Abstract
We applied an automated hippocampal segmentation technique based on adaptive boosting (AdaBoost) to the 1.5T MRI baseline and 1-year follow-up data of 243 subjects with mild cognitive impairment (MCI), 96 with Alzheimer's disease (AD) and 145 normal controls (NC) scanned as part of the Alzheimer's Disease Neuroimaging Initiative (ADNI). MCI subjects with positive maternal history of dementia had smaller hippocampal volumes at baseline and at follow-up, and greater 12-month atrophy rates than subjects with negative maternal history. 3D maps and volumetric multiple regression analyses demonstrated a significant effect of positive maternal history of dementia on hippocampal atrophy in MCI and AD after controlling for age, ApoE4 genotype and paternal history of dementia, resp. ApoE4 showed an independent effect on hippocampal atrophy in MCI and AD and in the pooled sample.
Keywords: Alzheimer's Disease, AD, Magnetic resonance imaging, MRI, Imaging, Maternal history, Hippocampus, Atrophy, Biomarker, Hereditary, Genetic
1. Introduction
As the world population continues to age, Alzheimer's disease (AD) is rapidly becoming a major, and increasing, health care concern. If novel successful therapeutic approaches do not become available, the number of patients with AD is expected to rise from 4.5 million in 2000 to 13.2 million in 2050 in the United States alone [Cummings, 2004; Hebert, et al., 2003]. The annual cost of care for AD patients worldwide is estimated at $83.9 billion (in 1996 US dollars) and will continue to increase with the rising prevalence of AD [Cummings, 2004; Wimo and Winblad, 2001]. The predictions of significant limitations in healthcare resources for elderly care in the near future underscore the pressing need for understanding the pathophysiology of AD and more effective therapies.
The rare variants of early-onset familial AD have been attributed to autosomal dominant mutations in the amyloid precursor protein genes, presenilin 1 and presenilin 2 [Bertram and Tanzi, 2008]. The genetics of late-onset, sporadic AD are much less clear. Family history is the second greatest risk factor for sporadic AD after age [Tanzi and Bertram, 2001]. Having a first-degree relative with AD increases one's risk for AD 4–10 fold [Cupples, et al., 2004; Green, et al., 2002; Mosconi, et al., 2009; Silverman, et al., 2005] suggesting a significant genetic component to the sporadic form of the disease. The apolipoprotein E4 (APOE4) gene has repeatedly demonstrated a strong association with late onset AD, but its prevalence of 40–70% in the AD population suggests that additional genetic risk factors will be discovered [Bertram and Tanzi, 2009; Seripa, et al., 2009; Slooter, et al., 1998]. Several genome-wide association studies have reported other candidate AD-associated genes that warrant further exploration [Bertram and Tanzi, 2009; Li, et al., 2006; Rogaeva, et al., 2007; Sundar, et al., 2007]. At the same time, epidemiologic studies suggest that subjects with maternal history of AD have increased risk of developing the disease [Edland, et al., 1996; Ehrenkrantz, et al., 1999; Green, et al., 2002]. The Framingham Offspring study reported that middle aged individuals with AD-affected mothers show deficient neuropsychological test performance compared to those with AD-affected fathers and those lacking parental history of AD [Wolf, et al., 2005]. Recently Mosconi et al. reported that relative to subjects with no paternal history of AD those with maternal history of AD not only show decreased glucose uptake in AD-vulnerable regions on positron emission tomography examination (PET) cross-sectionally [Mosconi, et al., 2007] but also exhibit progressive decline in glucose uptake in these regions over time [Mosconi, et al., 2009]. In addition, two recent structural MRI studies examined the effect of parental history of dementia on hippocampal and total brain volume [Debette, et al., 2009] and gray matter volume [Honea, et al.]. The larger study [Debette, et al., 2009] reported no effect of history of parental dementia on hippocampal and total brain volume at baseline, or on total brain volume and hippocampal atrophy rates, in cognitively normal subjects. No significant associations were detected after stratification for ApoE4 status as well as for maternal/paternal history of dementia. The smaller study [Honea, et al.] likewise studied the effect of maternal and paternal history of AD on brain structure in cognitively normal elderly. They found significantly smaller inferior frontal, middle frontal, inferior temporal and precuneal gray matter volumes, but no effect on hippocampal volume.
Here we wanted to further explore the effect of maternal history of dementia on the hippocampus, an area affected early by AD pathology. We hypothesized that maternal history of dementia would be associated with greater hippocampal atrophy relative to that seen in groups of subjects with paternal or no parental history of dementia.
2. Methods
2.1. Subjects
The Alzheimer Disease Neuroimaging Intitiative (ADNI) is a large scale multi-site longitudinal study collecting detailed clinical, imaging and laboratory data from 200 normal control (NC), 400 amnestic mild cognitive impairment (MCI) and 200 AD subjects at multiple time points over a 4-year period [Mueller, et al., 2005] (also see http://www.loni.ucla.edu/ADNI and ADNI-info. org). ADNI's goal is to establish risk factors associated with cognitive decline from normal aging to MCI and from MCI to AD, as well as to assist the development of sensitive disease-specific biomarkers for early diagnosis and the development of effective therapeutic agents for AD. All ADNI participants were 55–90 years old at the time of enrollment. Subjects were excluded if they had any significant neurologic disease other than AD, abnormal baseline magnetic resonance imaging (MRI) scan or contraindications to MRI, psychiatric disorder, alcohol or substance abuse or dependency within the last 2 years and medical illnesses that could effect cognition or protocol compliance. Other grounds for exclusion were residence in a skilled nursing facility, use of psychoactive medications other then certain antidepressants, warfarin, or investigational agents. The full list of inclusion/exclusion criteria can be accessed on pages 20–30 of the online ADNI protocol (http://www.adni-info.org/images/stories/Documentation/adni_protocol_9_19_08.pdf).
NC subjects scored within age- and education-adjusted norms on the Logical Memory II subscale from Wechsler Memory Scale-Revised (LM II) [Wechsler, 1987], between 24–30 on the Mini-Mental State Examination (MMSE) [Folstein, et al., 1975], and 0 on the global score of the Clinical Dementia Rating Scale (CDR) [Morris, 1993]. Subjects with MCI had memory complaints and scored below age- and education-adjusted norms on LM II. The MMSE scores of MCI subjects at baseline were between 24–30 and their global CDR was 0.5. General cognition and sufficiently preserved activities of daily living precluding diagnosis of AD were also required for the diagnosis of MCI. ADNI enrolled mildly affected AD subjects who met the National Institute of Neurologic and Communicative Disorders and Stroke/Alzheimer's Disease and Related Disorders Association (NINDS/ADRDA) criteria for probable AD [McKhann, et al., 1984]. AD subjects had MMSE scores between 20–26 and CDR of 0.5 or 1.
The present study used ADNI baseline and one-year follow-up cognitive and imaging data of 243 amnestic MCI, 96 AD and 145 NC subjects. Subjects were stratified based on their family history of dementia collected through self-report. Out of 484 subjects, 68 (14%) reported paternal (MHany/PH+) and 167 (35%) maternal history (MH+/PHany) of dementia. 149 subjects had maternal but not paternal history of dementia (MH+/PH−). 50 subjects had paternal but not maternal history of dementia (MH−/PH+). 18 subjects had both maternal and paternal history of dementia (MH+/PH+) and 267 had neither maternal nor paternal history of dementia (MH−/PH−).
