be used to determine ligand binding. Again, HNF4α serves as a useful example. Bacterially expressed HNF4 was found to bind a mixture of saturated and monounsaturated fatty acids, and in an essentially irreversible fashion (Dhe-Paganon et al., 2002; Wisely et al., 2002), but endogenous HNF4α isolated directly from liver tissue (or appropriately expressed in mammalian cells) bound only a single polyunsaturated fatty acid (LA) in a reversible fashion (Yuan et al., 2009). This was no doubt due in part to the fact that bacteria typically do not contain polyunsaturated fatty acids such as LA (Cronan and Rock, 1994). Therefore, expressing the receptor in the appropriate cellular environment is key to the identification of true ligands, although that might be difficult for NRs from organisms that are currently extinct. Fortuitous ligands – i.e., those that happen to bind simply because they can under the given circumstance, even if they are not physiologically relevant – are often found during crystallographic studies with bacterially expressed proteins (Ingraham and Redinbo, 2005; Potier et al., 2003) and must be followed up with additional assays. Another example of a NR that was found to bound one compound when expressed in bacteria but another when expressed in mammalian cells is SF-1 (NR5A1) (Krylova et al., 2005; Li et al., 2005; Urs et al., 2007; Urs et al., 2006). As analytical chemistry techniques (e.g., HPLC, gas chromatography and mass spectrometry) become more powerful, and highly specific antibodies to receptors are developed, this unbiased method of identifying ligands for NRs under physiological conditions (affinity isolation followed by mass spec, AIMS) should be more generally applicable (Bridgham et al., 2006; Lafont and Mathieu, 2007; Motola et al., 2006; Urs et al., 2006; Yuan et al., 2009). Variations on this method, such as using a tagged receptor when appropriate antibodies are not available, have also been used successfully. A recent study identified the endogenous ligand for an epitope-tagged PPARα in liver as 1-palmitoyl-2-oleoyl-sn-glycerol-3-phosphocholine (16:0/18:1-GPC) (Chakravarthy et al., 2009) (as opposed to free fatty acids that were previously proposed as ligands, see above). It should also be noted that the mass spec approach can replace previous approaches employing radiolabeled precursors that were so used to identify 9-cis retinoic acid the ligand for RXR (Heyman et al., 1992), but which we now know might have introduced a bias into the system by requiring that the ligand identified be derived from the starting compound (Markov et al., 2008). Finally, using a direct binding approach we may find that the concept of a given NR having multiple ligands, which is well established for PXR and CAR, may be more broadly applicable than was previously thought.
While the identification of ligands via direct binding to NRs is quite recent, the identification of ligands based on function, such as transactivation of a reporter gene in a transient transfection assay, has been used for nearly two decades. As powerful as that approach has been for identifying synthetic ligands and potential drugs, it has not been as successful for identifying endogenous ligands, and may even lead to false positives more frequently than we would like to admit. While the compounds thus identified may be hydrophobic ones and may have structures similar to known NR ligands, that alone does not necessarily mean that they are true ligands – i.e., that they directly bind the receptor. The compounds could, for example, alter a signal transduction pathway that in turn affects the phosphorylation state of the receptor, and it is that phosphostate, and not direct ligand binding, that affects function. Indeed, there are an increasing number of examples of NR ligands activating various kinase activities (e.g., (Cheskis et al., 2008; Gardner et al., 2005; Kim et al., 2009; Wu et al., 2007)). It is also possible in reporter gene assays that a compound may activate an endogenous receptor, which in turn affects the expression and/or activity of the ectopically expressed receptor being tested. There are many examples of intricate transcriptional networks among NRs in which one could envision such a cascade taking place (Bookout et al., 2006). Alternatively, a compound could be a precursor to the true ligand, as may be the case for the free fatty acids that were originally identified as ligands for PPARα by transfection assays (Chakravarthy et al., 2009). Requiring evidence of direct binding by the ligand to the receptor, under the appropriate physiological conditions, circumvents these, and other, problems (Markov et al., 2008).
