Abstract
Chemokine receptors direct T lymphocytes to the site of an infection by following coordinated chemokine gradients, which allow their recruitment to specific tissues. Although identification of receptors needed for homing to some mucosal sites, such as skin and gut, have been elucidated, the receptors that direct lymphocytes to the genital mucosa remain relatively uncharacterized. Here we identify that the chemokine receptors CXCR3 and CCR5 are pivotal for T lymphocyte access to the genital tract during Chlamydia trachomatis infection. Chlamydia specific CD4+ transgenic T cells that lack CXCR3 or CCR5 do not accumulate in the genital mucosa following infection. Loss of either CXCR3 or CCR5 impairs the protective capacity of Chlamydia specific T cells, while T cells lacking both receptors are completely non-protective. These results show that CXCR3 and CCR5 are the predominant chemokine receptors, which act cooperatively to promote homing to the genital mucosa during Chlamydia infection.
Keywords: Chlamydia, T cell trafficking, Chemokine receptors
Introduction
The ability of a host to respond to and clear infections depends on coordinated communication between the innate and adaptive immune responses. Infection stimulates innate immune sensing mechanisms, such as Toll-like receptors that secrete specific chemokines into the tissues1. Activated T cells follow these chemokine gradients by upregulating the expression of distinct chemokine receptors in order to exit the lymphatic system and traffic to the site of infection2.
Several previous reports have identified dominant chemokine receptors that are expressed by T cells in order for proper migration to mucosal tissues. For example, CCR9 is critical for recruitment to the intestinal tract under many conditions3, while CXCR3 directs T cell migration to the lungs following influenza virus infections4, 5. Despite the increase in concern over the public health impact of sexually transmitted infections (STIs)6, homing receptors for the genitourinary tract remain largely undefined. The most prevalent genital tract bacterial infection is caused by the obligate intracellular pathogen Chlamydia trachomatis. Clearance of C. trachomatis is primarily dependent on T cell secretion of IFNγ, which directly and indirectly targets the infected epithelial cells7–15. However, the immune response to C. trachomatis is also responsible for tissue damage and the induction of sequelae such as infertility and ectopic pregnancy7, 16. Understanding the trafficking of immune cells following an STI is key to the development of therapeutic interventions that lead to protection and prevent pathology.
In order to define how T cells respond to C. trachomatis, it is critical to elucidate what chemokine/chemokine receptor pairs promote proper activation and recruitment of T cells during infection. Reports examining the inflammatory conditions in the genital mucosa of mice following infection with the mouse-adapted species Chlamydia muridarum showed that the chemokines CCL5 and CXCL10 are upregulated and secreted at high levels from cells in the tissues17–19. Another study examined samples from human patients and demonstrated an increase in CCR5 expression following C. trachomatis genital infections20, while a different report showed mice deficient in CCR5 were unable to clear C. muridarum infections21. Even though these data provide insight into expression dynamics following infection, they do not directly reveal which subsets of immune cells, such as T cells, are activated and recruited to the genital mucosa through the action of chemokine receptors. Furthermore, these studies are unable to distinguish the response of C. trachomatis specific lymphocytes from infiltrating bystander lymphocytes that respond in a non-specific manner to the general inflammatory conditions in the genital mucosa. In this study, we monitored CD4+ C. trachomatis-specific T cells which harbor a fixed TCR specific for the C. trachomatis antigen Cta112. We were able to directly assess the impact of two chemokine receptors on the ability of Chlamydia-specific T cells to become activated and respond during infection of the genital tract. We find that concomitant expression of CXCR3 and CCR5 expression is required for antigen specific T cells to access, accumulate, and protect the genital tracts of mice from infection with C. trachomatis.
Results
CXCR3 and CCR5 are upregulated on lymphocytes during C. trachomatis infection
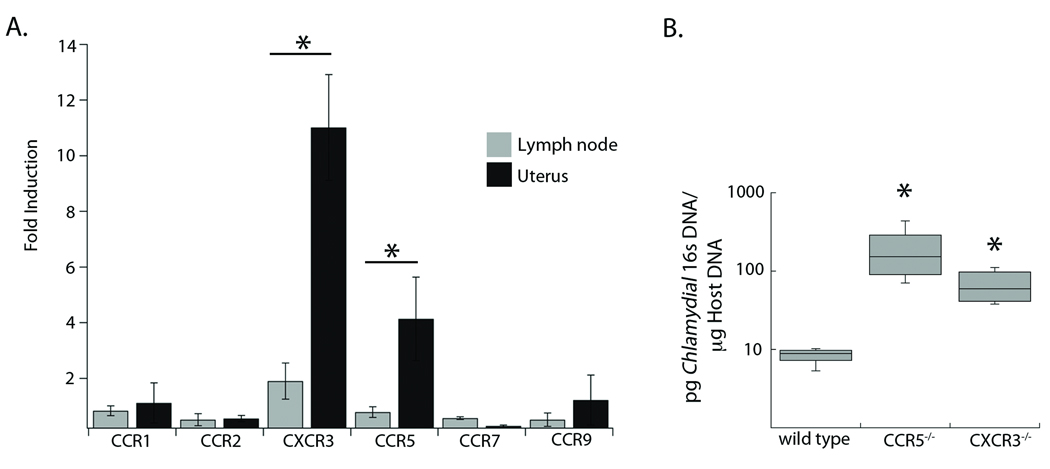
Previous studies have examined chemokines and chemokine receptors that are induced in whole tissue homogenates of the genital tract following C. trachomatis infection. Yet these studies could not differentiate non-specific homing, resulting from general inflammation, from the truly pathogen-specific lymphocytes responding to the genital mucosa. To determine which receptors are required for lymphocyte homing to the genital mucosa, we surveyed chemokine receptor mRNA expression in isolated lymphocytes from naïve vs. C. trachomatis infected mice. We extracted lymphocyte RNA isolated from the draining lymph node or genital tract in naïve mice or from mice infected in the uterus with 106 C. trachomatis. Using RT-PCR we examined the differences in expression of five different chemokine receptors on tissues from uninfected and infected mice. Analysis of the draining lymph node showed that only CXCR3 expression was increased following infection, showing a two-fold enhancement (Figure 1A). In contrast, the lymphocytes isolated from the genital tract showed that expression of both CXCR3 and CCR5 was significantly induced following C. trachomatis infection. Expression of CXCR3 was increased more than ten-fold after infection, while CCR5 was upregulated by more than four-fold. In order for lymphocytes to exit the lymph node, they down-regulate CCR7, consistent with our observation that the level of this marker decreased on lymphocytes in both the draining lymph node and genital tract of infected mice22. All other chemokine receptors examined showed no changes in expression levels following infection with C. trachomatis. Therefore, the expression of the chemokine receptors CXCR3 and CCR5 is upregulated in lymphocytes in response to a genital tract infection with C. trachomatis.
Figure 1. CXCR3 and CCR5 are upregulated in the genital tract during infection and necessary for clearance of C. trachomatis.