2.2 Image Acquisition and Preprocessing
All subjects were scanned at 1.5T magnetic field strength, on scanners from one of three manufacturers (General Electric Healthcare, Siemens Medical Solutions, and Philips Medical Systems) with a scanner-specific standardized MRI protocol (http://www.loni.ucla.edu/ADNI/Research/Cores/index.shtml) [Jack, et al., 2008; Leow, et al., 2006]. The TE/TR/TI (echo, repetition, and inversion time) parameters were set of optimize contrast to noise in a feasible acquisition time. The raw data had an acquisition matrix of 192×192×166 and voxel size 1.25×1.25×1.2 mm3 [Jack, et al., 2008]. Following an in-plane, zero-filled reconstruction (i.e., sinc interpolation) yielded a matrix of 256×256 and a voxel size of 0.9375×0.9375×1.2 mm3. Two T1-weighted MRI sessions were collected from each subject at each visit. The image with greater apparent signal to noise ratio was selected by the ADNI MRI quality control center at the Mayo Clinic (Rochester, MN, USA) [Jack, et al., 2008]. The ADNI MRI quality control center reviewed each scan (including T2-weighted and proton density scans) for structural abnormalities such as cortical or subcortical strokes, significant white matter ischemic changes, focal lesions, etc. Subjects in whom structural abnormalities were found were excluded from participation. All scans were subjected to additional image corrections including GradWarp correction for geometric distortions due to gradient non-linearity [Jovicich, et al., 2006], “B1-correction” for image intensity non-uniformity [Jack, et al., 2008] and “N3” bias field correction, for reducing intensity inhomogeneity [Sled, et al., 1998]. Both uncorrected and corrected imaging data were transmitted to the central repository site at the Laboratory of Neuro Imaging at University of California Los Angeles and are freely available for download at http://www.loni.ucla.edu/ADNI.
All data downloaded from the ADNI database reflects the state of the database at the point of download. The downloaded, corrected images were linearly registered to the International Consortium for Brain Mapping (ICBM-53) standard brain template [Mazziotta, et al., 2001] with a 9-parameter (9P) transformation (3 translations, 3 rotations, 3 scales) using the Minctracc algorithm [Collins, et al., 1994]. Globally aligned images were resampled in an isotropic space of 220 voxels along each axis (x, y, and z), with a final voxel size of 1 mm3.
2.3 Imaging Analyses
We applied a recently validated automated hippocampal segmentation technique based on a machine learning method called adaptive boosting (AdaBoost) to the baseline and 1-year follow-up images of the selected ADNI study sample. [Morra, et al., 2008b; Morra, et al., 2008a; Morra, et al., 2009; Schapire and Freund, 1998] The AdaBoost segmentation method is a machine learning algorithm that applies a pattern recognition approach to a training dataset consisting of small number of manually traced structures of interest to develop a set of segmentation rules for extracting the structures of interest (in our case the hippocampi) from unfamiliar images while minimizing global error. AdaBoost iteratively develops and tests a set of rules for combining thousands of voxel-wise imaging features such as intensity, stereotaxic positions, tissue classification, local curvatures, gradients and filters into a segmentation algorithm that can optimally segment the training dataset and ultimately unknown images. Our training dataset consisted of the manual hippocampal traces of 21 randomly chosen ADNI subjects (7 NC, 7MCI and 7AD) created by a single human expert (AEG, intra-rater reliability Cronbach's Alpha=0.98) following a widely used and extensively validated hippocampal tracing protocol [Narr, et al., 2004]. The final AdaBoost classification algorithm was then applied to the full imaging dataset consisting of 484 baseline and 484 follow-up scans. AdaBoost has been previously validated and its segmentation output agrees with human raters as well as human raters agree with each other [Morra, et al., 2008a].
After hippocampal segmentation the left and right hippocampal volumes were computed and retained for future statistical analyses. Hippocampal segmentations were converted into 3D parametric meshes. A medial core (a medial curve that runs through the center) for each hippocampal structure and the radial distance to the medial core from each surface point for each structure were computed [Thompson, et al., 2004]. The 3D hippocampal radial distance maps were used for future statistical analyses.
3. Statistical Methods
We used Analysis of Variance (ANOVA) to compare age, education and MMSE score between the diagnostic groups. A chi-squared test was used to compare the gender distribution between the groups. We used a two-tailed t-test to compare the baseline and follow-up volumes of all MH+/PHany subjects vs. all subjects with negative maternal history of dementia (MH−/PHany) and all subjects with positive PH of dementia (MHany/PH+) vs. all subjects with negative paternal diagnosis of dementia (MHany/PH−), as well as MH+/PH− vs. MH−/PH+ in the pooled sample and in each diagnostic group. Next we constructed multiple linear regression models with hippocampal volume or hippocampal radial distance at 12 months, respectively as the dependent variables and MH, PH as well as ApoE4 genotype (coded as ApoE4 allele present or absent) as predictor variables. Age and hippocampal volume at baseline were included as covariates in the volumetric and radial distance linear regression models. The 3D radial distance regression results were further corrected for multiple comparisons using permutation testing at a predifined threshold of p<0.01. Permutation analyses allow us to derive a single overall corrected p-value for each 3D statistical output based on the number of surface points surviving a particular a priori threshold (0.01 in our analyses). For more details on the permutation approach, please see [Thompson and Apostolova, 2007].
4. Results
The mean group demographic characteristics are presented in Table 1. There were statistically significant differences in age and the prevalence of the ApoE4 allele between the groups. There were no differences in sex, race, education or mean MMSE scores.
Table 1.
Demographic Characteristics
| Variable | MH+/PH− N=149 |
PH+/MH− N=50 |
MH+/PH+ N=18 |
MH+/PH− N=267 |
ANOVA/ Chi- Square p-value |
|---|---|---|---|---|---|
| Age | 74.1 (6.1) | 75.2 (6.8) | 76.3 (6.7) | 74.8 (7.4) | 0.016 |
| Education | 16.1 (2.7) | 15.4 (7.2) | 15.6 (3.1) | 16.5 (2.5) | 0.16 |
| Gender (M:F) | 78:71 | 27:23 | 11:7 | 166:101 | 0.24 |
| Race | 94.6% | 98% | 92.1% | 94.4% | 0.51 |
| ApoE4+, % | 61.1% | 48% | 76.5% | 40.1% | <0.0001 |
|
Diagnostic breakdown |
28% NC 48% MCI 24 % AD |
32 % NC 52% MCI 16 % AD |
17% NC 50% MCI 33% AD |
32% NC 51% MCI 17% AD |
0.4 |
| MMSE | 26.7 (2.6) | 27.0 (2.3) | 27.1 (2.5) | 26.0 (3.3) | 0.17 |
Note: Chi-Squared test for differences in diagnostic breakdown between MH+ and PH+ p=0.5
4.1. Volumetric results
Hippocampal volumetric data is provided in Table 2. In the pooled sample, MH+/PHany subjects had smaller right hippocampal volumes at both baseline (p=0.056; mean absolute difference 3.7 %) and follow-up (p=0.009; mean absolute difference 5.5%), and significantly greater right hippocampal atrophy rate (3.9 vs. 2.1 %, p=0.035) relative to MH−/PHany subjects. MH+/PH− showed trend-level smaller right hippocampal volume at follow-up relative to to MH−/PH− (p=0.057).
