Abstract
The PI3K/AKT signaling pathway is aberrant in a wide variety of cancers. Downstream effectors of AKT are involved in survival, growth, and metabolic-related pathways. In contrast, contradictory data relating to AKT effects on cell motility and invasion, crucial pro-metastatic processes, have been reported pointing to a potential cell type and isoform type-specific AKT driven function. By implication, study of AKT signaling should optimally be conducted in the appropriate intracellular environment. Prognosis in soft-tissue sarcoma (STS), aggressive malignancies of mesenchymal origin, is poor reflecting our modest abilities to control metastasis, an effort hampered by lack of insight into molecular mechanisms driving STS progression and dissemination. We examined the impact of the cancer progression relevant AKT pathway on the mesenchymal tumor cell internal milieu. We demonstrate that AKT1 activation induces STS cell motility and invasiveness at least partially via a novel interaction with the intermediate filament vimentin. The binding of AKT (tail region) to vimentin (head region) results in vimentin Ser39 phosphorylation enhancing the ability of vimentin to induce motility and invasion while protecting vimentin from caspase induced proteolysis. Moreover, vimentin phosphorylation was shown to enhance tumor and metastasis growth in vivo. Insights into this mesenchymal-related molecular mechanism may facilitate development of critically lacking therapeutic options for these devastating malignancies.
Keywords: Soft-Tissue Sarcoma, AKT1, Vimentin, phosphorylation, Migration/Invasion
Introduction
The AKT kinase is a convergence point for multiple extracellular and other upstream signals functioning as a master switch to generate a plethora of intracellular signals and responses. Much evidence exists to support an important role for AKT in human cancer(Sulis & Parsons, 2003). Downstream effectors of AKT are thought to be involved in survival, growth, and metabolic-related pathways(Phung et al., 2006). In contrast, AKT effects on cell motility and invasion, prerequisite processes for metastasis, while suggested, have been less well elucidated(Grille et al., 2003). AKT is a family of isoforms including AKT1, AKT2, and AKT3 sharing structural similarities. The expression patterns of the AKT isoforms differ, as do their apparent biological activities(Sumitani et al., 2002). Although AKT1 and AKT2 both play roles in cell motility and invasion, distinct and even opposing functions have been observed. Several studies demonstrated that AKT1 attenuates invasive migration of breast cancer cells (Irie et al., 2005; Liu et al., 2006; Yoeli-Lerner et al., 2005) while AKT2 promoted migration in these cells in a growth factor-dependent manner(Irie et al., 2005). Similar findings were seen in vivo in breast and ovarian cancer models(Arboleda et al., 2003; Hutchinson et al., 2004; Maroulakou et al., 2007). In contrast, a recent study identified AKT1 as inducing the migration of mammary epithelial tumor cells; AKT1 deficiency reduced migration and inhibited metastasis in an ErbB2-induced breast cancer mouse model(Ju et al., 2007). Both AKT1 and AKT2 were found to enhance the invasiveness of human pancreatic cancer cells(Tanno et al., 2001). Several potential direct and indirect AKT targets have been identified as participatory in regulation of actin cytoskeleton organization, cellular interaction with the extracellular matrix, expression of motility genes, and establishment of cellular polarity(Miyazaki et al., 2007). These studies point to the potential tumor/cell and AKT isoform type-specific function as relevant to regulation of tumor cell migration/invasion. By implication, study of signaling pathways should optimally be conducted in the appropriate intracellular environment to best understand AKT complex effects.
Mesenchymal-derived soft-tissue sarcomas (STS) are rare compared to epithelial-origin tumors and comprise more than 50 distinct histological subtypes. Genetically, STS are classified into two major subgroups: those with simple karyotypes and specific genetic alterations (translocations or point mutations; e.g. GIST), and a more prevalent cohort that has complex, unbalanced aneuploid karyotypes (e.g. leiomyosarcoma). STS disseminate hematogenously, particularly to the lungs; these metastases are a major determinant of the 5-year 50% overall STS survival rate, a survivorship now stagnant for nearly 50 years. Chemotherapy is used to control dissemination, but few drugs have efficacy in STS(Clark et al., 2005). The lack of effective systemic therapy is thus the major unresolved STS clinical dilemma.