A) RNA was isolated from tissues of uninfected and infected mice. RT-PCR was used to quantify the expression of five chemokine receptors relative to a GAPDH control. Fold-induction was calculated (relative expression infected)/(relative expression uninfected). The data shown are representative of three independent experiments. Each bar represents the average fold-induction ± SD of two pools of five mice for each condition B) Genomic DNA was isolated from the genital tract of wild-type, CXCR3−/−, or CCR5−/− mice six days following infection. Quantitative PCR was used to calculate the levels of C. trachomatis 16s DNA relative to levels of host GAPDH. Shown is a box and whisker plot of two combined experiments each conducted with three mice per genotype. * = p<0.05
Mice deficient in CXCR3 or CCR5 have increased C. trachomatis burden following genital tract infection
The increase in the expression of CXCR3 and CCR5 on lymphocytes following genital tract infection with C. trachomatis suggested that both receptors might be important for the resolution of infection. We therefore assessed the efficiency of C. trachomatis clearance from the genital tracts of mice deficient in either CXCR3 or CCR5 as compared to wild-type mice. Wild-type, CXCR3−/−, and CCR5−/− mice were infected in the uterus with 106 IFU of C. trachomatis, and levels of bacteria in the genital tract were evaluated six days later by qPCR (Figure 1B)12. All wild-type mice had under 10 pg of bacterial 16s DNA per µg of host DNA, whereas both the CXCR3−/− and CCR5−/− mice had almost 8-fold higher levels of bacterial 16s DNA in the genital tract. Since there was no difference in bacterial load between CXCR3−/− or CCR5−/− mice, it suggests that both receptors may be needed for the proper resolution of genital tract infection with Chlamydia.
Chlamydia specific CD4+ cells become activated and traffic normally to the genital tract in chemokine receptor-deficient hosts
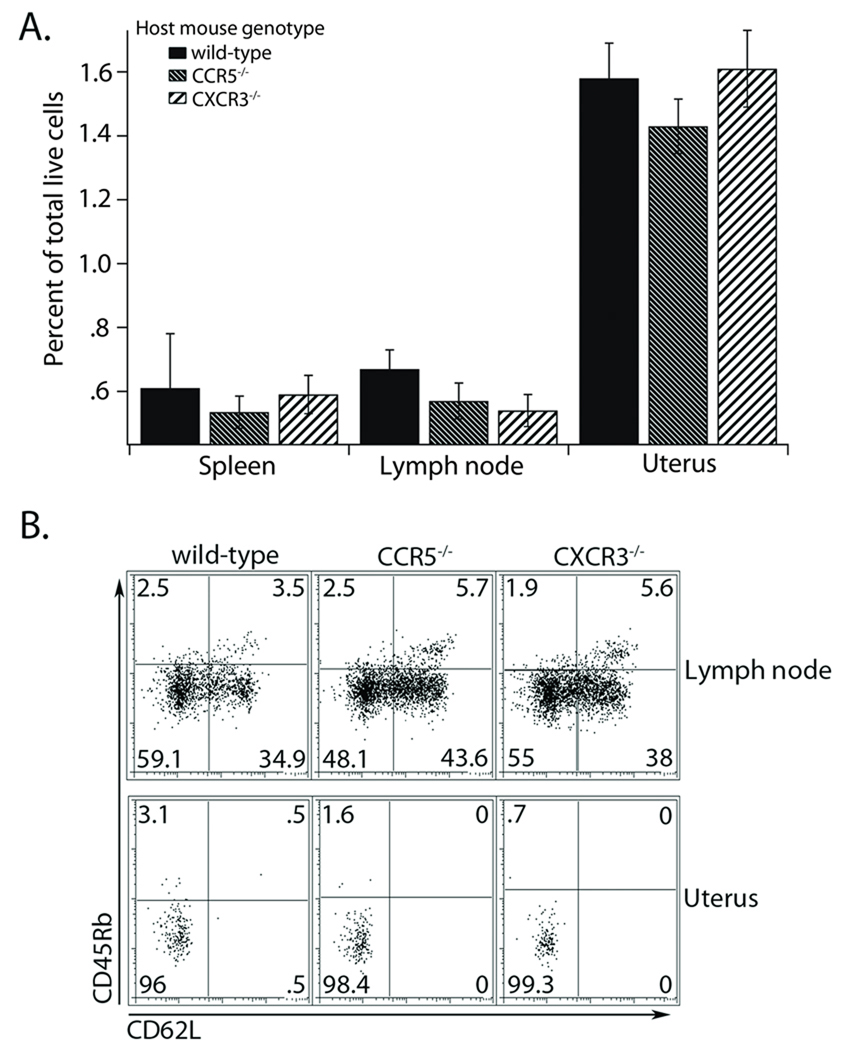
The high bacterial loads observed in the CXCR3 and CCR5 deficient mice could be due to an inability to properly prime an immune response 23, 24. To test this, we monitored wild-type Chlamydia specific TCR transgenic T cells during infection of wild-type, CXCR3−/−, and CCR5−/− host mice inoculated with C. trachomatis. Seven days after infection, we found that wild-type pathogen-specific T cells showed no defect in trafficking to the spleen, draining lymph nodes, or the genital tract in hosts deficient for CXCR3 or CCR5 (Figure 2A). Thus, the recruitment and/or retention of pathogen specific T cells in the genital tract is independent of chemokine receptor expression in the host. To ensure that the Chlamydia specific T cells were activated normally, we stained the isolated T cells for CD62L, CD45Rb, and CD25 as markers of early and late activation (Figure 2B, and data not shown). When we compared cells isolated from mice expressing the chemokine receptors with those lacking expression, we found that the cells showed identical activation. Furthermore, these cells also secrete IFNγ to similar levels upon restimulation in vitro (data not shown). Most importantly, all the Chlamydia-specific T cells that were present in the genital tract were completely activated, indicating that even when the host bystander population is deficient in CXCR3 or CCR5 there is no deficiency in the priming of Chlamydia specific T cells.
Figure 2. Wild-type Chlamydia specific CD4+ T cells are recruited to the genital tract and become activated in CXCR3 or CCR5 deficient mice.
A) Wild-type CD90.1+ transgenic CD4+ T cells were transferred into CD90.2 wild-type, CXCR3−/−, or CCR5−/− host mice. The following day, hosts were challenged in the genital tract with C. trachomatis. Seven days after infection the tissues were harvested and cells prepared for flow cytometry. Antigen specific CD4+ T cells were identified as both CD4 and CD90.1 positive. The graph shows the percent of total live cells in each tissue examined and is representative of one experiment of two conducted. B) Surface levels of the activation markers CD45Rb and CD62L were examined by flow cytometry. All cells were pre-gated on CD4 and CD90.1 positive T cells. A representative plot from one of two independent experiments is shown.