Summary and Future Directions
While the anthropocentric, pharmacological approach of the past 25 years since the cloning of the first NR has without a doubt been instrumental in characterizing in great detail this family of key transcriptional regulators, it is now time to consider NRs and their ligands from an evolutionary perspective. Remarkable advances have been made in our understanding of the evolutionary origins of life on Earth in recent years, in terms of both traditional paleontology and paleobiology (e.g., molecular fossils, genomic evolution). These advances, combined with those in the NR field, allow a new view of NRs and their ligands to emerge; a view in which the NRs have been constantly sampling their external environment as sensors since they first evolved ~635 Myr ago, an environment that was, and still is, quite large and diverse. As the NRs and the organisms that expressed them evolved, new receptors arose that were able to bind compounds synthesized in the host, and allowed for the communication between organs in an endocrine fashion, and hence the development of even more complex organisms. The modern endocrine receptors, we know, have high affinity ligands, where as many of the modern receptors that act as environmental or metabolic sensors bind ligands with relatively low affinity. There is at least one example of the ancestral NR (estrogen receptor) binding xenobiotics with low affinity (Thornton, 2001); the affinity of additional ancient NR ligands remains to be determined. This view of the modern day endocrine NRs evolving from more ancient receptors that originally sensed their environment is completely consistent not only with early events in the history of the Earth, but also with the prevalence of environmental endocrine disrupters acting via NRs. Continued investigation into the evolution of NRs and their ligands (defined by rigorous standards) will hopefully shed additional light on these and other aspects of NR and their ligands.
Once ligands for ancient NRs have been identified, we will be able to address the next set of questions, answers to which may impact our understanding of evolution of life on Earth itself. For example, even though there were clearly many hundreds of lipophilic molecules that could have been ligands for NRs, why were the first ligands, whatever they may be, used by the first NRs? Was there something special about those compounds that gave the host organism an evolutionary advantage, or could any other compound have served the role equally well? Similarly, why were more of those compounds not used as NR ligands? Why are there relatively few NRs compared to the Cyp genes? And why did the NRs take so long to appear on the evolutionary scene, when Cyp genes and potential ligands were present well in advance of these transcription factors? The only thing that is certain at this point is that the exciting field of NRs and their ligands will keep us as busy in the next 25 years exploring their evolutionary origins as they did in the first 25 years identifying and characterizing them.
Acknowledgments
Special thanks to M. Maduro for discussions on organismal development and B. Fang for drawing ligand structures. The Sladek lab is supported by NIH grants R01 DK053892 and R21 MH087397.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abad P, Gouzy J, Aury JM, Castagnone-Sereno P, Danchin EG, Deleury E, Perfus-Barbeoch L, Anthouard V, Artiguenave F, Blok VC, et al. Genome sequence of the metazoan plant-parasitic nematode Meloidogyne incognita. Nat Biotechnol. 2008;26:909–915. doi: 10.1038/nbt.1482. [DOI] [PubMed] [Google Scholar]
- Al-Moghrabi S, Allemand D, Couret JM, Jaubert J. Fatty acids of the scleractinian coral Galaxea fascicularis: effect of light and feeding. J Comp Physiol B. 1995;165:183–192. [Google Scholar]
- Baker ME. Trichoplax, the simplest known animal, contains an estrogen-related receptor but no estrogen receptor: Implications for estrogen receptor evolution. Biochem Biophys Res Commun. 2008;375:623–627. doi: 10.1016/j.bbrc.2008.08.047. [DOI] [PubMed] [Google Scholar]