Table 2.
Hippocampal volumetric data
| Side | MH+/PH− | PH+/MH− | MH+/PH+ | MH−/PH− | |
|---|---|---|---|---|---|
| Pooled Sample (N) | N=149 | N=50 | N=18 | N=267 | |
| Baseline volume, mm (SD) |
Left | 3787 (725) | 4039(639) | 3928 (693) | 3796 (831) |
| Right | 3572 (776) | 3888 (656) | 3502 (799) | 3669 (760) | |
| Follow-up volume, mm (SD) |
Left | 3654 (775) | 3949 (675) | 3804 (752) | 3708 (852) |
| Right | 3441 (801) | 3796 (682) | 3348 (902) | 3598 (800) | |
| NC (N) | N=41 | N=16 | N=3 | N=85 | |
| Baseline volume, mm (SD) |
Left | 4019 (696) | 4309 (675) | 4557 (487) | 4088 (628) |
| Right | 3933 (552) | 4222 (670) | 3746 (494) | 3946 (709) | |
| Follow-up volume, mm (SD) |
Left | 3909 799) | 4237 (725) | 4363 (640) | 4054 (591) |
| Right | 3922 (536) | 4128 (717) | 3661 (431) | 3971 (697) | |
| MCI (N) | N=72 | N=26 | N=9 | N=136 | |
| Baseline volume, mm (SD) |
Left | 3700 (747) | 4013 (617) | 3765 (765) | 3806 (802) |
| Right | 3422 (774) | 3797 (642) | 3479 (106) | 3620 (743) | |
| Follow-up volume, mm (SD) |
Left | 3586 (790) | 3929 (633) | 3737 (657) | 3721 (789) |
| Right | 3233 (801) | 3714 (664) | 3350 (117) | 3525 (766) | |
| AD (N) | N=36 | N=8 | N=6 | N=46 | |
| Baseline volume, mm (SD) |
Left | 3700 (669) | 3586 (345) | 3858 (737) | 3231 (962) |
| Right | 3465 (878) | 3516 (365) | 3416 (470) | 3303 (725) | |
| Follow-up volume, mm (SD) |
Left | 3500 (656) | 3435 (368) | 3626 (913) | 3027 (103) |
| Right | 3311 (838) | 3402 (364) | 3191 (656) | 3123 (783) |
Relative to MH−/PHany MCI subjects, MH+/PHany MCI subjects had smaller right hippocampal volumes at both baseline (p=0.032; mean absolute difference 6 %) and follow-up (p=0.04; mean absolute difference 9.5%) and significantly greater mean atrophy rate on the right (5.7 vs. 2.7 %, p=0.01). MH+/PH− MCI subjects showed trend-level smaller hippocampal volumes relative to MH−/PH− MCI subjects at baseline (p=0.072), significantly smaller right hippocampal volume in follow-up (p=0.011) and significantly greater right atrophy rate (5.7 vs. 2.8%, p=0.027). In both the pooled sample and the MCI group, the left-sided atrophy rates among MH+/PHany subjects were higher than in MH−/PHany subjects (pooled sample: 3.7% vs. 2.6%; MCI 3% vs. 2.2%) but the group difference failed to reach statistical significance.
In AD, MH+/PHany subjects had generally larger volumes than MH−/PHany subjects. This difference reached statistical significance on the left at both time points (baseline p=0.01; mean absolute difference 9%; at follow-up, p=0.013; mean absolute difference 14 %); however the atrophy rates were not statistically different. When comparing MH+/PH− to MH−/PH− AD subjects the findings remained unchanged.
There were no significant volumetric or atrophy rate differences between MH+/PHany and MH−/PHany or between MH+/PH− and MH−/PH− in the NC group.
MHany/PH+ subjects had consistently larger hippocampal volumes than MHany/PH− subjects across all groups, but the difference reached significance only in the pooled sample on the left (left hippocampal baseline volume p=0.033, mean absolute difference 6.7%; left follow-up volume p=0.036, mean absolute difference 6%). When comparing MH−/PH− to MH−/PH+ subjects, the difference was no longer statistically significant.
When directly comparing MH+/PH− vs. MH−/ PH+ subjects, we found significantly smaller hippocampal volumes at baseline (left p=0.03, mean absolute difference 6.7%; right p=0.011, mean absolute difference 8.8%) and in follow-up (left p=0.017, mean absolute difference 8.1%; right p=0.005, mean absolute difference 10.3%) in the pooled sample. Next we examined these differences in each diagnostic group. The MCI comparison included 72 MH+/PH− and 26 MH−/PH+ MCI subjects. We found significantly smaller right hippocampal volume at baseline (left p=0.06, mean absolute difference 8.5%; right p=0.029, mean absolute difference 11%) and in follow-up (left p=0.049, mean absolute difference 9.6%; right p=0.007, mean absolute difference 14.9%) in MH+/PH− MCI vs. MH−/PH+ MCI subjects. For the NC comparison we had 41 MH+/PH− and 16 MH−/PH+ subjects. While we found smaller hippocampal volumes in MH+/PH− NC vs. MH−/PH+ NC subjects both at baseline and follow-up (baseline mean absolute difference 7.2% on the left and 7.4% on the right; follow-up mean absolute difference 8.4% on the left and 5.3% on the right), these differences failed to reach statistical significance. For the AD comparison we had 36 MH+/PH− and 8 MH−/PH+ AD subjects. MH+/PH− AD subjects had 3.2% larger left and 1.5% smaller right hippocampal volumes at baseline and 1.4% larger left and 2.8% smaller right hippocampal volumes at follow-up relative to MH−/PH+ AD subjects. These differences failed to reach statistical significance, again most likely because the available sample sizes in the subdivided categories were small. MH+/PH− subjects had nonsignificantly greater atrophy rates relative to MH−/PH+ both in the NC (left 3.2% vs. 2%; right 5.6% vs. 2.2%) and in the AD stage (left 5.2% vs. 4.2%; right 4% vs. 3.2%).
In the pooled sample ApoE4+ subjects had significantly smaller hippocampal volume at follow-up relative to ApoE4- subjects (left p=0.01, mean absolute difference 5.2%; right p=0.008, absolute difference 5.5%). ApoE4+ subjects also showed nonsignificantly smaller hippocampal volumes at baseline (left mean absolute difference 2.2%; right mean absolute difference 3.3%). There were no significant differences between baseline and follow-up hippocampal volumes between ApoE4+ and ApoE4- subjects in any diagnostic subgroup. The ApoE4 effects on hippocampal atrophy rates were reported in a previous publication [Morra, et al., 2009].