Both CXCR3 and CCR5 are upregulated on Chlamydia specific T cells present in the genital mucosa
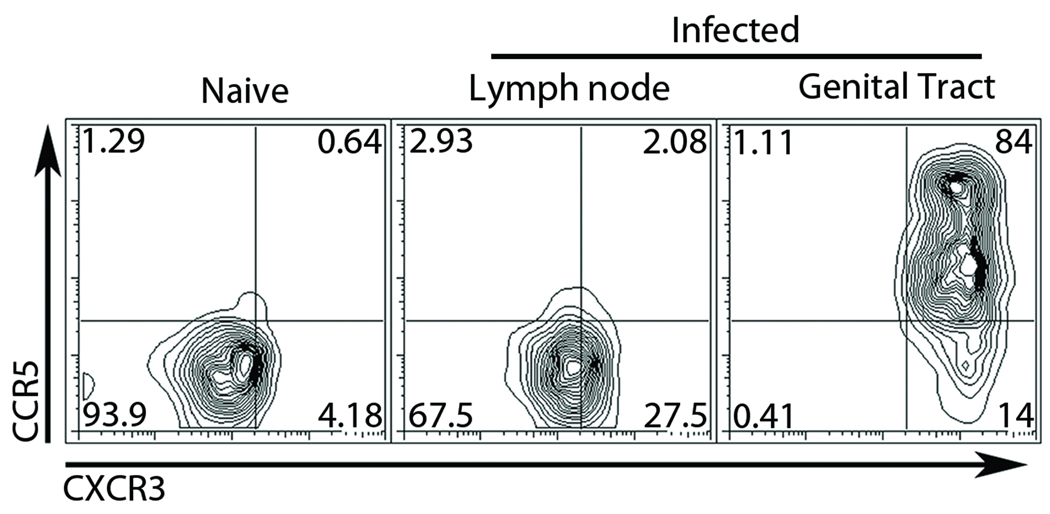
Because host deficiency in CXCR3 or CCR5 did not alter the priming or homing of wild type antigen specific T cells, we hypothesized that T cells responding to C. trachomatis antigens in the genital tract might be enriched for CXCR3 and/or CCR5. To test this, we examined the presence of CXCR3 and CCR5 on the surface of antigen specific transgenic T cells during infection. Naïve Chlamydia specific T cells were isolated from TCR transgenic mice and stained for CXCR3 and CCR5 (Figure 3). All naïve cells examined expressed low levels of both CXCR3 and CCR5. We then transferred these antigen specific T cells into mice that were infected in the uterus with C. trachomatis. Seven days after infection we isolated lymphocytes from the draining lymph nodes and genital mucosa and stained them for CXCR3 and CCR5. Chlamydia specific T cells from the lymph nodes were more than twenty percent positive for CXCR3 while all cells remained low for CCR5. However, almost all of the antigen specific T cells isolated from the genital tract were CXCR3 high and over 80 percent were now CCR5 positive. We observed two distinct CCR5 positive subsets in the genital tract, but the significance and phenotypic difference of these distinct populations is still unclear. These data suggest that in response to infection Chlamydia specific T-cells upregulate CXCR3 on their surface beginning in the draining lymph node. Upon exiting the lymph node, CXCR3 is further upregulated, and CCR5 now becomes expressed on the surface at high levels, resulting in their recruitment to or their retention in the genital tract.
Figure 3. CXCR3 and CCR5 are upregulated on the surface of antigen specific CD4+ T cells in a stepwise fashion following C. trachomatis infection.
Wild-type CD90.1 Chlamydia specific CD4+ T cells were transferred intravenously into CD90.2 hosts. Mice were challenged in the genital tract with C. trachomatis the following day. Seven days after infection the tissues were isolated and analyzed by flow cytometry. Chlamydia antigen specific cells were identified by CD4+CD90.1+ gating and assessed for surface expression of CXCR3 and CCR5 Shown is a representative plot from three independent experiments.
Chlamydia specific CD4+ T cells deficient in CXCR3 or CCR5 are unable to traffic normally to the genital tract
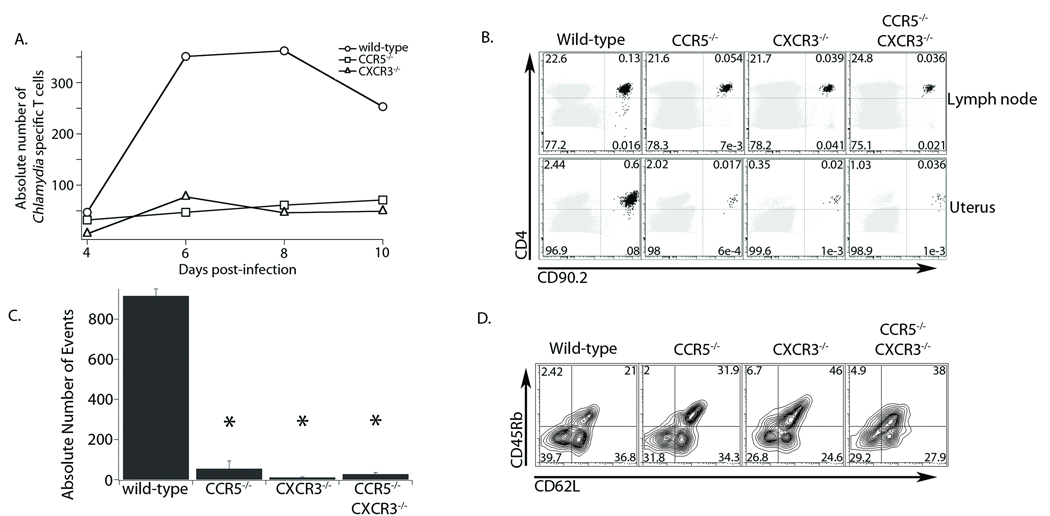
As described above we found that CXCR3 and CCR5 are upregulated on C. trachomatis specific T cells in response to genital tract infection and that pathogen specific T cells activate normally in CXCR3−/− or CCR5−/− hosts. This led us to test whether the high C. trachomatis burden seen in mice deficient in CXCR3 or CCR5 is due to an inability of pathogen specific T cells to traffic to the genital mucosa during infection. We obtained Chlamydia specific T cells that are deficient in CXCR3 or CCR5 and monitored their trafficking in mice during C. trachomatis genital tract infection. We transferred 105 wild-type, CXCR3−/−, or CCR5−/− antigen specific T cells into CD90.1 wild-type hosts. The mice were then infected with C. trachomatis and the number of Chlamydia specific T cells in the genital tract was assessed at four time points (Figure 4A). Four days after infection there was no significant trafficking of any T cells into the genital tract. However, by six days following infection, wild-type Chlamydia specific T cells amassed in the genital tract, while cells from either CXCR3−/− or CCR5−/− mice failed to migrate to the site of infection. Eight days after infection the recruitment of wild-type cells to the genital tract continued to increase, whereas cells from CXCR3−/− and CCR5−/− mice showed no accumulation. Ten days following infection, when C. trachomatis is mostly cleared (data not shown), the number of wild-type cells in the genital tract began to contract, whereas pathogen specific T cells from the chemokine receptor deficient mice showed no signs of trafficking to the genital tract. These data illustrate that lack of either CXCR3 or CCR5 does not delay recruitment to the genital tract, rather it completely prevents homing to the site of infection.
Figure 4. Chlamydia specific CD4+ cells lacking CXCR3 and/or CCR5 are unable to traffic to the genital mucosa.