Recent molecular insights have led to new therapy for some simple-karyotype STS. In contrast, the molecular diversity and intricacy of genetically complex STS has impeded a comparable awareness of molecular deregulations driving sarcomagenesis, progression, and metastasis needed for therapeutic progress in this latter group. Studies of epithelial cancers indicate that both invasion and metastasis may depend on the acquisition of a mesenchymal phenotype by the incipient cancer cell(Yang & Weinberg, 2008). Therefore, it is logical to propose that STS, originating from mesenchymal cells, might possess inherent capacities for enhanced migration/invasion. Identifying the molecular basis for these processes could provide a heretofore unexploited basis for development of critically needed therapies for STS.
While not extensively explored, evidence points to possible involvement of AKT in STS development and progression. Increased pAKT expression in a large panel of complex karyotype STS has recently been reported(Hernando et al., 2007) and a correlation between pAKT expression and subsequent tumor recurrence and patient survival was described(Tomita et al., 2006). Using a conditional PTEN knockout mouse model, a critical role for AKT in leiomyosarcoma development was shown(Hernando et al., 2007). We have recently demonstrated a significant impact of AKT inhibition on STS growth in vitro and in vivo(Zhu et al., 2008). However, little is known about the contributions of AKT to STS migration and invasion, or about possible AKT downstream targets that are affected in the unique mesenchymal STS intracellular milieu. To bridge this knowledge gap, we evaluated the contribution of AKT, specifically the AKT1 isoform, to complex-karyotype STS cell migration and invasion. We identified vimentin (Vim), an intermediate filament highly expressed in mesenchymal cells, as a novel AKT1 downstream target. AKT1 binds to Vim, resulting in Vim phosphorylation and activation, findings of potential significance in establishing novel therapeutics for STS.
Results
AKT1 signaling contributes to STS migration and invasion
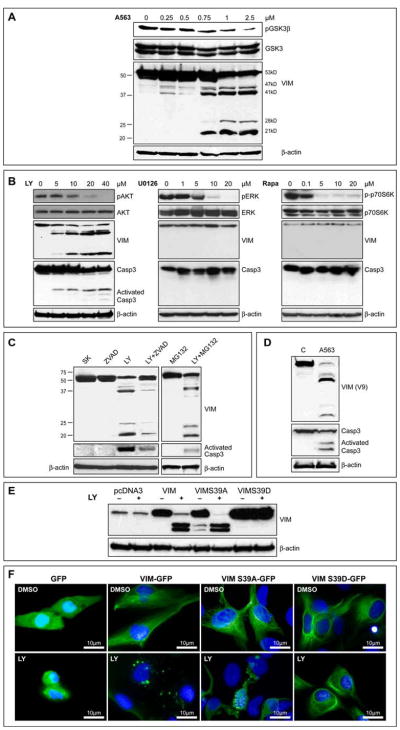
The effect of AKT on STS cell migration/invasion was first determined. Activation of AKT (via EGF stimulation) induced a significant increase in migration/invasion (p<0.05; Figure 1A and S1A). However, this approach is not specific, in that EGFR stimulation induces multiple downstream signaling pathways. Seeking more specificity, we determined the impact of pAKT blockade on these processes. STS cells were treated for 2h with the PI3K inhibitor LY294002 (10μM), the AKT inhibitor A674563 (A563; 1μM), or DMSO as a control. After discontinuation of the drugs viable cells were examined in migration/invasion assays; a significant decrease was observed (p<0.05; Figure 1A and S1A). In contrast, no significant effect on migration/invasion was observed with a MEK inhibitor (UO126; Figures S1A and S1B).