- Baldauf SL, Bhattacharya D, Cockrill J, Hugenholtz P, Pawlowski J, Simpson AGB. In: The Tree of Life: A Overview. Craycraft J, Donoghue MJ, editors. Assembling the Tree of Life Oxford University Press; 2004. pp. 43–75. [Google Scholar]
- Benoit G, Malewicz M, Perlmann T. Digging deep into the pockets of orphan nuclear receptors: insights from structural studies. Trends Cell Biol. 2004;14:369–376. doi: 10.1016/j.tcb.2004.05.007. [DOI] [PubMed] [Google Scholar]
- Bertrand S, Brunet FG, Escriva H, Parmentier G, Laudet V, Robinson-Rechavi M. Evolutionary genomics of nuclear receptors: from twenty-five ancestral genes to derived endocrine systems. Mol Biol Evol. 2004;21:1923–1937. doi: 10.1093/molbev/msh200. [DOI] [PubMed] [Google Scholar]
- Bertrand S, Thisse B, Tavares R, Sachs L, Chaumot A, Bardet PL, Escriva H, Duffraisse M, Marchand O, Safi R, et al. Unexpected novel relational links uncovered by extensive developmental profiling of nuclear receptor expression. PLoS Genet. 2007;3:e188. doi: 10.1371/journal.pgen.0030188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogan AA, Dallas-Yang Q, Ruse MD, Jr, Maeda Y, Jiang G, Nepomuceno L, Scanlan TS, Cohen FE, Sladek FM. Analysis of protein dimerization and ligand binding of orphan receptor HNF4alpha. J Mol Biol. 2000;302:831–851. doi: 10.1006/jmbi.2000.4099. [DOI] [PubMed] [Google Scholar]
- Bolotin E, Liao H, Ta TC, Yang C, Hwang-Verslues W, Evans JR, Jiang T, Sladek FM. Integrated approach for the identification of human hepatocyte nuclear factor 4alpha target genes using protein binding microarrays. Hepatology. 2009 doi: 10.1002/hep.23357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bookout AL, Jeong Y, Downes M, Yu RT, Evans RM, Mangelsdorf DJ. Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell. 2006;126:789–799. doi: 10.1016/j.cell.2006.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridgham JT, Carroll SM, Thornton JW. Evolution of hormone-receptor complexity by molecular exploitation. Science. 2006;312:97–101. doi: 10.1681/01.asn.0000926836.46869.e5. [DOI] [PubMed] [Google Scholar]
- Brocks JJ, Banfield J. Unravelling ancient microbial history with community proteogenomics and lipid geochemistry. Nat Rev Microbiol. 2009;7:601–609. doi: 10.1038/nrmicro2167. [DOI] [PubMed] [Google Scholar]
- Brocks JJ, Logan GA, Buick R, Summons RE. Archean molecular fossils and the early rise of eukaryotes. Science. 1999;285:1033–1036. doi: 10.1126/science.285.5430.1033. [DOI] [PubMed] [Google Scholar]
- Burr GO, Burr MM. A new deficiency disease produced by rigid exclusion of fat from diet. J Biol Chem. 1929;89:345–367. [Google Scholar]
- Burr GO, Burr MM. The nature and role of the fatty acids essential in nutrition. J Biol Chem. 1930;86:587–621. [Google Scholar]
- Burris TP. Nuclear hormone receptors for heme: REV-ERBalpha and REV-ERBbeta are ligand-regulated components of the mammalian clock. Mol Endocrinol. 2008;22:1509–1520. doi: 10.1210/me.2007-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SM, Bridgham JT, Thornton JW. Evolution of hormone signaling in elasmobranchs by exploitation of promiscuous receptors. Mol Biol Evol. 2008;25:2643–2652. doi: 10.1093/molbev/msn204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarthy MV, Lodhi IJ, Yin L, Malapaka RR, Xu HE, Turk J, Semenkovich CF. Identification of a physiologically relevant endogenous ligand for PPARalpha in liver. Cell. 2009;138:476–488. doi: 10.1016/j.cell.2009.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T. Nuclear receptor drug discovery. Curr Opin Chem Biol. 2008;12:418–426. doi: 10.1016/j.cbpa.2008.07.001. [DOI] [PubMed] [Google Scholar]
- Cheskis BJ, Greger J, Cooch N, McNally C, McLarney S, Lam HS, Rutledge S, Mekonnen B, Hauze D, Nagpal S, Freedman LP. MNAR plays an important role in ERa activation of Src/MAPK and PI3K/Akt signaling pathways. Steroids. 2008;73:901–905. doi: 10.1016/j.steroids.2007.12.028. [DOI] [PubMed] [Google Scholar]