A) Wild-type, CXCR3−/− or CCR5−/− CD90.2+ Chlamydia specific T cells were transferred into CD90.1+ host mice. Mice were challenged the following day with C. trachomatis in the genital tract. At the timepoints indicated, the genital tract was harvested and cells prepared for flow cytometry. The total number of antigen specific T cells was also quantified. Each data point represents the total number of cells from three mice for each genotype. The figure represents one experiment of two conducted. B) CD90.2+ wild-type, CXCR3−/−, CCR5−/−, or CXCR3−/− CCR5−/− transgenic CD4+ T cells were transferred into CD90.1+ host mice and infected the following day with C. trachomatis. Eight days following infection the tissues were harvested and prepared for flow cytometry. Shown is a representative cytometry plot from one experiment of three conducted of the draining lymph node (top panel) and genital tract (bottom panel). The host CD4+ T cell population is shown in grey and the antigen specific CD4+ T cells are shown in black. The values presented are a percent of total live cells. C) Absolute total numbers of Chlamydia specific CD4+ T cells in the genital tract were quantified from the experiment in B. D) Cells isolated from the draining lymph node were pre-gated for CD90.2+ and CD4+ positive populations and examined for surface staining of the activation markers CD45Rb and CD62L. Shown is a representative plot from one of two independent experiments. *=p<0.05
Since our time-course data suggested that the loss of either CXCR3 or CCR5 blocks recruitment to the genital tract, we next wanted to assess whether a loss of both CXCR3 and CCR5 would further impede homing to the genital tract. We bred the TCR transgenic onto the CXCR3/CCR5 double knockout mouse background. We then transferred these antigen specific CXCR3−/− CCR5−/− T cells (as well as wild-type, CXCR3-deficient, and CCR5-deficient T cells) into CD90.1 wild-type hosts that were then infected with C. trachomatis. Eight days following infection, a time when we found the greatest difference in homing between wild-type and receptor-deficient cells, Chlamydia specific cells of all four genotypes accumulated equivalently in the draining lymph nodes of infected mice (Figure 4B). However, in the genital tract, wild-type cells accumulated at levels more than twenty-fold higher than chemokine receptor-deficient cells (Figure 4B,C). All variants of the chemokine receptor-deficient Chlamydia specific T cells were unable to home correctly to the genital tract following infection, and the double knockout phenotype was identical to that of either single knockout. These data show that both CXCR3 and CCR5 are needed to properly direct antigen specific T cells following C. trachomatis infection in the genital mucosa.
While these data suggest that CD4+ cells deficient for CXCR3 or CCR5 are not recruited efficiently to the genital tract following infection, we wanted to confirm they were activated normally in order to rule out any deficiency in T cell priming. We assessed the levels of CD62L and CD45Rb on all of the chemokine receptor-deficient Chlamydia specific T cells in the draining lymph node eight days after infection. Surface staining showed no significant difference in activation pattern between the chemokine receptor-deficient T cells and wild-type T cells even though there is a slight decrease in activation of chemokine receptor deficient T cells (Figure 4D). Taken together, loss of CXCR3, CCR5, or both severely inhibits trafficking of pathogen specific T cells to the genital mucosa even though these cells accumulate equivalently in the draining lymph node and are activated similarly to wild-type T cells.
Chlamydia-specific CD4+ cells deficient for CCR5 or CXCR3 are unable to compete with WT cells in trafficking to the genital tract
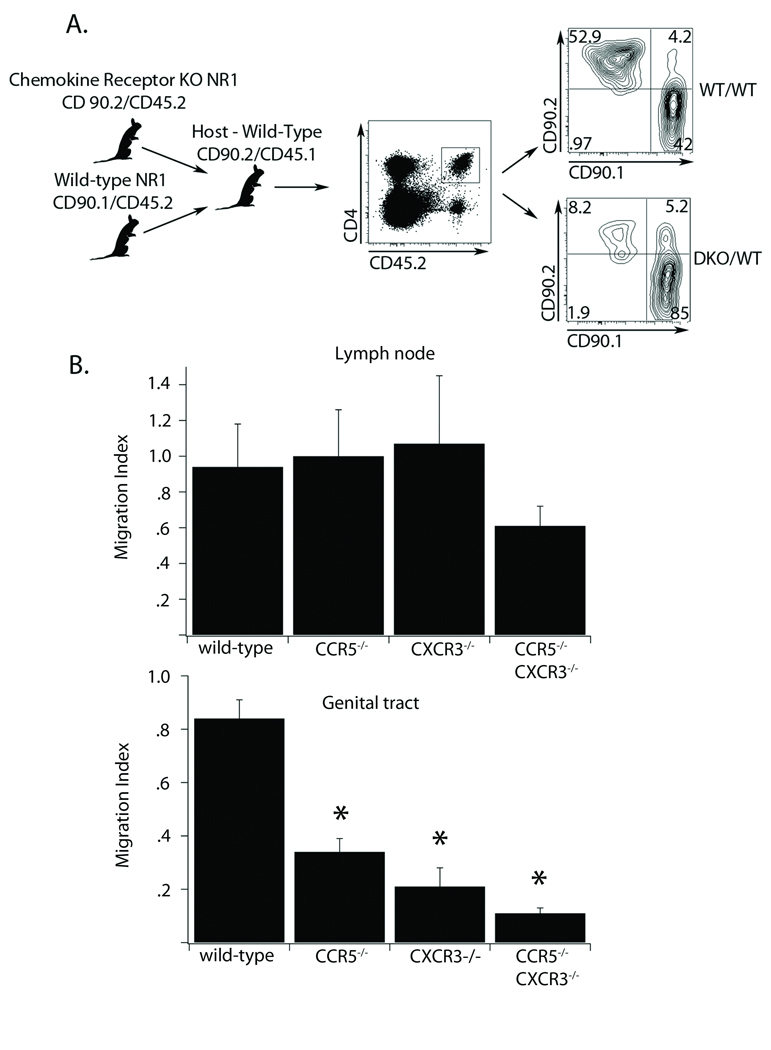
Since trafficking to the genital tract was completely ablated in transfer studies using single chemokine receptor knockouts, we were unable to determine if the CXCR3−/− CCR5−/− T cells exacerbated the homing deficiency to the genital mucosa. To test this we developed a competitive homing assay that would be more sensitive to subtle differences in T cell numbers homing to the genital mucosa. This approach allowed us to directly monitor the ability of wild-type and chemokine receptor deficient T cells to home to the genital tract under identical conditions in the same host (Figure 5A). We first transferred an equal number of CD90.1 chemokine receptor sufficient and CD90.2 chemokine receptor deficient antigen specific T cells into CD45.1 hosts, which we then infected in the genital tract with C. trachomatis. Seven days after infection, we calculated a migration index [(% CD90.1)/(% CD90.2)] for all the T cells transferred in both the draining lymph node and the genital tract (Figure 5B). T cells of all genotypes showed no statistical difference in the migration index in their draining lymph node. In contrast, when we calculated the migration index for T cells in the genital tract, only the wild-type T cell transfer yielded a migration index near 1. Pathogen specific T cells that were deficient in CCR5 migrated 30 percent less efficiently than wild-type cells, while T cells lacking CXCR3 were 20 percent less efficient. Interestingly, the antigen specific T cells lacking both chemokine receptors migrated ten fold less efficiently than wild-type cells suggesting an additive impact of both CXCR3 and CCR5. This indicates that while both single chemokine receptor knockouts are impaired in their ability to compete with wild-type T cells and be recruited to the genital tract, loss of both receptors may be more detrimental to their ability to access the genital mucosa.
Figure 5. Chemokine receptor-deficient Chlamydia specific CD4+ T cells are unable to compete with wild-type cells in homing to the genital tract.