FIGURE 1. AKT phosphorylation enhances STS cell migration and invasion.
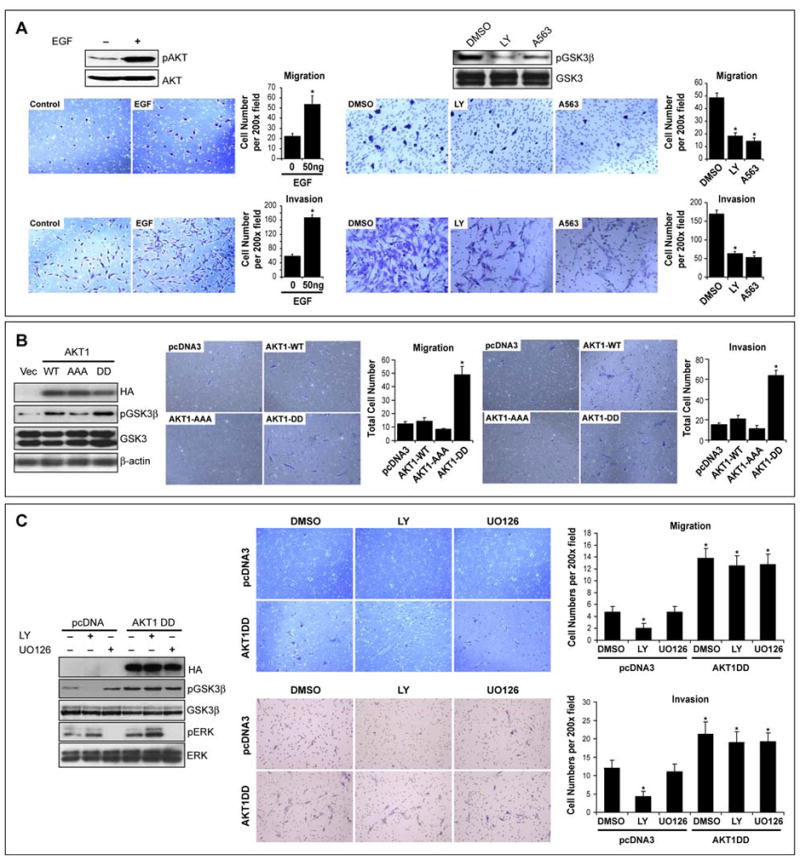
A. EGF induces AKT phosphorylation (WB for pS473) in SKLMS1 cells and enhances migration/invasion. Inhibitors of PI3K (LY=LY294002) and AKT (A563) block AKT downstream signaling (evaluated by pGSK3β) and abrogate EGF-induced SKLMS1 migration/invasion; B. Normal human smooth muscle cells (HC-SMC) were transfected to overexpress HA-tagged wild type AKT, dominant negative AKT (AAA), and activated AKT (D); pGSK3β levels confirm the function of the overexpressed proteins. Overexpression of AKT-DD significantly increased the number of migrating and invading cells (p<0.001); C. Effects of AKT-DD on HC-SMC downstream signaling and cell migration and invasion is independent of PI3K or MEK blockade. [Graphs represent the average of three repeated experiments ±SEM; * depict statistically significant effects (p<0.05); pAKT WB in all panels refer to pS473] (See also Figure S1)
Next, primary cultures of normal smooth muscle cells and normal human dermal fibroblasts (mesenchymal cells exhibiting minimal constitutive AKT activity, thus enabling the study of AKT mutants’ effects independent of endogenous AKT impact; Figure S1C) were transiently transfected to express WT-AKT, dominant-negative AKT (AAA), and constitutively active AKT (DD; transfection efficiency was >70%), functional impact were confirmed by pGSK3β (Figures 1B and S1D). Control (pcDNA3-transfected) cells exhibited only minimal migration/invasion capacity. Overexpression of WT-AKT induced a minimal increase in cell migration/invasion; AKT-AAA inhibited cell motility/invasion. In contrast, AKT-DD increased the number of migrating and invading cells approximately five- and six fold, respectively (p<0.001; Figures 1C and S1D). The effects of activated AKT on GSK3β phosphorylation and cell migration/invasion were not sensitive to PI3K or MEK/ERK pharmacological blockade (Figure 1C).