- Cornell W, Nam K. Steroid hormone binding receptors: application of homology modeling, induced fit docking, and molecular dynamics to study structure-function relationships. Curr Top Med Chem. 2009;9:844–853. doi: 10.2174/156802609789207109. [DOI] [PubMed] [Google Scholar]
- Cronan JE, Jr, Rock CO. The presence of linoleic acid in Escherichia coli cannot be confirmed. J Bacteriol. 1994;176:3069–3071. doi: 10.1128/jb.176.10.3069-3071.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielson PB. The cytochrome P450 superfamily: biochemistry, evolution and drug metabolism in humans. Curr Drug Metab. 2002;3:561–597. doi: 10.2174/1389200023337054. [DOI] [PubMed] [Google Scholar]
- Degnan BM, Vervoort M, Larroux C, Richards GS. Early evolution of metazoan transcription factors. Curr Opin Genet Dev. 2009;19:591–599. doi: 10.1016/j.gde.2009.09.008. [DOI] [PubMed] [Google Scholar]
- Dhe-Paganon S, Duda K, Iwamoto M, Chi YI, Shoelson SE. Crystal structure of the HNF4 alpha ligand binding domain in complex with endogenous fatty acid ligand. J Biol Chem. 2002;277:37973–37976. doi: 10.1074/jbc.C200420200. [DOI] [PubMed] [Google Scholar]
- Duda K, Chi YI, Shoelson SE. Structural basis for HNF-4alpha activation by ligand and coactivator binding. J Biol Chem. 2004;279:23311–23316. doi: 10.1074/jbc.M400864200. [DOI] [PubMed] [Google Scholar]
- Escriva H, Bertrand S, Laudet V. The evolution of the nuclear receptor superfamily. Essays Biochem. 2004;40:11–26. doi: 10.1042/bse0400011. [DOI] [PubMed] [Google Scholar]
- Escriva H, Delaunay F, Laudet V. Ligand binding and nuclear receptor evolution. Bioessays. 2000;22:717–727. doi: 10.1002/1521-1878(200008)22:8<717::AID-BIES5>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Evans RM. The steroid and thyroid hormone receptor superfamily. Science. 1988;240:889–895. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forman BM, Chen J, Evans RM. Hypolipidemic drugs, polyunsaturated fatty acids, and eicosanoids are ligands for peroxisome proliferator-activated receptors alpha and delta. Proc Natl Acad Sci U S A. 1997;94:4312–4317. doi: 10.1073/pnas.94.9.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner OS, Dewar BJ, Graves LM. Activation of mitogen-activated protein kinases by peroxisome proliferator-activated receptor ligands: an example of nongenomic signaling. Mol Pharmacol. 2005;68:933–941. doi: 10.1124/mol.105.012260. [DOI] [PubMed] [Google Scholar]
- Ghedin E, Wang S, Spiro D, Caler E, Zhao Q, Crabtree J, Allen JE, Delcher AL, Guiliano DB, Miranda-Saavedra D, et al. Draft genome of the filarial nematode parasite Brugia malayi. Science. 2007;317:1756–1760. doi: 10.1126/science.1145406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giguere V, Ong ES, Segui P, Evans RM. Identification of a receptor for the morphogen retinoic acid. Nature. 1987;330:624–629. doi: 10.1038/330624a0. [DOI] [PubMed] [Google Scholar]
- Gilman AG, Mayer SE, Melmon KL. Pharmacodynamics: Mechanisms of Drug Action and the Relationship Between Drug Concentration and Effect. In: Gilman AG, Goodman LS, Gilman A, editors. Goodman and Gilman’s The Pharmacological Basis of Therapeutics. Macmillan Publiching Co., Inc; New York: 1980. pp. 28–39. [Google Scholar]
- Gottlicher M, Widmark E, Li Q, Gustafsson JA. Fatty acids activate a chimera of the clofibric acid-activated receptor and the glucocorticoid receptor. Proc Natl Acad Sci U S A. 1992;89:4653–4657. doi: 10.1073/pnas.89.10.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant PA, Duggan L, Cote J, Roberts SM, Brownell JE, Candau R, Ohba R, Owen-Hughes T, Allis CD, Winston F, et al. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- Grasso LC, Hayward DC, Trueman JW, Hardie KM, Janssens PA, Ball EE. The evolution of nuclear receptors: evidence from the coral Acropora. Mol Phylogenet Evol. 2001;21:93–102. doi: 10.1006/mpev.2001.0994. [DOI] [PubMed] [Google Scholar]