A) A schematic of the competitive homing assay is shown. Equal numbers of antigen specific CD4+ T cells from CD45.2/CD90.1 wild type mice and CD45.2/CD90.2 wild-type, CXCR3−/−, CCR5−/−, or CXCR3−/− CCR5−/− mice were transferred into CD45.1/CD90.2 host mice. The following day mice were infected in the genital tract with C. trachomatis. Seven days after infection tissues were harvested and prepared for flow cytometry. The strategy for gating is illustrated. We first gated CD4 and CD45.2 positive cells. Then we compared the percent of the gated cells that were CD90.1 or CD90.2 positive. Shown is a representative plot comparing the homing to the genital tract between the wild-type:wild-type cell mix and CXCR3−/−CCR5−/−:wild-type mix. Shown is one representative of four independent experiments. B) We calculated a migration index for each cell mix using the percent of CD90.2/CD90.1 positive cells present in the draining lymph node (top) and the percent of the CD90.2/CD90.1 positive cells in the genital tract (bottom). Error bars represent standard deviation and *=p<0.05
Chlamydia-specific CD4+ cells deficient for CCR5 or CXCR3 are unable to protect mice from genital tract infection
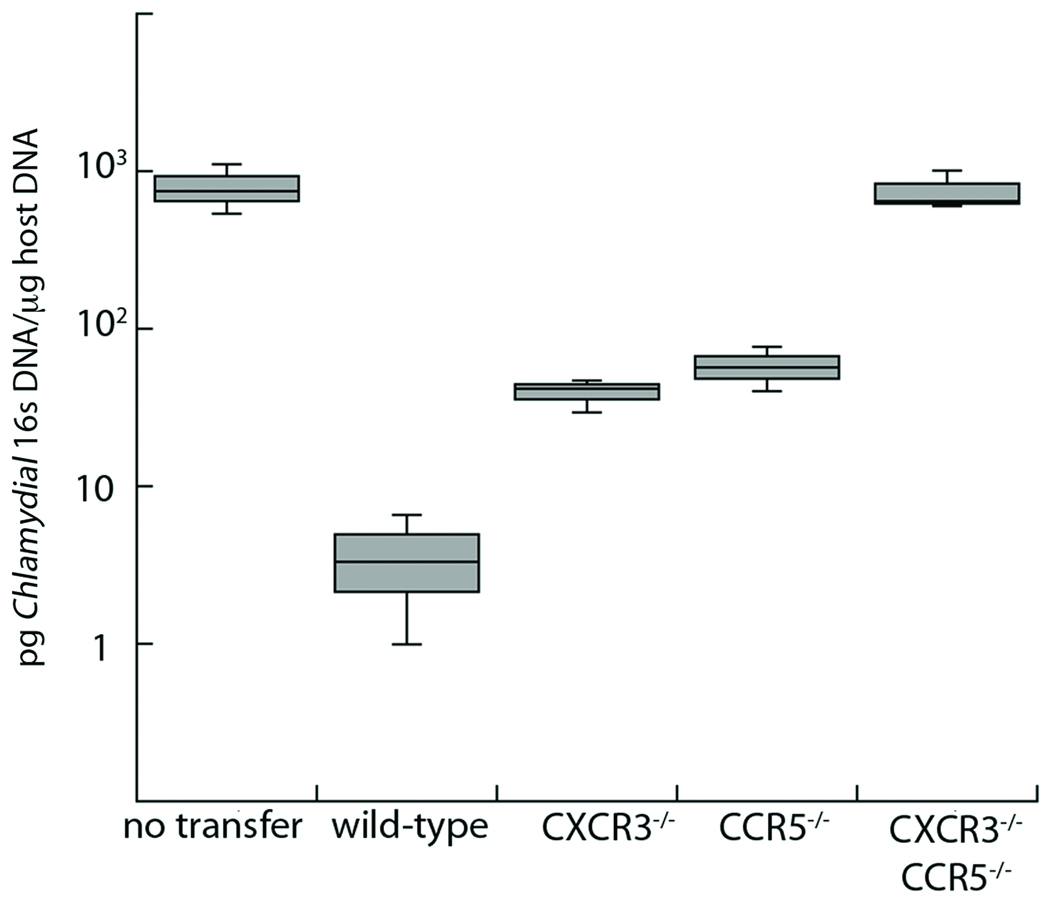
The experiments above show that loss of CXCR3 and/or CCR5 impairs the recruitment of Chlamydia specific T cells to the genital mucosa following infection. Therefore we predicted these T cells would not be able to protect mice from infection as well as wild-type T cells. To assess their protective capacity, we pre-activated wild-type, CXCR3−/−, CCR5−/− and double deficient T cells in vitro, polarizing them to the Th1 phenotype and then transferred them into IFNγ-deficient hosts. Since the clearance of C. trachomatis in mice is dependent on IFNγ, and the only source of IFNγ was the transferred population of cells, we were able to directly ascertain the capacity of Chlamydia specific T cells to traffic to the site of infection, locally produce IFNγ, and reduce bacterial burden in the genital tract8–10. Following in vitro TH1 skewing, all the chemokine receptor deficient T cells were competent at secreting IFNγ equivalently to wild-type T cells (data not shown). Activated cells were transferred into mice, which were subsequently challenged with C. trachomatis. After 6 days of infection, the peak of antigen specific T cells recruitment to the site of infection (Figure 4A), we isolated the genital tract and determined the levels of C. trachomatis by qPCR (Figure 6). Mice that received wild-type T cells had more than two logs less C. trachomatis relative to mice receiving no T cells. However, mice receiving either CXCR3 or CCR5 single knockout T cells had ten-fold higher bacterial levels compared to wild-type cells. This showed that single receptor knockout cells partially protected the host, decreasing Chlamydia burden by over one log as compared to mice receiving no transfer. Therefore, both CXCR3−/− and CCR5−/− antigen specific T cells are capable of protecting host mice from C. trachomatis infection to some degree, albeit much less than wild-type. Surprisingly, mice that received CXCR3−/− CCR5−/− T cells did not protect host mice whatsoever and had levels of C. trachomatis in the genital tract identical to mice that received no transfer. These data indicate that, although loss of CXCR3 or CCR5 in antigen specific T cells leads to a deficiency in protecting mice from infection, loss of both receptors in concert results in total ablation of protection against C. trachomatis. This illustrates that optimal trafficking and protective function of Chlamydia specific T cells in the genital tract requires a cooperative interaction between CXCR3 and CCR5.
Figure 6. Antigen specific TH1 cells deficient in both CXCR3 and CCR5, have no protective capacity in vivo.
Wild-type, CXCR3−/−, CCR5−/−, or CXCR3−/− CCR5−/− Chlamydia specific CD4+ T cells were skewed in vitro to the Th1 phenotype for five days. These pre-activated T cells were then transferred into IFNγ−/− host mice. The following day the mice were challenged with C. trachomatis. Six days after infection the genital tract was harvested and DNA was isolated. We used quantitative PCR to compare the levels of Chlamydia 16s DNA to host GAPDH DNA. Levels of C. trachomatis 16s DNA were converted into pg/µl. Shown is a box and whisker plot from one representative experiment of two conducted. In the graph shown all samples are significantly different (p<0.05) except for the comparison between the CXCR3−/− and CCR5−/− samples and CXCR3−/− CCR5−/− compared to “no transfer”.
Discussion
The ability of immune cells to traffic to sites of inflammation and infection is necessary for efficient clearance of invading pathogens. While chemokine receptors that are critical for directing T cells to the lungs and intestinal tract have been described previously1, 4, 5, characterization of receptors that are necessary for recruitment to the genital tract has remained elusive. In this report, we show that a combination of CXCR3 and CCR5 is necessary for pathogen-specific CD4+ T cells to respond to and clear C. trachomatis infection of the genital mucosa.