The expression of AKT isoforms was examined in a panel of human STS cell lines and primary cultures (see Figure S2A for primary culture details). We have previously demonstrated that STS express pAKT under serum and serum free conditions through a multitude of potential upstream mechanisms(Zhu et al., 2008). Here we show that STS cells express pAKT (Ser473) and all three AKT isoforms to varying levels (Figure 2A); however, only AKT1 was found to be constitutively phosphorylated (Figure 2B). Furthermore, EGF stimulation resulted in marked AKT1 phosphorylation (Figure 2C); AKT3 phosphorylation was also seen to a lesser extent but there was no evidence for AKT2 phosphorylation. This selective effect was not specific for EGFR activation and was also seen after HGF stimulation (data not shown). In contrast, EGF stimulation induced AKT2 phosphorylation in the epithelial-origin MDA-231 cell line. This differential effect was further confirmed via nonradioactive AKT1 and AKT2 kinase assays (Figure 2C). AKT1 knockdown significantly inhibited the migration/invasion of STS cells (p<0.05); no significant comparable effects were seen after AKT2 knockdown (Figure 2D). Interestingly, forced expression of AKT2 into STS cells does result in its constitutive and inducible phosphorylation (Figure S2B) and is able to rescue the effects of AKT1 knockdown on STS cell migration/invasion (Figure S2C). Transfection of AKT2-DD into STS cells results in enhanced migration/invasion (p<0.05; Figure S2D). Together, these data suggest that AKT2 expression as compared to AKT1 is relatively low in STS.
FIGURE 2. AKT1 is constitutively and inducibly activated in STS cells; its inhibition abrogates migration and invasion.
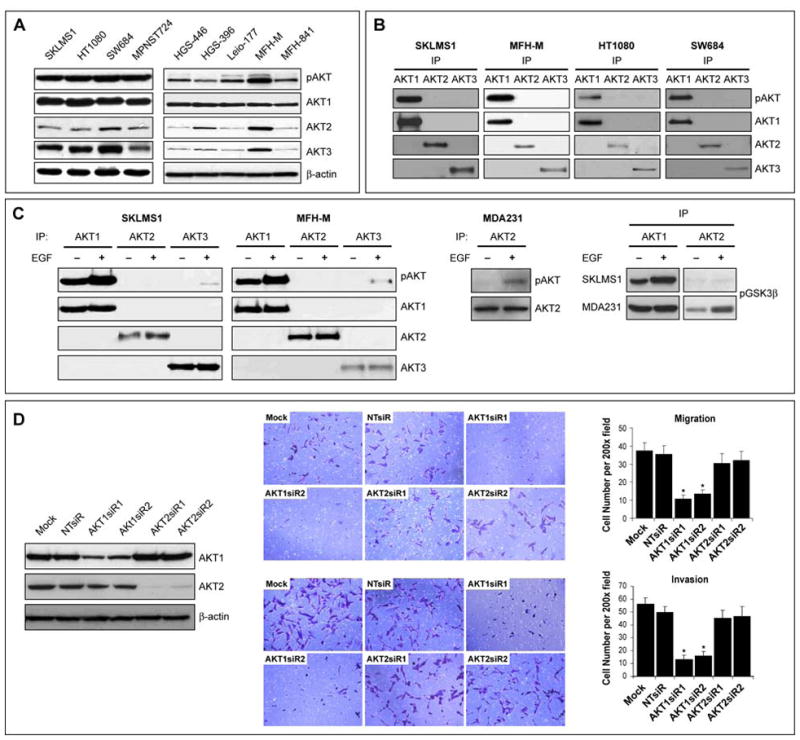
A. STS cell lines and high-grade human STS primary cultures express pAKT (pS473) and all three AKT isoforms to varying levels (WB); B. IP for AKT1, AKT2 and AKT3 and corresponding WB demonstrate that only AKT1 is constitutively phosphorylated in STS cells; C. IP of AKT1, 2, and 3 in STS cells after or without treatment with EGF demonstrates induction of AKT1 and AKT3 phosphorylation but not of AKT2. In contrast, EGF stimulation induces the phosphorylation of AKT2 in MDA231 cells. This differential effect was further demonstrated via an AKT kinase assay using GSK-3 fusion