- Green S, Walter P, Kumar V, Krust A, Bornert JM, Argos P, Chambon P. Human oestrogen receptor cDNA: sequence, expression and homology to v-erb-A. Nature. 1986;320:134–139. doi: 10.1038/320134a0. [DOI] [PubMed] [Google Scholar]
- Gronemeyer H, Gustafsson JA, Laudet V. Principles for modulation of the nuclear receptor superfamily. Nat Rev Drug Discov. 2004;3:950–964. doi: 10.1038/nrd1551. [DOI] [PubMed] [Google Scholar]
- Gunstone F. Fatty Acid and Lipid Chemistry. 1. Blackie Academic & Professional; London: 1996. [Google Scholar]
- Hertz R, Magenheim J, Berman I, Bar-Tana J. Fatty acyl-CoA thioesters are ligands of hepatic nuclear factor-4alpha. Nature. 1998;392:512–516. doi: 10.1038/33185. [DOI] [PubMed] [Google Scholar]
- Heyman RA, Mangelsdorf DJ, Dyck JA, Stein RB, Eichele G, Evans RM, Thaller C. 9-cis retinoic acid is a high affinity ligand for the retinoid X receptor. Cell. 1992;68:397–406. doi: 10.1016/0092-8674(92)90479-v. [DOI] [PubMed] [Google Scholar]
- Ingraham HA, Redinbo MR. Orphan nuclear receptors adopted by crystallography. Curr Opin Struct Biol. 2005;15:708–715. doi: 10.1016/j.sbi.2005.10.009. [DOI] [PubMed] [Google Scholar]
- Jaillon O, Aury JM, Brunet F, Petit JL, Stange-Thomann N, Mauceli E, Bouneau L, Fischer C, Ozouf-Costaz C, Bernot A, et al. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature. 2004;431:946–957. doi: 10.1038/nature03025. [DOI] [PubMed] [Google Scholar]
- Jover R, Moya M, Gomez-Lechon MJ. Transcriptional regulation of cytochrome p450 genes by the nuclear receptor hepatocyte nuclear factor 4-alpha. Curr Drug Metab. 2009;10:508–519. doi: 10.2174/138920009788898000. [DOI] [PubMed] [Google Scholar]
- Kim MH, Kim MO, Kim YH, Kim JS, Han HJ. Linoleic acid induces mouse embryonic stem cell proliferation via Ca2+/PKC, PI3K/Akt, and MAPKs. Cell Physiol Biochem. 2009;23:53–64. doi: 10.1159/000204090. [DOI] [PubMed] [Google Scholar]
- King N, Westbrook MJ, Young SL, Kuo A, Abedin M, Chapman J, Fairclough S, Hellsten U, Isogai Y, Letunic I, et al. The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature. 2008;451:783–788. doi: 10.1038/nature06617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovac J, Husse J, Oster H. A time to fast, a time to feast: the crosstalk between metabolism and the circadian clock. Mol Cells. 2009;28:75–80. doi: 10.1007/s10059-009-0113-0. [DOI] [PubMed] [Google Scholar]
- Kruse SW, Suino-Powell K, Zhou XE, Kretschman JE, Reynolds R, Vonrhein C, Xu Y, Wang L, Tsai SY, Tsai MJ, Xu HE. Identification of COUP-TFII orphan nuclear receptor as a retinoic acid-activated receptor. PLoS Biol. 2008;6:e227. doi: 10.1371/journal.pbio.0060227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krylova IN, Sablin EP, Moore J, Xu RX, Waitt GM, MacKay JA, Juzumiene D, Bynum JM, Madauss K, Montana V, et al. Structural analyses reveal phosphatidyl inositols as ligands for the NR5 orphan receptors SF-1 and LRH-1. Cell. 2005;120:343–355. doi: 10.1016/j.cell.2005.01.024. [DOI] [PubMed] [Google Scholar]
- Lafont R, Mathieu M. Steroids in aquatic invertebrates. Ecotoxicology. 2007;16:109–130. doi: 10.1007/s10646-006-0113-1. [DOI] [PubMed] [Google Scholar]
- Larroux C, Fahey B, Liubicich D, Hinman VF, Gauthier M, Gongora M, Green K, Worheide G, Leys SP, Degnan BM. Developmental expression of transcription factor genes in a demosponge: insights into the origin of metazoan multicellularity. Evol Dev. 2006;8:150–173. doi: 10.1111/j.1525-142X.2006.00086.x. [DOI] [PubMed] [Google Scholar]
- Latyshev NA, Naumenko NV, Svetashev VI, Latypov YY. Fatty acids of reef-building corals. MArine Ecology Progress Series. 1991;76:295–301. [Google Scholar]
- Laudet V. Evolution of the nuclear receptor superfamily: early diversification from an ancestral orphan receptor. J Mol Endocrinol. 1997;19:207–226. doi: 10.1677/jme.0.0190207. [DOI] [PubMed] [Google Scholar]