Previous studies showed that infection with the mouse specific pathogen C. muridarum led to elevated levels of the chemokines CXCL10 and CCL5 in the genital tract tissue17. These chemokines, specific for the receptors CXCR3 and CCR5, promote the recruitment of immune cells to resolve infection. CCR5 has been implicated in genital mucosa homing because CCR5 deficient mice are unable to clear C. muridarum infections efficiently21. However, one recent study suggested that CXCR3 may be important for CD8+ T cells to respond to the vaginal tissue during herpes simplex virus infection25. Therefore, we sought to determine the chemokine receptors that are necessary for pathogen specific T cells to clear a murine genital infection with the human bacterial pathogen C. trachomatis. Although mice clear C. trachomatis infection more efficiently than humans, we are able to demonstrate that T cells respond and migrate in the mouse following C. trachomatis infection. Therefore we could use C. trachomatis-specific TCR transgenic T cells to facilitate these experiments. It is possible that the rapid clearance of C. trachomatis from mice relative to humans affects the molecules responsible for T cell migration, but we have no evidence that this is the case.
Following stimulation and activation, pathogen specific T cells upregulate specific chemokine receptors to egress from the lymph node and respond rapidly to the infected tissue. Indeed, we saw a trend in CXCR3 and CCR5 expression at both the transcriptional level as well as expression through surface staining. In the draining lymph node there was a distinct upregulation of only CXCR3. Meanwhile, in the genital mucosa there was a dramatic upregulation of both CXCR3 and CCR5, indicating that these receptors may be regulated in a step-wise fashion. Our current model proposes that the initial upregulation of CXCR3 in the draining lymph node allows efficient egress following T cell activation. Upon exiting the lymph node, antigen specific T cells upregulate CCR5 in order to promote migration to the genital mucosa.
In this report we show that mice deficient in either CXCR3 or CCR5 are unable to clear genital tract infections with C. trachomatis as efficiently as wild-type mice. One possible hypothesis is that the loss of chemokine receptors may alter the development of a Th1 response that is critical for Chlamydia clearance. However, transfer of wild-type antigen specific T cells into CXCR3 or CCR5 deficient hosts resulted in no defect in T cell activation or accumulation in the draining lymph node. These T cells also arrive in the genital mucosa in similar numbers and are fully activated, showing that the chemokine receptor profile of any bystander T cells in the host does not affect the capacity of wild-type Chlamydia specific T cells to respond to infection normally. Furthermore these data suggest that the higher pathogen load seen in chemokine receptor-deficient mice is not due to a loss in T cell priming but rather a defect in trafficking to the infected tissue.
No previous reports had assessed whether a lack of CCR5 or CXCR3 impairs recruitment of antigen specific T cells to the genital mucosa during C. trachomatis infections. Our current study demonstrates that Chlamydia-specific CD4+ T cells deficient for either CXCR3 or CCR5 are unable to accumulate in the genital tract over the course of infection. This highlights the importance of CXCR3 and CCR5, as loss of either chemokine receptor does not delay recruitment but rather, blocks trafficking to the genital mucosa altogether. Even though receptor-deficient T cells could not home to the genital mucosa, these cells trafficked normally to the draining lymph node, where they were properly primed with antigen. We did see a minor decrease in activation among all receptor deficient T cells that was not statistically significant in our studies. It is possible however that this slight reduction in activation may have a small effect on the severe blockage of T cell recruitment, but it does not appear to be a major contributor. The more sensitive competitive homing assay bolstered our time course findings by showing that antigen specific chemokine receptor-deficient cells are significantly less efficient at trafficking to the genital tract than wild-type cells when they are both present in the same mouse. Furthermore, when mice received pre-activated antigen specific T cells deficient for either CXCR3 or CCR5, the T cells could not protect recipient mice to the same extent as wild-type cells, but each population was still able to reduce the C. trachomatis load in the genital tract 10-fold. Ultimately, these approaches identified both CXCR3 and CCR5 as necessary for proper homing of Chlamydia specific T cells to the genital mucosa and protecting mice from infection.
Previous reports have also examined a double deficiency of CXCR3 and CCR5 during virus infections of the lung and central nervous system4, 26. These investigators concluded that CXCR3 and CCR5 might function antagonistically on T cells. However, in our study the data support a cooperative interaction between CXCR3 and CCR5 following a genital tract infection with C. trachomatis. Our single transfer and competitive homing studies showed that T cells lacking both CXCR3 and CCR5 were unable to be recruited to or retained in the genital mucosa after infection. Furthermore, transfer of pre-activated antigen specific T cells lacking both CXCR3 and CCR5 did not protect recipient mice at all. Together these data indicate that loss of both CXCR3 and CCR5 together is more detrimental than loss of either receptor individually. This finding does not result from a drastic T cell priming defect because all chemokine receptor deficient cells were able to accumulate in the draining lymph node and be activated similarly. Therefore we conclude that the inability of CXCR3−/− CCR5−/− antigen specific T cells to traffic to and be retained in the genital tract following infection with C. trachomatis eliminates all protective capacity of these cells.
In contrast to the generally accepted view of CXCR3 and CCR5 as “general TH1 inflammation chemokine receptors”, recent evidence in lung infection models have indicated antigen specific cells do not utilize both receptors. Moreover, the expression of CCR5 was deleterious to TH1 T cell homing to the lung4. This incongruity in the “general inflammation” model of homing suggests that monitoring antigen specific responses is required to elucidate lymphocyte homing to infections at disparate mucosal sites. It remains a possibility that the host bystander T cell population, that is present in all experiments, does play some role during the clearance of a genital tract infection. Our data suggests however, that these non-antigen specific T cells are not critical for the priming of Chlamydia specific T cells (Figure 2) and not needed for antigen specific T cells to protect host mice from infection (Figure 6). Our studies, characterizing homing to the genital tract, indicate that an antigen specific CD4 population requires both CXCR3 and CCR5 for homing and protection of the genital mucosa.
The distinct chemokine receptor profiles for each mucosal site, such as CCR9 for homing to the intestine or CCR4/10 for homing to the skin, illustrate a coordinated specificity by the host to ensure that activated T cells arrive in the correct tissues efficiently. Understanding the chemokine receptor repertoire required to access distinct mucosal sites may allow for the development of tissue specific therapeutics. With the identification of CCR5 as a major player in genital tract homing however, it is tempting to speculate that co-infection of patients with C. trachomatis would also lead to higher rates of HIV infection, where CCR5 is a major receptor for entry into T cells27, 28. CCR5 has been proposed as a possible drug target to alter HIV infections, but it will be necessary to test these drugs for their impact on other infections of the genital tract to ensure that these HIV therapeutics do not promote other STIs29.
With additional experimentation it will be possible to determine whether T cells responding to other pathogens of the genital tract depend on the same chemokine receptors we have identified during C. trachomatis infection. By characterizing the phenotypes of other STIs, as well as the immune responses that are necessary to properly clear these infections, therapeutics can be designed that not only control infections but also obviate disease pathologies.
Methods
Mice
C57BL/6, B6.PL-Thy1a (CD90.1 congenic), B6.129S7-IFNgtm1Agt (IFNγ−/−), C57BL/6, B6.SJL-Ptprca Pep3/BoyJ (CD45.1 congenic) B6.Ccr5−/−B6.129P2-Ccr5tm1kuz/J (CCR5−/−) B6.129P2-Cxcr3tm1Dgen/J (CXCR3−/−) were purchased from The Jackson Laboratory (Bar Harbor, ME). We constructed C. trachomatis specific chemokine receptor deficient mice by breeding NR1 transgenic mice with either CCR5−/− or CXCR3−/− mice. All mice were maintained and cared for within the Harvard Medical School Center for Animal Resources and Comparative Medicine. All experiments were approved by Harvard's Institutional Animal Care and Use Committee.