protein as a substrate; D. SKLMS1 cells were transiently transfected with SMARTpool siRNA (20nM; two different pools were used) targeting AKT1 or AKT2; a target-specific knockdown was observed (WB). siRNA Knockdown of AKT1 but not of AKT2 significantly inhibits STS cell migration/invasion (p<0.05). [Graphs represent the average of three repeated experiments ±SEM; * depict statistically significant effects (p<0.05); pAKT WB in all panels refer to pS473] (See also Figure S2)
Vimentin is a novel AKT1 binding partner
Next, we sought to identify novel STS-related AKT1 binding proteins. A proteomics-based approach was utilized; AKT1 coimmunoprecipitated (co-IP) proteins were analyzed by mass spectrometry to identify putative AKT1 protein binding partners. Several bands from AKT1 immunoprecipitate were selected for further sequencing and 10 proteins were identified (Figure 3A, Table S1). All identifications, done primarily by MALDI-TOF mass spectrometry, were validated by sequencing with MS-MS. Interaction with AKT1 was further confirmed via IP/WB for three of the proteins: DDX5, GRP75, and Vim as well as for the known AKT1 binding partners MDM2 and GSK3β (Figure 3A).
FIGURE 3. Vimentin Is a Novel AKT1 Binding Partner.
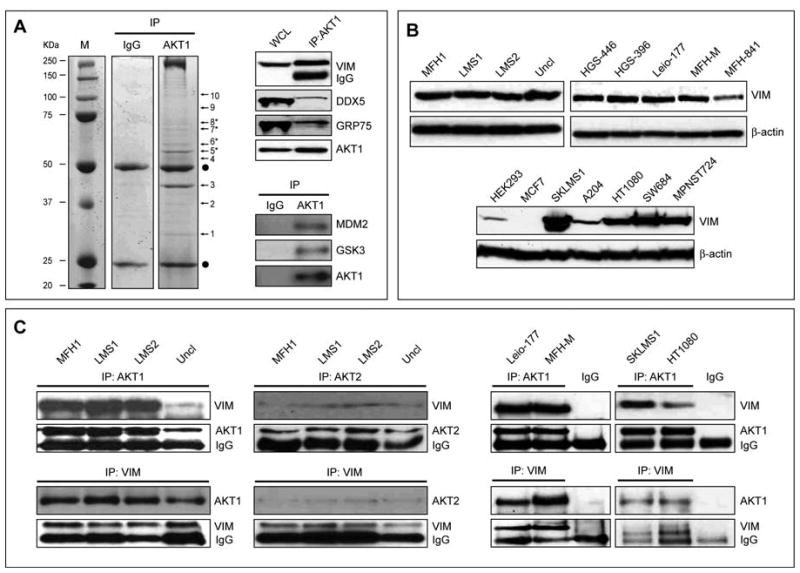
A. SDS-PAGE gel obtained for AKT1 co-immunopreciptiates (normal mouse IgG was used as control). Marked bands were excised, and subjected to identification by mass spectrometry (see protein details in Table S1). Interaction of proteins 5*, (Vim), 7* (DDX5), and 8* (GRP75) with AKT1 (band 6*) were further confirmed by AKT1 IP/WB. Interaction of AKT1 with the its known partners MDM2 and GSK3β in STS cells is shown as control; B. Human STS samples (right upper panel), primary cell cultures (left upper panel), and cell lines (lower panel) highly express Vim; C. IP/WB and reverse IP/WB further confirmed the interaction between AKT1 (and to a lesser extent AKT2, most probably due to the lower expression level of this AKT isoform in STS) and Vim in human STS tissues (left panels), primary cell cultures (mmiddle panel), and cell lines (right panel). (MFH1 = malignant fibrous histiocytoma; LMS1&2= leiomyosarcoma; uncl = unclassified pleomorphic sarcoma). (See also Figure S3)