- Li Y, Choi M, Cavey G, Daugherty J, Suino K, Kovach A, Bingham NC, Kliewer SA, Xu HE. Crystallographic identification and functional characterization of phospholipids as ligands for the orphan nuclear receptor steroidogenic factor-1. Mol Cell. 2005;17:491–502. doi: 10.1016/j.molcel.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Love GD, Grosjean E, Stalvies C, Fike DA, Grotzinger JP, Bradley AS, Kelly AE, Bhatia M, Meredith W, Snape CE, et al. Fossil steroids record the appearance of Demospongiae during the Cryogenian period. Nature. 2009;457:718–721. doi: 10.1038/nature07673. [DOI] [PubMed] [Google Scholar]
- Maglich JM, Caravella JA, Lambert MH, Willson TM, Moore JT, Ramamurthy L. The first completed genome sequence from a teleost fish (Fugu rubripes) adds significant diversity to the nuclear receptor superfamily. Nucleic Acids Res. 2003;31:4051–4058. doi: 10.1093/nar/gkg444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makishima M, Lu TT, Xie W, Whitfield GK, Domoto H, Evans RM, Haussler MR, Mangelsdorf DJ. Vitamin D receptor as an intestinal bile acid sensor. Science. 2002;296:1313–1316. doi: 10.1126/science.1070477. [DOI] [PubMed] [Google Scholar]
- Markov GV, Bonneton F, Laudet V. What does evolution teach us about nuclear receptors? In: Campbell CMBaMJ., editor. Nuclear Receptors: Current Concepts and Future Challenges. Springer; Netherlands: 2010. pp. 15–29. [Google Scholar]
- Markov GV, Paris M, Bertrand S, Laudet V. The evolution of the ligand/receptor couple: a long road from comparative endocrinology to comparative genomics. Mol Cell Endocrinol. 2008;293:5–16. doi: 10.1016/j.mce.2008.06.011. [DOI] [PubMed] [Google Scholar]
- Markov GV, Tavares R, Dauphin-Villemant C, Demeneix BA, Baker ME, Laudet V. Independent elaboration of steroid hormone signaling pathways in metazoans. Proc Natl Acad Sci U S A. 2009;106:11913–11918. doi: 10.1073/pnas.0812138106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez E. Multi-protein complexes in eukaryotic gene transcription. Plant Mol Biol. 2002;50:925–947. doi: 10.1023/a:1021258713850. [DOI] [PubMed] [Google Scholar]
- Michalik L, Auwerx J, Berger JP, Chatterjee VK, Glass CK, Gonzalez FJ, Grimaldi PA, Kadowaki T, Lazar MA, O’Rahilly S, et al. International Union of Pharmacology. LXI. Peroxisome proliferator-activated receptors. Pharmacol Rev. 2006;58:726–741. doi: 10.1124/pr.58.4.5. [DOI] [PubMed] [Google Scholar]
- Miyachi H. Design, synthesis, and structure-activity relationship study of peroxisome proliferator-activated receptor (PPAR) delta-selective ligands. Curr Med Chem. 2007;14:2335–2343. doi: 10.2174/092986707781745587. [DOI] [PubMed] [Google Scholar]
- Motola DL, Cummins CL, Rottiers V, Sharma KK, Li T, Li Y, Suino-Powell K, Xu HE, Auchus RJ, Antebi A, Mangelsdorf DJ. Identification of ligands for DAF-12 that govern dauer formation and reproduction in C. elegans. Cell. 2006;124:1209–1223. doi: 10.1016/j.cell.2006.01.037. [DOI] [PubMed] [Google Scholar]
- Nawaz Z, O’Malley BW. Urban renewal in the nucleus: is protein turnover by proteasomes absolutely required for nuclear receptor-regulated transcription? Mol Endocrinol. 2004;18:493–499. doi: 10.1210/me.2003-0388. [DOI] [PubMed] [Google Scholar]
- Nebert DW, Nelson DR, Feyereisen R. Evolution of the cytochrome P450 genes. Xenobiotica. 1989;19:1149–1160. doi: 10.3109/00498258909043167. [DOI] [PubMed] [Google Scholar]
- Nelson DR. Updated The Cytochrome P450 Homepage. [Accessed January 15, 2010];From the Human Genomics. 4:59–65. doi: 10.1186/1479-7364-4-1-59. http://drnelson.utmem.edu/CytochromeP450.html. [DOI] [PMC free article] [PubMed]
- O’Malley BW, Conneely OM. Orphan receptors: in search of a unifying hypothesis for activation. Mol Endocrinol. 1992;6:1359–1361. doi: 10.1210/mend.6.9.1331771. [DOI] [PubMed] [Google Scholar]
- Ortlund EA, Bridgham JT, Redinbo MR, Thornton JW. Crystal structure of an ancient protein: evolution by conformational epistasis. Science. 2007;317:1544–1548. doi: 10.1126/science.1142819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottow E, Weinmann H, editors. Nuclear Receptors as Drug Targets. Wiley-VCH; Weinheim, Germany: 2008. [Google Scholar]
- Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nat Rev Drug Discov. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- Parks DJ, Blanchard SG, Bledsoe RK, Chandra G, Consler TG, Kliewer SA, Stimmel JB, Willson TM, Zavacki AM, Moore DD, Lehmann JM. Bile acids: natural ligands for an orphan nuclear receptor. Science. 1999;284:1365–1368. doi: 10.1126/science.284.5418.1365. [DOI] [PubMed] [Google Scholar]
- Potier N, Billas IM, Steinmetz A, Schaeffer C, van Dorsselaer A, Moras D, Renaud JP. Using nondenaturing mass spectrometry to detect fortuitous ligands in orphan nuclear receptors. Protein Sci. 2003;12:725–733. doi: 10.1110/ps.0232503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuram S, Stayrook KR, Huang P, Rogers PM, Nosie AK, McClure DB, Burris LL, Khorasanizadeh S, Burris TP, Rastinejad F. Identification of heme as the ligand for the orphan nuclear receptors REV-ERBalpha and REV-ERBbeta. Nat Struct Mol Biol. 2007;14:1207–1213. doi: 10.1038/nsmb1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhard CT, Raiswell R, Scott C, Anbar AD, Lyons TW. A late Archean sulfidic sea stimulated by early oxidative weathering of the continents. Science. 2009;326:713–716. doi: 10.1126/science.1176711. [DOI] [PubMed] [Google Scholar]
- Reitzel AM, Tarrant AM. Nuclear receptor complement of the cnidarian Nematostella vectensis: phylogenetic relationships and developmental expression patterns. BMC Evol Biol. 2009;9:230. doi: 10.1186/1471-2148-9-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renaud JP, Moras D. Structural studies on nuclear receptors. Cell Mol Life Sci. 2000;57:1748–1769. doi: 10.1007/PL00000656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson-Rechavi M, Maina CV, Gissendanner CR, Laudet V, Sluder A. Explosive lineage-specific expansion of the orphan nuclear receptor HNF4 in nematodes. J Mol Evol. 2005;60:577–586. doi: 10.1007/s00239-004-0175-8. [DOI] [PubMed] [Google Scholar]
- Rosenfeld MG, Lunyak VV, Glass CK. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 2006;20:1405–1428. doi: 10.1101/gad.1424806. [DOI] [PubMed] [Google Scholar]
- Schierwater B, Eitel M, Jakob W, Osigus HJ, Hadrys H, Dellaporta SL, Kolokotronis SO, Desalle R. Concatenated analysis sheds light on early metazoan evolution and fuels a modern “urmetazoon” hypothesis. PLoS Biol. 2009;7:e20. doi: 10.1371/journal.pbio.1000020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman IG, Heyman RA. The flip side: Identifying small molecule regulators of nuclear receptors. Chem Biol. 2004;11:639–646. doi: 10.1016/j.chembiol.2003.12.021. [DOI] [PubMed] [Google Scholar]
- Shiau AK, Barstad D, Loria PM, Cheng L, Kushner PJ, Agard DA, Greene GL. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell. 1998;95:927–937. doi: 10.1016/s0092-8674(00)81717-1. [DOI] [PubMed] [Google Scholar]
- Sladek F. Desperately seeking…something. Mol Cell. 2002;10:219–221. doi: 10.1016/s1097-2765(02)00605-6. [DOI] [PubMed] [Google Scholar]
- Sladek FM, Ruse MD, Jr, Nepomuceno L, Huang SM, Stallcup MR. Modulation of transcriptional activation and coactivator interaction by a splicing variation in the F domain of nuclear receptor hepatocyte nuclear factor 4alpha1. Mol Cell Biol. 1999;19:6509–6522. doi: 10.1128/mcb.19.10.6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sladek FM, Zhong WM, Lai E, Darnell JE., Jr Liver-enriched transcription factor HNF-4 is a novel member of the steroid hormone receptor superfamily. Genes Dev. 1990;4:2353–2365. doi: 10.1101/gad.4.12b.2353. [DOI] [PubMed] [Google Scholar]
- Sluder AE, Maina CV. Nuclear receptors in nematodes: themes and variations. Trends Genet. 2001;17:206–213. doi: 10.1016/s0168-9525(01)02242-9. [DOI] [PubMed] [Google Scholar]
- Sperling EA, Pisnai D, Peterson KJ. Poriferan paraphyly and its implications for Precambrian palaeobiology. In: Vicker-Ricd PKP, editor. The Rise and Fall of the Ediacaran Biota. Geological Scoeity; London: 2007. pp. 355–368. [Google Scholar]