Growth, isolation and detection of bacteria
C. trachomatis serovar L2 (434/Bu) was propagated within McCoy cell monolayers grown in Eagle’s MEM (Invitrogen, Grand Island NY) supplemented with 10% FCS, 1.5 g/L sodium bicarbonate, 0.1 M nonessential amino acids, and 1 mM sodium pyruvate. Infected monolayers were disassociated from plates using .05% trypsin/EDTA and sonicated to disrupt the inclusion. EBs were purified by density gradient centrifugation as described30. Aliquots were stored at −80°C in medium containing 250 mM sucrose, 10 mM sodium phosphate, and 5 mM L-glutamic acid and thawed immediately prior to use.
Flow cytometry
Tissues were mechanically disaggregated and immediately stained for activation markers. Cells were pre-incubated with anti-FcRγ (Bio X-Cell, West Lebanon, NH) before staining with anti-CD4 Pacific Blue (Biolegend, San Diego, CA), anti-CD90.1 peridinin chlorophyll-a protein (BD Bioscience, San Jose, CA), anti-CD90.2 (phycoerythrin or peridinin chlorophyll-a protein), and anti-CD45.2 (allophycocyanin). We used anti-CD45Rb phycoerythrin-cychrome 7 (Biolegend) anti-CD62L allophycocyanin-Alexa 750 (Ebioscience, San Diego, CA) in order to examine the activation levels of antigen specific T cells following infection. Data were collected on a modified FACSCalibur (Cytek Development, Fremont CA) or an LSRII (BD Bioscience) and analyzed using FlowJo (Tree Star Industries, Ashland OR).
Transfer of Transgenic T cells, infection of mice, and preparation of tissue
Prior to transfer, C. trachomatis (Cta1133–152)-specific CD4+ T cells12 were isolated from the peripheral lymphoid tissues of donor NR1 mice. Recipient mice were injected intravenously with either 105 or 106 cells from the donors. To infect the genital tract, mice were treated with 2.5 mg of medroxyprogestrone acetate subcutaneously and then infected one week later in the uterus with 106 IFU of C. trachomatis L28, 12. Intrauterine infection was done using the NSET device (http://paratechs.com/nset.htm). In short, 5µl of SPG containing 106 C. trachomatis was deposited using the NSET pipet tip through the NSET speculum. Infection occurs at the base of the uterine horns (including the cervix). Five days following infection, the lymph nodes, spleen, and uterine horns were collected. The uterus was digested with 1 mg/ml of type XI collagenase (Sigma) and 50 Kunitz/ml of DNase (Sigma, St Louis MO) for 45 min at 37 C. Single cell suspensions were prepared for staining via mechanical disaggregation.
Real time PCR
Tissue from two pools of five mice infected in the uterus with 106 IFU of C. trachomatis L2 (and corresponding tissue from uninfected controls) were harvested and processed as above. Isolated cells were resuspended in TRIzol (Invitrogen) and RNA was isolated using an RNeasy mini kit (Qiagen, Valencia CA). RNA was quantified and diluted to 5 ng/ul for real-time analysis. Primers for chemokine receptors were CCR1-Forward-5’-TCTGGACCCCCTACAATCTG-3’, Reverse-5’-GGCAATCACCTCAGTCACCT3’ CCR2-Forward-5’ACACCCTGTTTCGCTGTAGG-3’, Reverse-5’-CCTGGAAGGTGGTCAAGAAG, CXCR3-Forward-5’GCCCTCACCTGCATAGTTGT-3’, Reverse-5’-ATTGAGGCGCTGATCGTAGT-3’, CCR5-Forward-5’-CGAAAACACATGGTCAAACG-3’, Reverse-5’-GTTCTCCTGTGGATCGGGTA-3’, CCR7-Forward-5’-AACGGGCTGGTGATACTGAC-3’, Reverse-5’-TAGGCCCAGAAGGGAAGAAT-3’, CCR9-Forward-5’-TGACTCCACTGCTTCCACAG-3’, Reverse-5’-GTGCCCACAATGAACACAAG-3’, GAPDH-Forward-5’-GGTGCTGAGTATGTCGTGGA-3’, Reverse-5’-CGGAGATGATGACCCTTTTG-3’. Real-time PCR was conducted using the Quantitect SYBR Green RT-PCR kit (Qiagen) and run on an ABI prism 7000 sequence detection system (Applied Biosystems). Resulting Ct values were normalized against GAPDH for each sample resulting in a relative expression for each tissue. Fold induction was calculated by dividing relative expression of infected tissue by relative expression of uninfected tissue.
Competitive homing
One million wild-type NR1 cells (CD45.2/CD90.1) were mixed with an equivalent number of wild-type, CCR5−/−, CXCR3−/−, or CCR5−/− CXCR3−/− NR1 cells (CD45.2/CD90.2) and transferred into CD45.1/CD90.2 hosts. Twenty-four hours following transfer, 106 IFU of C. trachomatis L2 were inoculated into the uterus. Tissues were harvested seven days post-infection and processed as described above.
Skewing of NR1 cells
CD4+ T cells were purified from NR1 mice using a mouse CD4 negative isolation kit (Invitrogen) per the manufacturers directions. The T cells were cultured in RPMI-1640 (Invitrogen) supplemented with 10% FCS, L-glutamine, HEPES, 50 µM 2-mercaptoethanol, 50 U/ml penicillin, and 50 ug/ml streptomycin. To stimulate the T cells, irradiated feeder splenocytes were pulsed with 5 µM of Cta1133–152 peptide and cocultured with the CD4-enriched NR1 cells at a stimulator to T cell ratio of 4:1. To polarize T cells towards the Th1 phenotype cells were incubated with 10 ng/ml of IL-12 (Peprotech, Rocky Hill NJ) and 10 µg/ml of anti-IL4 (Biolegend). Cells were stimulated for five days then 106 cells were transfered into naïve CD90.2+ host mice. Twenty-four hours following transfer, mice were challenged in the uterus with 106 IFU of C. trachomatis L2.
Quantitative PCR
To assess the levels of C. trachomatis present, we used a previously described qPCR method31. Briefly we isolated nucleic acid from genital tract homogenates using the QIAamp DNA mini kit from Qiagen. Mouse GAPDH DNA and Chlamydia 16S DNA were quantified by qPCR on an ABI Prism 7000 sequence detection system using primer pairs and dual-labeled probes. Using standard curves from known amounts of Chlamydia and mouse DNA, we calculated the amount (in picograms) of Chlamydia DNA per unit weight (in micrograms) of mouse DNA in the samples.
Statistical analysis
All groups were evaluated for statistical significance through the use of unpaired two-tailed t tests. Where it appeared necessary to highlight significant differences between data points, the level of significance is depicted as * = p<0.05
Acknowledgments
We thank Stephanie Jehl for critical reading of the manuscript. This work was supported by National Institutes of Health grants AI39558 and AI062827.