All regression models were adjusted for age, education and baseline hippocampal volume. After controlling for paternal history of dementia and ApoE4 genotype, postive MH was significantly associated with smaller right hippocampal volume at follow-up in the pooled sample (p=0.04) and in MCI (p=0.007). Positive MH showed no significant association with hippocampal volume at follow-up in NC and AD. Positive PH showed no statistically significant associations with hippocampal volume in the pooled sample or in any of the diagnostic groups.
After controlling for maternal and paternal diagnosis of dementia, ApoE4+ showed significant association with smaller hippocampal volume at follow-up in the pooled sample bilaterally (left p<0.0001; right p=0.005) and on the left in MCI (p=0.04). A trend-level effect was present in AD on the left (p=0.07).
4.2. 3D imaging results
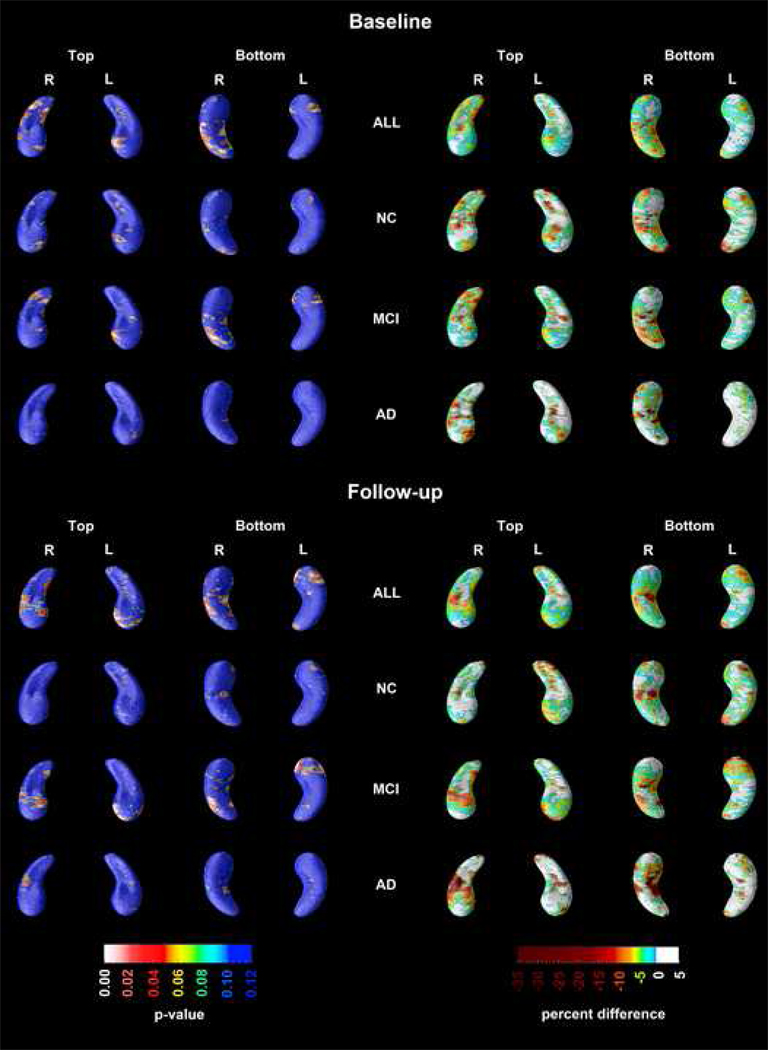
All 3D regression models (see Figure 1) were adjusted for age, education and baseline hippocampal volume. After controlling for PH, ApoE4 genotype, age and baseline hippocampal volume in our linear regression model and applying stringent permutation-based correction for multiple comparisons MH showed a significant negative association with hippocampal radial distance on the left in subjects with MCI while showing a trend-level effect on the right (left pcorrected=0.033; right pcorrected=0.08). In some areas these effects were positive and in others negative. In AD, a significant negative effect of MH was seen on the right only (pcorrected=0.023). There was no effect of MH on hippocampal radial distance in NC. PH did not show a significant effect on hippocampal radial distance in any of the statistical models after controlling for ApoE4 genotype and MH. ApoE4+ had a statistically significant negative effect on hippocampal radial distance independent of dementia history in any parent on the left in MCI (left pcorrected=0.02) and bilaterally in AD (left pcorrected=0.05; right pcorrected=0.043) as well as in the pooled sample (left pcorrected=0.0003; right pcorrected=0.0015). ApoE4 showed no effect on hippocampal radial distance in NC after controlling for age and parental history of dementia (pcorrected>0.4).
Figure 1.
3D maps showing the regionally significant effects of ApoE4 genotype and parental history of AD on hippocampal radial distance (Areas in red and white show significant associations at pcorrected<0.05; areas in blue are nonsignificant, pcorrected>0.1).
Direct comparisons of MH+/PH− vs. MH−/PH+ at baseline and at follow-up are shown in Figure 2. Regionally significant areas showed 10–35% smaller radial distance in MH+/PH− vs. MH−/PH+ in the pooled sample. After permutation-based correction for multiple comparisons, we found significant differences between the groups in the pooled sample on the right at baseline (pcorrected=0.028) and on the left at follow-up (pcorrected=0.048). The only diagnostic subgroup where the direct MH+/PH− vs. MH−/PH+ comparison resulted in significant permutation-corrected findings was MCI at follow-up (left pcorrected=0.03; right pcorrected=0.09). Despite observing regionally significant differences between MH+/PH− and MH−/PH+ in the NC and AD groups these statistical maps did not survive stringent permutation-based correction for multiple comparisons.
Figure 2.
3D comparison of hippocampal radial distance at baseline and in follow-up between MH+ and PH+ subjects in the pooled sample and in each diagnostic group (Areas in red and white show significant associations at pcorrected<0.05; areas in blue are nonsignificant, pcorrected>0.1; % difference =(radial distance at follow-up/radialdistance at baseline)*100).
5. Discussion
A recently published PET study demonstrated significant baseline hypometabolism in the posterior association cortices - known to be affected early in AD - in cognitively normal subjects with maternal history of dementia, while such an effect was absent in those with a paternal history of AD [Mosconi, et al., 2007]. In a follow-up study, the authors demonstrated progressive decline in posterior cortical glucose uptake in those with a maternal history of AD [Mosconi, et al., 2009]. The authors hypothesized that the observed differences may potentially reflect increased AD vulnerability in the offspring of female AD subjects. This theory can be tested only after prospective follow-up of large cohorts of subjects with and without parental history of AD and after assessing progression to AD. Other approaches using pre-existing longitudinal datasets such as the ADNI data as we have done here can examine evidence to support or refute this hypothesis.