- Srivastava M, Begovic E, Chapman J, Putnam NH, Hellsten U, Kawashima T, Kuo A, Mitros T, Salamov A, Carpenter ML, et al. The Trichoplax genome and the nature of placozoans. Nature. 2008;454:955–960. doi: 10.1038/nature07191. [DOI] [PubMed] [Google Scholar]
- Staudinger JL, Goodwin B, Jones SA, Hawkins-Brown D, MacKenzie KI, LaTour A, Liu Y, Klaassen CD, Brown KK, Reinhard J, et al. The nuclear receptor PXR is a lithocholic acid sensor that protects against liver toxicity. Proc Natl Acad Sci U S A. 2001;98:3369–3374. doi: 10.1073/pnas.051551698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ta TC, Sladek FM. HNF4: From orphan receptor to orphan ligand. Molecular Endocrinology. 2010 (in preparation) [Google Scholar]
- Tarrant AM, Cortés J, Atkinson M, Atkinson S, Johanning K, Chiang T, Vargas JA, Mclachlan JA. Three orphan nuclear receptors in the scleractinian coral Pocillopora damicornis from the Pacific coast of Costa Rica. Rev Biol Trop. 2008;56:39–48. [Google Scholar]
- Thornton JW. Evolution of vertebrate steroid receptors from an ancestral estrogen receptor by ligand exploitation and serial genome expansions. Proc Natl Acad Sci U S A. 2001;98:5671–5676. doi: 10.1073/pnas.091553298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwary B, Li WH. Parallel evolution between aromatase and androgen receptor in the animal kingdom. Mol Biol Evol. 2009;26:123–129. doi: 10.1093/molbev/msn233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urs AN, Dammer E, Kelly S, Wang E, Merrill AH, Jr, Sewer MB. Steroidogenic factor-1 is a sphingolipid binding protein. Mol Cell Endocrinol. 2007;265–266:174–178. doi: 10.1016/j.mce.2006.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urs AN, Dammer E, Sewer MB. Sphingosine regulates the transcription of CYP17 by binding to steroidogenic factor-1. Endocrinology. 2006;147:5249–5258. doi: 10.1210/en.2006-0355. [DOI] [PubMed] [Google Scholar]
- Webster NJ, Green S, Jin JR, Chambon P. The hormone-binding domains of the estrogen and glucocorticoid receptors contain an inducible transcription activation function. Cell. 1988;54:199–207. doi: 10.1016/0092-8674(88)90552-1. [DOI] [PubMed] [Google Scholar]
- Wickramashighe RH, Villee CA. Early role during chemical evolution for cytochrome P450 in oxygen detoxification. Nature. 1975;256:509–510. doi: 10.1038/256509a0. [DOI] [PubMed] [Google Scholar]
- Wiens M, Batel R, Korzhev M, Muller WE. Retinoid X receptor and retinoic acid response in the marine sponge Suberites domuncula. J Exp Biol. 2003;206:3261–3271. doi: 10.1242/jeb.00541. [DOI] [PubMed] [Google Scholar]
- Wisely GB, Miller AB, Davis RG, Thornquest AD, Jr, Johnson R, Spitzer T, Sefler A, Shearer B, Moore JT, Miller AB, et al. Hepatocyte nuclear factor 4 is a transcription factor that constitutively binds fatty acids. Structure. 2002;10:1225–1234. doi: 10.1016/s0969-2126(02)00829-8. [DOI] [PubMed] [Google Scholar]
- Wu W, Zhang X, Zanello LP. 1alpha, 25-Dihydroxyvitamin D(3) antiproliferative actions involve vitamin D receptor-mediated activation of MAPK pathways and AP-1/p21(waf1) upregulation in human osteosarcoma. Cancer Lett. 2007;254:75–86. doi: 10.1016/j.canlet.2007.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Downes M, Yu RT, Bookout AL, He W, Straume M, Mangelsdorf DJ, Evans RM. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126:801–810. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- Yin L, Wu N, Curtin JC, Qatanani M, Szwergold NR, Reid RA, Waitt GM, Parks DJ, Pearce KH, Wisely GB, Lazar MA. Rev-erbalpha, a heme sensor that coordinates metabolic and circadian pathways. Science. 2007;318:1786–1789. doi: 10.1126/science.1150179. [DOI] [PubMed] [Google Scholar]
- Yuan X, Ta TC, Lin M, Evans JR, Dong Y, Bolotin E, Sherman MA, Forman BM, Sladek FM. Identification of an endogenous ligand bound to a native orphan nuclear receptor. PLoS One. 2009;4:e5609. doi: 10.1371/journal.pone.0005609. [DOI] [PMC free article] [PubMed] [Google Scholar]