We focused our investigations on Vim being the major intermediate filament in mesenchymal cells. As expected, Vim expression was seen in all evaluated human STS samples, primary cell cultures, and established cell lines (Figure 3B). Interaction between AKT1 and Vim was identified in all STS immunoprecipitates tested (Figure 3C). AKT2:Vim interaction was also identified although to a lower degree most probably reflecting low AKT2 expression levels. To demonstrate the interaction of AKT1 and Vim within the context of the intact STS cell we have utilized three assays: 1) double immunoflorescence, demonstrating co-localization of endogenous AKT1 and Vim; 2) co-transfection of DsRed-AKT1 and Vim-GFP, demonstrating co-localization of the transfected proteins via live cell microscopy; and, 3) protein-fragment complementation assay (RePCA): protein-protein interaction was demonstrated by positive fluorescent expression evaluated by microscopy and flow cytometry (Figure S3A–C). This interaction was further confirmed in an experimental model by overexpressing Vim and AKT1 in cells expressing low (HEK293) or no (MCF7) Vim (see Figure S3D&E). Lastly, using a an in vitro co-sedimentation assay we found that AKT1 co-sendiments with the polymerized vimentin fraction (Fig S3F).
To identify the potential interacting sites on AKT1 and Vim, GST-fused full-length AKT1 and specific AKT1 domains as well as full-length Vim and specific Vim domains were produced (Figure 4A). GST-AKT1-FL was able to pull down Vim from crude STS cell lysates (Figure 4B). Similarly, GST-AKT1-TAIL, but not other domains, resulted in Vim pull-down. Similarly, AKT1 was pulled down by GST-VIM-FL and GST-VIM-HEAD, but not by other Vim domains (Figure 4C). However, data obtained from intact cells do not confirm direct AKT-Vim binding, as intermediary proteins might be involved and AKT-Vim co-IP could be a result of their dual interaction with other proteins. To confirm direct AKT- Vim interaction, an in vitro protein binding assay was conducted. GST- Vim proteins, purified as above, were evaluated for binding to a commercially available AKT1 protein. GST-AKT1 proteins were also incubated with recombinant Vim protein. These in vitro protein binding assays confirmed the direct interaction between AKT1 tail domain and Vim head domain (Figure 4D).
FIGURE 4. AKT1 C-Terminal tail domain interacts with vimentin head domain.