Hippocampal atrophy is a well-established early imaging marker of AD [Apostolova, et al., 2006a; Apostolova, et al., 2006b; De Leon, et al., 1997; Henneman, et al., 2009; Jack, et al., 2000; Jack, et al., 1998]. The factors that determine the rate of hippocampal atrophy are not yet completely understood. Recently several computational anatomy techniques have been successfully employed in predicting and tracking AD progression in vivo [Apostolova, et al., 2006b; Apostolova, et al., 2009; Apostolova and Thompson, 2008; Csernansky, et al., 2000; Morra, et al., 2009; Thompson, et al., 2004; Wang, et al., 2003]. Here we used the radial distance mapping technique and conventional volumetric analyses to investigate the effects of MH and PH on hippocampal atrophy in a large ADNI cohort spanning the spectrum from normal aging to mild AD. As hypothesized, MH was associated with greater hippocampal atrophy in both the volumetric and 3D radial distance analyses independently of ApoE4 genotype and PH. As others have reported, we found no effect of maternal history of dementia on hippocampal volume in NC [Debette, et al., 2009; Honea, et al.]. PH showed no effect on hippocampal volume or radial distance. In agreement with previous reports we also found significant independent effects of ApoE4 genotype on hippocampal volume in MCI and AD [Fleisher, et al., 2005; Mueller, et al., 2008; Mueller and Weiner, 2009; van de Pol, et al., 2007].
We observed a negative MH effect in the pooled sample, and in MCI, but not in AD and NC. Others have also found no effect of maternal history of dementia on hippocampal volume in NC [Debette, et al., 2009; Honea, et al.], yet metabolic differences have been seen in the NC stage [Mosconi, et al., 2007] [Mosconi, et al., 2009]. It is now well accepted that metabolic changes generally precede structural ones [Jack, et al., 2010]. The fact that we observe no effect in the AD group may be a true observation but it may also result from the smaller sample size of AD subjects (AD N=96 as opposed to MCI N=243). The latter possibility is also supported by the fact that absolute differences of approximately the same magnitude are present in AD subjects both at baseline and in follow-up, as demonstrated by the percent maps in Figure 2.
Our findings suggest that MH indirectly influences brain structure and that this effect is independent of PH and ApoE4 genotype. This unique predisposition may be mediated by X chromosome [Kehoe, et al., 1999; Zubenko, et al., 1998] or mitochondrial DNA susceptability genes [Mancuso, et al., 2008; Onyango, et al., 2006; Schapira, 2006]. The theory of causative mitochondrial DNA alterations is especially compelling as a decline in mitochondrial respiratory function invariably leads to metabolic disruption, increased generation of reactive oxygen species, enhanced apopotosis [Lin and Beal, 2006], cell loss and brain atrophy all of which occur in AD [Markesbery, 1997].
Although the main effect of MH on hippocampal volume was in the negative direction the 3D maps showed some areas with positive association. 3D hippocampal mapping is an advanced method that allows us to examine disease affects in far greater detail than hippocampal volumetry and has the capability of demonstrating subtle regional effects. Diseases, genetic predisposition, environmental factors, etc., can affect the hippocampal subregions differentially and can show an anterior/posterior effect gradient (reflecting the very complex hippocampal structure and connectivity). Such complex interactions can not be visualized with simple volumetric measures but are easily discernible with more complex imaging protocols.
To our knowledge, this is the first report of parental history affecting hippocampal atrophy in MCI and AD subjects. Several strengths and limitations of our study need to be recognized. The strengths include the selection of a well-established, large, prospectively collected and exceptionally well-characterized longitudinal subject sample for our analyses and the use of an advanced hippocampal analytic technique. Several limitations of the study should be considered. First, in ADNI, parental history of dementia is provided by subject self-report. This ascertainment method is inferior to definite neuropathologic diagnosis, although more practical to perform. Prevalence rates recorded via self-report are frequently lower then the true prevalence due to poor recollection, uncertainty about parental medical history, denial, or data censoring due to early parental death. We chose to use the parental history of dementia variable as opposed to the parental history of AD variable, as in many instances the etiology of parental dementia was never recorded. This likely led to the inclusion of some subjects with parental non-AD dementia such as vascular dementia, dementia with Lewy Bodies or Parkinson's disease dementia. Both limitations may have influenced our analyses by reducing our power to detect the presence or true extent of the parental history effect in AD. It could be argued that focusing on history of AD might give clearer and more interpretable results than combining people at risk for different dementia subtypes, including forms of dementia not so strongly associated with hippocampal atrophy. For example, there might be a grouping bias, as men tend to develop strokes, vascular damage and atherosclerosis more frequently than women, which may lead to PH more frequently reflecting vascular dementia than MH. If this were the case, then the lack of PH effects on hippocampal atrophy may be considered a false negative attributable to groupings based on non-specific risk factors. Even so, the rationale for using “family history of dementia” instead of “family history of AD” was based on the number of subjects with available positive parental history in each diagnostic group. For example, in the MCI category there were 35 subjects with paternal history of dementia but only 16 with paternal history of AD. Similarly there were 81 MCI subjects with maternal history of dementia but only 46 with maternal history of AD. In our center, and perhaps in general, the variable parental diagnosis of AD is marked as positive only when the diagnosis has been pathologically ascertained. As AD is the commonest dementia etiology - found in about 3/4 of dementia subjects postmortem - we would estimate that around 26 out of 35 subjects’ fathers with dementia and 61 out of 81 subjects’ mothers with dementia, would have AD pathology. As such, the “family history of dementia” variable allows us to develop a sample large enough to have sufficient statistical power, based on potentially more reliable information. This is because the level of ascertainment (pathological validation vs. diagnosis rendered by neurologist/geriatric psychiatrist/geriatrician vs. diagnosis rendered by internal medicine/family practice clinician) is likely highly variable for multicenter data.
As only a portion of MCI patients have underlying AD as the cause of their cognitive impairment it remains to be determined if those MCI patients with a maternal history of dementia are more likely to progress to AD. The documented parentally determined difference in AD predisposition in offspring of parents with AD is inetersing and warrants further expoloration.
Acknowledgments
Data used in preparing this article were obtained from the Alzheimer's Disease Neuroimaging Initiative database (www.loni.ucla.edu/ADNI). Many ADNI investigators therefore contributed to the design and implementation of ADNI or provided data but did not participate in the analysis or writing of this report. A complete listing of ADNI investigators is available at www.loni.ucla.edu/ADNI/Collaboration/ADNI_Citation.shtml. All data collection was funded by the following ADNI funding sources (Principal Investigator: Michael Weiner; NIH grant number U01 AG024904): National Institute of Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Foundation for the National Institutes of Health, Pfizer Inc., Wyeth Research, Bristol-Myers Squibb, Eli Lilly and Company, GlaxoSmithKline, Merck & Co. Inc., AstraZeneca AB, Novartis Pharmaceuticals Corporation, the Alzheimer’s Association, Eisai Global Clinical Development, Elan Corporation plc, Forest Laboratories, and the Institute for the Study of Aging (ISOA), with participation from the U.S. Food and Drug Administration. The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. We thank the members of the ADNI Imaging Core for their contributions to the image pre-processing and the ADNI project.
All study analyses were funded by NIA K23 AG026803 (jointly sponsored by NIA, AFAR, The John A. Hartford Foundation, The Atlantic Philanthropies, The Starr Foundation and an anonymous donor; to LGA), the Easton Consortium for Alzheimer Drug Discovery and Biomarker Development (to LGA and JLC), the Turken Foundation (to LGA); NIA P 50 AG16570 (to LGA, JLC and PMT); NIBIB EB01651, NLM LM05639, NCRR RR019771 (to PMT); and NIMH R01 MH071940, NCRR P41 RR013642 and NIH U54 RR021813 (to AWT).