Footnotes
Conflict of Interest: None
References
- 1.Sigmundsdottir H, Butcher EC. Environmental cues, dendritic cells and the programming of tissue-selective lymphocyte trafficking. Nat Immunol. 2008;9:981–987. doi: 10.1038/ni.f.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thelen M. Dancing to the tune of chemokines. Nat Immunol. 2001;2:129–134. doi: 10.1038/84224. [DOI] [PubMed] [Google Scholar]
- 3.Stenstad H, et al. Gut-associated lymphoid tissue-primed CD4+ T cells display CCR9-dependent and -independent homing to the small intestine. Blood. 2006;107:3447–3454. doi: 10.1182/blood-2005-07-2860. [DOI] [PubMed] [Google Scholar]
- 4.Fadel SA, Bromley SK, Medoff BD, Luster AD. CXCR3-deficiency protects influenza-infected CCR5-deficient mice from mortality. Eur J Immunol. 2008;38:3376–3387. doi: 10.1002/eji.200838628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kohlmeier JE, et al. CXCR3 directs antigen-specific effector CD4+ T cell migration to the lung during parainfluenza virus infection. J Immunol. 2009;183:4378–4384. doi: 10.4049/jimmunol.0902022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Starnbach MN, Roan NR. Conquering sexually transmitted diseases. Nat Rev Immunol. 2008;8:313–317. doi: 10.1038/nri2272. [DOI] [PubMed] [Google Scholar]
- 7.Brunham RC, Rey-Ladino J. Immunology of Chlamydia infection: implications for a Chlamydia trachomatis vaccine. Nat Rev Immunol. 2005;5:149–161. doi: 10.1038/nri1551. [DOI] [PubMed] [Google Scholar]
- 8.Gondek DC, Roan NR, Starnbach MN. T cell responses in the absence of IFN-gamma exacerbate uterine infection with Chlamydia trachomatis. J Immunol. 2009;183:1313–1319. doi: 10.4049/jimmunol.0900295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johansson M, Schon K, Ward M, Lycke N. Genital tract infection with Chlamydia trachomatis fails to induce protective immunity in gamma interferon receptor-deficient mice despite a strong local immunoglobulin A response. Infect Immun. 1997;65:1032–1044. doi: 10.1128/iai.65.3.1032-1044.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perry LL, et al. Differential sensitivity of distinct Chlamydia trachomatis isolates to IFN-gamma-mediated inhibition. J Immunol. 1999;162:3541–3548. [PubMed] [Google Scholar]
- 11.Rank RG, Soderberg LS, Barron AL. Chronic chlamydial genital infection in congenitally athymic nude mice. Infect Immun. 1985;48:847–849. doi: 10.1128/iai.48.3.847-849.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roan NR, Gierahn TM, Higgins DE, Starnbach MN. Monitoring the T cell response to genital tract infection. Proc Natl Acad Sci U S A. 2006;103:12069–12074. doi: 10.1073/pnas.0603866103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roan NR, Starnbach MN. Antigen-specific CD8+ T cells respond to Chlamydia trachomatis in the genital mucosa. J Immunol. 2006;177:7974–7979. doi: 10.4049/jimmunol.177.11.7974. [DOI] [PubMed] [Google Scholar]
- 14.Starnbach MN, Bevan MJ, Lampe MF. Protective cytotoxic T lymphocytes are induced during murine infection with Chlamydia trachomatis. J Immunol. 1994;153:5183–5189. [PubMed] [Google Scholar]
- 15.Williams DM, Grubbs BG, Pack E, Kelly K, Rank RG. Humoral and cellular immunity in secondary infection due to murine Chlamydia trachomatis. Infect Immun. 1997;65:2876–2882. doi: 10.1128/iai.65.7.2876-2882.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rekart ML, Brunham RC. Epidemiology of chlamydial infection: are we losing ground? Sex Transm Infect. 2008;84:87–91. doi: 10.1136/sti.2007.027938. [DOI] [PubMed] [Google Scholar]
- 17.Belay T, et al. Chemokine and chemokine receptor dynamics during genital chlamydial infection. Infect Immun. 2002;70:844–850. doi: 10.1128/IAI.70.2.844-850.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sakthivel SK, et al. CCL5 regulation of mucosal chlamydial immunity and infection. BMC Microbiol. 2008;8:136. doi: 10.1186/1471-2180-8-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shibahara H, et al. Chemokine bioactivity of RANTES is elevated in the sera of infertile women with past Chlamydia trachomatis infection. Am J Reprod Immunol. 2003;49:169–173. doi: 10.1034/j.1600-0897.2003.01154.x. [DOI] [PubMed] [Google Scholar]
- 20.Ficarra M, et al. A distinct cellular profile is seen in the human endocervix during Chlamydia trachomatis infection. Am J Reprod Immunol. 2008;60:415–425. doi: 10.1111/j.1600-0897.2008.00639.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barr EL, et al. Host inflammatory response and development of complications of Chlamydia trachomatis genital infection in CCR5-deficient mice and subfertile women with the CCR5delta32 gene deletion. J Microbiol Immunol Infect. 2005;38:244–254. [PubMed] [Google Scholar]
- 22.Bromley SK, Thomas SY, Luster AD. Chemokine receptor CCR7 guides T cell exit from peripheral tissues and entry into afferent lymphatics. Nat Immunol. 2005;6:895–901. doi: 10.1038/ni1240. [DOI] [PubMed] [Google Scholar]
- 23.Chakravarty SD, et al. The chemokine receptor CXCR3 attenuates the control of chronic Mycobacterium tuberculosis infection in BALB/c mice. J Immunol. 2007;178:1723–1735. doi: 10.4049/jimmunol.178.3.1723. [DOI] [PubMed] [Google Scholar]
- 24.Manicone AM, Burkhart KM, Lu B, Clark JG. CXCR3 ligands contribute to Th1-induced inflammation but not to homing of Th1 cells into the lung. Exp Lung Res. 2008;34:391–407. doi: 10.1080/01902140802221987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakanishi Y, Lu B, Gerard C, Iwasaki A. CD8(+) T lymphocyte mobilization to virus-infected tissue requires CD4(+) T-cell help. Nature. 2009;462:510–513. doi: 10.1038/nature08511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Lemos C, et al. Opposing effects of CXCR3 and CCR5 deficiency on CD8+ T cell-mediated inflammation in the central nervous system of virus-infected mice. J Immunol. 2005;175:1767–1775. doi: 10.4049/jimmunol.175.3.1767. [DOI] [PubMed] [Google Scholar]
- 27.Granelli-Piperno A, et al. Efficient interaction of HIV-1 with purified dendritic cells via multiple chemokine coreceptors. J Exp Med. 1996;184:2433–2438. doi: 10.1084/jem.184.6.2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patterson BK, et al. Repertoire of chemokine receptor expression in the female genital tract: implications for human immunodeficiency virus transmission. Am J Pathol. 1998;153:481–490. doi: 10.1016/S0002-9440(10)65591-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Telenti A. Safety concerns about CCR5 as an antiviral target. Curr Opin HIV AIDS. 2009;4:131–135. doi: 10.1097/COH.0b013e3283223d76. [DOI] [PubMed] [Google Scholar]
- 30.Howard L, Orenstein NS, King NW. Purification on renografin density gradients of Chlamydia trachomatis grown in the yolk sac of eggs. Appl Microbiol. 1974;27:102–106. doi: 10.1128/am.27.1.102-106.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bernstein-Hanley I, et al. Genetic analysis of susceptibility to Chlamydia trachomatis in mouse. Genes Immun. 2006;7:122–129. doi: 10.1038/sj.gene.6364285. [DOI] [PubMed] [Google Scholar]