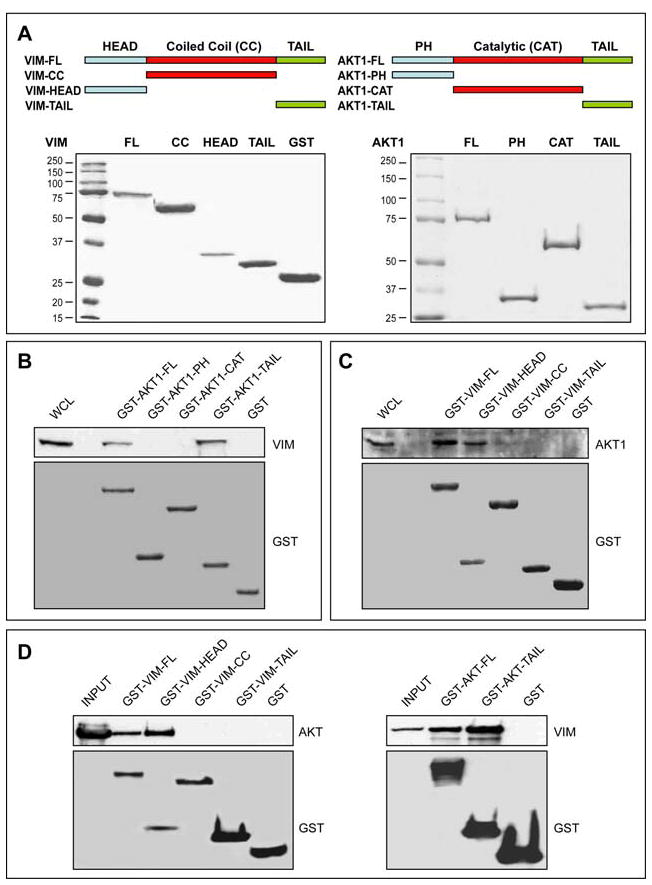
A. full-length AKT1 (AKT1-FL), AKT1 pleckstrin homology domain (AKT1-PH), AKT1 catalytic domain (AKT1-CAT), AKT1 C-terminal tail domain (AKT1-TAIL), full-length Vim (VIM-FL), Vim head domain (VIM-HEAD), Vim coiled-coil domain (VIM-CC), and Vim tail domain (VIM-TAIL) cDNA fragments were cloned into the pGEX4T1 vector to produce GST fusion proteins in E. coli BL21; B. GST-pull down assay indicated that the GST-AKT1-FL and GST-AKT1-TAIL domains interact with Vim in SKLMS1 cells; C. GST-pull down assay indicated that GST-VIM-FL and GST-VIM-HEAD domain interact with AKT1 in SKLMS1 cells; D. In vitro protein binding assays indicated that the GST-VIM-HEAD and the GST-AKT1-TAIL domains directly interact.
Vimentin is a novel AKT1 kinase target
Next, we evaluated whether AKT activation results in vimentin phosphorylation. We first evaluated protein extracts from primary human STS samples (see Figure S4A); all samples expressed pAKT to varying degrees (Figure 5A). Protein extracts were immunoprecipitated for Vim and a WB for anti-phosphorylated AKT substrate (PAS) antibody was conducted. This antibody, raised against an AKT phosphorylation consensus sequence, detects phosphorylated AKT target sites. Vim was recognized by the PAS antibody in all samples to varying degrees (Figure 5A). Vim immunoprecipitates from STS cells stimulated with EGF, with or without pretreatment with LY294002 or rapamycin (an mTORC1 inhibitor), were subjected to WB using PAS antibody (Figure 5B). Vim was recognized by the PAS antibody, and Vim phosphorylation increased in a time dependent manner after AKT activation and decreased after PI3K blockade but not after the inhibition of mTORC1 and S6 kinase. Similarly, double immunoflorescence studies suggested colocalization of Vim and PAS in STS cells. This colocalization was abrogated after short treatment (4h) with LY294002 (Figure S4B). Furthermore, AKT1 knockdown markedly decreased Vim phosphorylation while AKT2 did not (Fig 5B). In contrast to STS cells, no constitutive AKT-induced Vim phoshorylation was observed in normal mesenchymal cells (Figure S1E). However, overexpression of AKT1-DD in normal cells did induce Vim phosphorylation (Figure S1F).
FIGURE 5. Vimentin is a novel AKT1 Kinase downstream target.