The authors would like to thank Mr. Anthony Ramirez for his help with some of the statistical analyses.
List of important abbreviations
- MH
maternal history
- PH
paternal history
- MH+/PHany
positive maternal history of dementia (including subjects with positive and negative paternal history of dementia)
- MHany/PH+
positive paternal history of dementia (including subjects with positive and negative maternal history of dementia)
- MH−/PHany
negative maternal history of dementia (including subjects with positive and negative paternal history of dementia)
- MHany/PH−
negative paternal history of dementia (including subjects with positive and negative maternal history of dementia)
- MH+/PH+
positive maternal and paternal history of dementia
- MH+/PH−
positive maternal and negative paternal history of dementia
- MH−/PH−
no maternal and paternal history of dementia
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure Statement: The authors have no potential financial or personal conflicts of interest including relationships with other people or organizations within three years of beginning the work submitted that could inappropriately influence their work.
References
- Apostolova LG, Dinov ID, Dutton RA, Hayashi KM, Toga AW, Cummings JL, Thompson PM. 3D comparison of hippocampal atrophy in amnestic mild cognitive impairment and Alzheimer's disease. Brain. 2006a;129:2867–2873. doi: 10.1093/brain/awl274. [DOI] [PubMed] [Google Scholar]
- Apostolova LG, Dutton RA, Dinov ID, Hayashi KM, Toga AW, Cummings JL, Thompson PM. Conversion of mild cognitive impairment to Alzheimer disease predicted by hippocampal atrophy maps. Arch Neurol. 2006b;63:693–699. doi: 10.1001/archneur.63.5.693. [DOI] [PubMed] [Google Scholar]
- Apostolova LG, Mosconi L, Thompson PM, Green AE, Mistur R, Tsui WH, de Leon MJ. Subregional hippocampal atrophy predicts future decline to Alzheimer's dementia in cognitively normal subjects. Neurobiol Aging. 2009 doi: 10.1016/j.neurobiolaging.2008.08.008. in press; doi:10.1016/j.neurobiolaging.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolova LG, Thompson PM. Mapping progressive brain structural changes in early Alzheimer's disease and mild cognitive impairment. Neuropsychologia. 2008;46:1597–1612. doi: 10.1016/j.neuropsychologia.2007.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram L, Tanzi RE. Genome-wide association studies in Alzheimer's disease. Hum Mol Genet. 2009;18:R137–R145. doi: 10.1093/hmg/ddp406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram L, Tanzi RE. Thirty years of Alzheimer's disease genetics: the implications of systematic meta-analyses. Nat Rev Neurosci. 2008;9:768–778. doi: 10.1038/nrn2494. [DOI] [PubMed] [Google Scholar]
- Collins DL, Neelin P, Peters TM, Evans AC. Automatic 3D intersubject registration of MR volumetric data in standardized Talairach space. J Comput Assist Tomogr. 1994;18:192–205. [PubMed] [Google Scholar]
- Csernansky JG, Wang L, Joshi S, Miller JP, Gado M, Kido D, McKeel D, Morris JC, Miller MI. Early DAT is distinguished from aging by high-dimensional mapping of the hippocampus. Dementia of the Alzheimer type. Neurology. 2000;55:1636–1643. doi: 10.1212/wnl.55.11.1636. [DOI] [PubMed] [Google Scholar]
- Cummings JL. Alzheimer's disease. N Engl J Med. 2004;351:56–67. doi: 10.1056/NEJMra040223. [DOI] [PubMed] [Google Scholar]
- Cupples LA, Farrer LA, Sadovnick AD, Relkin N, Whitehouse P, Green RC. Estimating risk curves for first-degree relatives of patients with Alzheimer's disease: the REVEAL study. Genet Med. 2004;6:192–196. doi: 10.1097/01.gim.0000132679.92238.58. [DOI] [PubMed] [Google Scholar]
- De Leon MJ, George AE, Golomb J, Tarshish C, Convit A, Kluger A, De Santi S, McRae T, Ferris SH, Reisberg B, Ince C, Rusinek H, Bobinski M, Quinn B, Miller DC, Wisniewski HM. Frequency of hippocampal formation atrophy in normal aging and Alzheimer's disease. Neurobiol Aging. 1997;18:1–11. doi: 10.1016/s0197-4580(96)00213-8. [DOI] [PubMed] [Google Scholar]
- Debette S, Wolf PA, Beiser A, Au R, Himali JJ, Pikula A, Auerbach S, Decarli C, Seshadri S. Association of parental dementia with cognitive and brain MRI measures in middle-aged adults. Neurology. 2009;73:2071–2078. doi: 10.1212/WNL.0b013e3181c67833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edland SD, Silverman JM, Peskind ER, Tsuang D, Wijsman E, Morris JC. Increased risk of dementia in mothers of Alzheimer's disease cases: evidence for maternal inheritance. Neurology. 1996;47:254–256. doi: 10.1212/wnl.47.1.254. [DOI] [PubMed] [Google Scholar]
- Ehrenkrantz D, Silverman JM, Smith CJ, Birstein S, Marin D, Mohs RC, Davis KL. Genetic epidemiological study of maternal and paternal transmission of Alzheimer's disease. Am J Med Genet. 1999;88:378–382. doi: 10.1002/(sici)1096-8628(19990820)88:4<378::aid-ajmg15>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- Fleisher A, Grundman M, Jack CR, Jr, Petersen RC, Taylor C, Kim HT, Schiller DH, Bagwell V, Sencakova D, Weiner MF, DeCarli C, DeKosky ST, van Dyck CH, Thal LJ. Sex, apolipoprotein E epsilon 4 status, and hippocampal volume in mild cognitive impairment. Arch Neurol. 2005;62:953–957. doi: 10.1001/archneur.62.6.953. [DOI] [PubMed] [Google Scholar]
- Folstein MF, Folstein SE, McHugh PR. Mini-Mental State: a practical method for grading the cogntive state of patients for the clinician. J Psychiatric Res. 1975;12:189–198. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- Green RC, Cupples LA, Go R, Benke KS, Edeki T, Griffith PA, Williams M, Hipps Y, Graff-Radford N, Bachman D, Farrer LA. Risk of dementia among white and African American relatives of patients with Alzheimer disease. JAMA. 2002;287:329–336. doi: 10.1001/jama.287.3.329. [DOI] [PubMed] [Google Scholar]
- Hebert LE, Scherr PA, Bienias JL, Bennett DA, Evans DA. Alzheimer disease in the US population: prevalence estimates using the 2000 census. Arch Neurol. 2003;60:1119–1122. doi: 10.1001/archneur.60.8.1119. [DOI] [PubMed] [Google Scholar]
- Henneman WJ, Sluimer JD, Barnes J, van der Flier WM, Sluimer IC, Fox NC, Scheltens P, Vrenken H, Barkhof F. Hippocampal atrophy rates in Alzheimer disease: added value over whole brain volume measures. Neurology. 2009;72:999–1007. doi: 10.1212/01.wnl.0000344568.09360.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honea RA, Swerdlow RH, Vidoni ED, Goodwin J, Burns JM. Reduced gray matter volume in normal adults with a maternal family history of Alzheimer disease. Neurology. 74:113–120. doi: 10.1212/WNL.0b013e3181c918cb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Jr, Bernstein MA, Fox NC, Thompson P, Alexander G, Harvey D, Borowski B, Britson PJ, J L W, Ward C, Dale AM, Felmlee JP, Gunter JL, Hill DL, Killiany R, Schuff N, Fox-Bosetti S, Lin C, Studholme C, DeCarli CS, Krueger G, Ward HA, Metzger GJ, Scott KT, Mallozzi R, Blezek D, Levy J, Debbins JP, Fleisher AS, Albert M, Green R, Bartzokis G, Glover G, Mugler J, Weiner MW. The Alzheimer's Disease Neuroimaging Initiative (ADNI): MRI methods. J Magn Reson Imaging. 2008;27:685–691. doi: 10.1002/jmri.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Jr, Knopman DS, Jagust WJ, Shaw LM, Aisen PS, Weiner MW, Petersen RC, Trojanowski JQ. Hypothetical model of dynamic biomarkers of the Alzheimer's pathological cascade. Lancet Neurol. 2010;9:119–128. doi: 10.1016/S1474-4422(09)70299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Jr, Petersen RC, Xu Y, O'Brien PC, Smith GE, Ivnik RJ, Boeve BF, Tangalos EG, Kokmen E. Rates of hippocampal atrophy correlate with change in clinical status in aging and AD. Neurology. 2000;55:484–489. doi: 10.1212/wnl.55.4.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Jr, Petersen RC, Xu Y, O'Brien PC, Smith GE, Ivnik RJ, Tangalos EG, Kokmen E. Rate of medial temporal lobe atrophy in typical aging and Alzheimer's disease. Neurology. 1998;51:993–999. doi: 10.1212/wnl.51.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovicich J, Czanner S, Greve D, Haley E, van der Kouwe A, Gollub R, Kennedy D, Schmitt F, Brown G, Macfall J, Fischl B, Dale A. Reliability in multi-site structural MRI studies: effects of gradient non-linearity correction on phantom and human data. Neuroimage. 2006;30:436–443. doi: 10.1016/j.neuroimage.2005.09.046. [DOI] [PubMed] [Google Scholar]
- Kehoe P, Wavrant-De Vrieze F, Crook R, Wu WS, Holmans P, Fenton I, Spurlock G, Norton N, Williams H, Williams N, Lovestone S, Perez-Tur J, Hutton M, Chartier-Harlin MC, Shears S, Roehl K, Booth J, Van Voorst W, Ramic D, Williams J, Goate A, Hardy J, Owen MJ. A full genome scan for late onset Alzheimer's disease. Hum Mol Genet. 1999;8:237–245. doi: 10.1093/hmg/8.2.237. [DOI] [PubMed] [Google Scholar]
- Leow AD, Klunder AD, Jack CR, Jr, Toga AW, Dale AM, Bernstein MA, Britson PJ, Gunter JL, Ward CP, Whitwell JL, Borowski BJ, Fleisher AS, Fox NC, Harvey D, Kornak J, Schuff N, Studholme C, Alexander GE, Weiner MW, Thompson PM. Longitudinal stability of MRI for mapping brain change using tensor-based morphometry. Neuroimage. 2006;31:627–640. doi: 10.1016/j.neuroimage.2005.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Grupe A, Rowland C, Nowotny P, Kauwe JS, Smemo S, Hinrichs A, Tacey K, Toombs TA, Kwok S, Catanese J, White TJ, Maxwell TJ, Hollingworth P, Abraham R, Rubinsztein DC, Brayne C, Wavrant-De Vrieze F, Hardy J, O'Donovan M, Lovestone S, Morris JC, Thal LJ, Owen M, Williams J, Goate A. DAPK1 variants are associated with Alzheimer's disease and allele-specific expression. Hum Mol Genet. 2006;15:2560–2568. doi: 10.1093/hmg/ddl178. [DOI] [PubMed] [Google Scholar]
- Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- Mancuso M, Orsucci D, Siciliano G, Murri L. Mitochondria, mitochondrial DNA and Alzheimer's disease. What comes first? Curr Alzheimer Res. 2008;5:457–468. doi: 10.2174/156720508785908946. [DOI] [PubMed] [Google Scholar]
- Markesbery WR. Oxidative stress hypothesis in Alzheimer's disease. Free Radic Biol Med. 1997;23:134–147. doi: 10.1016/s0891-5849(96)00629-6. [DOI] [PubMed] [Google Scholar]
- Mazziotta J, Toga A, Evans A, Fox P, Lancaster J, Zilles K, Woods R, Paus T, Simpson G, Pike B, Holmes C, Collins L, Thompson P, MacDonald D, Iacoboni M, Schormann T, Amunts K, Palomero-Gallagher N, Geyer S, Parsons L, Narr K, Kabani N, Le Goualher G, Boomsma D, Cannon T, Kawashima R, Mazoyer B. A probabilistic atlas and reference system for the human brain: International Consortium for Brain Mapping (ICBM) Philos Trans R Soc Lond B Biol Sci. 2001;356:1293–1322. doi: 10.1098/rstb.2001.0915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology. 1984;34:939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- Morra JH, Tu Z, Apostolova LG, Green AE, Avedissian C, Madsen SK, Parikshak N, Hua X, Toga AW, Jack CR, Jr, Weiner MW, Thompson PM. Validation of a fully automated 3D hippocampal segmentation method using subjects with Alzheimer's disease mild cognitive impairment, and elderly controls. Neuroimage. 2008a;43:59–68. doi: 10.1016/j.neuroimage.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morra JH, Tu Z, Apostolova LG, Green AE, Avedissian C, Madsen SK, Parikshak N, Hua X, Toga AW, Jack CR, Schuff N, Weiner MW, Thompson PM. Mapping hippocampal degeneration in 400 subjects with a novel automated segmentation approach. ISBI. 2008b in press. [Google Scholar]
- Morra JH, Tu Z, Apostolova LG, Green AE, Avedissian C, Madsen SK, Parikshak N, Toga AW, Jack CR, Jr, Schuff N, Weiner MW, Thompson PM. Automated mapping of hippocampal atrophy in 1-year repeat MRI data from 490 subjects with Alzheimer's disease, mild cognitive impairment, and elderly controls. Neuroimage. 2009;45:S3–S15. doi: 10.1016/j.neuroimage.2008.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JC. The Clinical Dementia Rating (CDR): current version and scoring rules. Neurology. 1993;43:2412–2414. doi: 10.1212/wnl.43.11.2412-a. [DOI] [PubMed] [Google Scholar]
- Mosconi L, Brys M, Switalski R, Mistur R, Glodzik L, Pirraglia E, Tsui W, De Santi S, de Leon MJ. Maternal family history of Alzheimer's disease predisposes to reduced brain glucose metabolism. Proc Natl Acad Sci U S A. 2007;104:19067–19072. doi: 10.1073/pnas.0705036104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosconi L, Mistur R, Switalski R, Brys M, Glodzik L, Rich K, Pirraglia E, Tsui W, De Santi S, de Leon MJ. Declining brain glucose metabolism in normal individuals with a maternal history of Alzheimer disease. Neurology. 2009;72:513–520. doi: 10.1212/01.wnl.0000333247.51383.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller SG, Schuff N, Raptentsetsang S, Elman J, Weiner MW. Selective effect of Apo e4 on CA3 and dentate in normal aging and Alzheimer's disease using high resolution MRI at 4 T. Neuroimage. 2008;42:42–48. doi: 10.1016/j.neuroimage.2008.04.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller SG, Weiner MW. Selective effect of age, Apo e4, and Alzheimer's disease on hippocampal subfields. Hippocampus. 2009;19:558–564. doi: 10.1002/hipo.20614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller SG, Weiner MW, Thal LJ, Petersen RC, Jack C, Jagust W, Trojanowski JQ, Toga AW, Beckett L. The Alzheimer's disease neuroimaging initiative. Neuroimaging Clin N Am. 2005;15:869–877. xi–xii. doi: 10.1016/j.nic.2005.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narr KL, Thompson PM, Szeszko P, Robinson D, Jang S, Woods RP, Kim S, Hayashi KM, Asunction D, Toga AW, Bilder RM. Regional specificity of hippocampal volume reductions in first-episode schizophrenia. Neuroimage. 2004;21:1563–1575. doi: 10.1016/j.neuroimage.2003.11.011. [DOI] [PubMed] [Google Scholar]
- Onyango I, Khan S, Miller B, Swerdlow R, Trimmer P, Bennett P., Jr Mitochondrial genomic contribution to mitochondrial dysfunction in Alzheimer's disease. J Alzheimers Dis. 2006;9:183–193. doi: 10.3233/jad-2006-9210. [DOI] [PubMed] [Google Scholar]
- Rogaeva E, Meng Y, Lee JH, Gu Y, Kawarai T, Zou F, Katayama T, Baldwin CT, Cheng R, Hasegawa H, Chen F, Shibata N, Lunetta KL, Pardossi-Piquard R, Bohm C, Wakutani Y, Cupples LA, Cuenco KT, Green RC, Pinessi L, Rainero I, Sorbi S, Bruni A, Duara R, Friedland RP, Inzelberg R, Hampe W, Bujo H, Song YQ, Andersen OM, Willnow TE, Graff-Radford N, Petersen RC, Dickson D, Der SD, Fraser PE, Schmitt-Ulms G, Younkin S, Mayeux R, Farrer LA, St George-Hyslop P. The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer disease. Nat Genet. 2007;39:168–177. doi: 10.1038/ng1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schapira AH. Mitochondrial disease. Lancet. 2006;368:70–82. doi: 10.1016/S0140-6736(06)68970-8. [DOI] [PubMed] [Google Scholar]
- Schapire R, Freund Y. Boosting the margin: a new explanation for the effectiveness of voting methods. Annals of Statistics. 1998;26:1651–1686. [Google Scholar]
- Seripa D, Panza F, Franceschi M, D'Onofrio G, Solfrizzi V, Dallapiccola B, Pilotto A. Non-apolipoprotein E and apolipoprotein E genetics of sporadic Alzheimer's disease. Ageing Res Rev. 2009;8:214–236. doi: 10.1016/j.arr.2008.12.003. [DOI] [PubMed] [Google Scholar]
- Silverman JM, Ciresi G, Smith CJ, Marin DB, Schnaider-Beeri M. Variability of familial risk of Alzheimer disease across the late life span. Arch Gen Psychiatry. 2005;62:565–573. doi: 10.1001/archpsyc.62.5.565. [DOI] [PubMed] [Google Scholar]
- Sled JG, Zijdenbos AP, Evans AC. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imaging. 1998;17:87–97. doi: 10.1109/42.668698. [DOI] [PubMed] [Google Scholar]
- Slooter AJ, Cruts M, Kalmijn S, Hofman A, Breteler MM, Van Broeckhoven C, van Duijn CM. Risk estimates of dementia by apolipoprotein E genotypes from a population-based incidence study: the Rotterdam Study. Arch Neurol. 1998;55:964–968. doi: 10.1001/archneur.55.7.964. [DOI] [PubMed] [Google Scholar]
- Sundar PD, Feingold E, Minster RL, DeKosky ST, Kamboh MI. Gender-specific association of ATP-binding cassette transporter 1 (ABCA1) polymorphisms with the risk of late-onset Alzheimer's disease. Neurobiol Aging. 2007;28:856–862. doi: 10.1016/j.neurobiolaging.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Tanzi RE, Bertram L. New frontiers in Alzheimer's disease genetics. Neuron. 2001;32:181–184. doi: 10.1016/s0896-6273(01)00476-7. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Apostolova LG. Computational anatomical methods as applied to ageing and dementia. Br J Radiol. 2007;80(Spec No 2):S78–S91. doi: 10.1259/BJR/20005470. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Hayashi KM, De Zubicaray GI, Janke AL, Rose SE, Semple J, Hong MS, Herman DH, Gravano D, Doddrell DM, Toga AW. Mapping hippocampal and ventricular change in Alzheimer disease. Neuroimage. 2004;22:1754–1766. doi: 10.1016/j.neuroimage.2004.03.040. [DOI] [PubMed] [Google Scholar]
- van de Pol LA, van der Flier WM, Korf ES, Fox NC, Barkhof F, Scheltens P. Baseline predictors of rates of hippocampal atrophy in mild cognitive impairment. Neurology. 2007;69:1491–1497. doi: 10.1212/01.wnl.0000277458.26846.96. [DOI] [PubMed] [Google Scholar]
- Wang L, Swank JS, Glick IE, Gado MH, Miller MI, Morris JC, Csernansky JG. Changes in hippocampal volume and shape across time distinguish dementia of the Alzheimer type from healthy aging. Neuroimage. 2003;20:667–682. doi: 10.1016/S1053-8119(03)00361-6. [DOI] [PubMed] [Google Scholar]
- Wechsler D. Wechsler Memory Scale - Revised. San Antonio, TX: The Psycholocical Corporation; 1987. [Google Scholar]
- Wimo A, Winblad B. Health economical aspects of Alzheimer disease and its treatment. Psychogeriatrics. 2001;1:189–193. [Google Scholar]
- Wolf P, Beiser A, Au R, Auerbach S, DeCarli C. Parental Occurrence of Dementia Linked to Lower Cognitive Function in the Framingham Offspring Study. Neurology. 2005;64 Suppl 1:A267–A268. [Google Scholar]
- Zubenko GS, Stiffler JS, Hughes HB, Hurtt MR, Kaplan BB. Initial results of a genome survey for novel Alzheimer's disease risk genes: association with a locus on the X chromosome. Am J Med Genet. 1998;81:196–205. [PubMed] [Google Scholar]