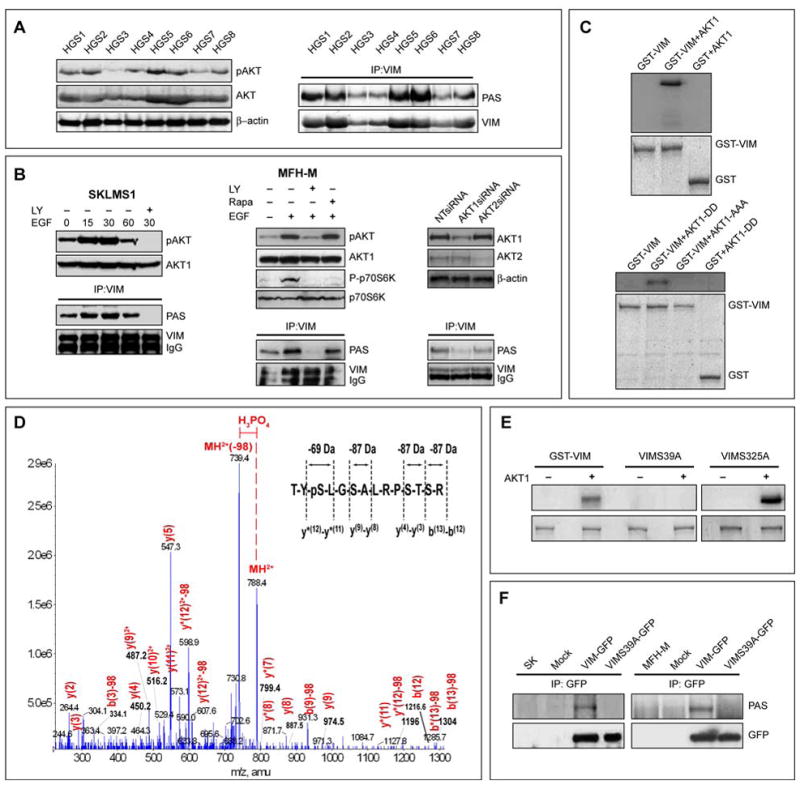
A. Primary human high grade STS (HGS 1-8) express pAKT (pS473; WB); Vim in these samples is recognized by the phospho-AKT substrate antibody (PAS antibody #9611, Cell Signaling; IP/WB) suggesting AKT-induced Vim phosphorylation; B. EGF induced pAKT enhances Vim phosphorylation in a time dependent manner in STS cells as indicated by increase in PAS detection on Vim. This effect was abrogated upon treatment with LY294002 but not by rapamycin. siRNA knockdown of AKT1 but to a lesser degree of AKT2 results in decreased Vim phosphorylation (demonstrated by Vim IP/PAS WB); C. An in vitro AKT1 kinase assay demonstrates the incorporation of [γ-32P]ATP into GST-VIM in the presence of activated AKT1 kinase but not AKT1 kinase dead protein (AKT1-AAA); D. An in vitro AKT1 kinase assay was performed in the presence of cold ATP and two bands corresponding to Vim treated with AKT1 or without AKT1 were subjected to mass spectrometry; shown is the product ion spectra for the of 2+ charge state of the TYSLGSALRPSTSR Vim peptide (788.39). LC/MS/MS data-dependent acquisition; the product ion spectra were searched against the Swissprot database using Mascot (Table S2). The 49 Da (2+) change designated in red indicates that a hydrogen phosphate group (HPO42-) and a water molecule (H2O) were lost from the peptide. y and b ions are indicated in red; the inset reveals a 69 Da change associated with the loss of a phospho-serine versus the 87 Da changes in ion mass associated with the loss of a non-phospho serine: E. In vitro AKT1 kinase assay demonstrated that constitutively activated AKT1 induces the phosphorylation of GST-VIM and GST-VIM-S325A but not GST-VIM-S39A; F. Anti-GFP IP after transfection of STS cells with either VIM-GFP or VIMS39A-GFP constructs demonstrated that Ser39 mutation abrogates AKT-induced Vim phosphorylation. [pAKT WB in all panels refer to pS473] (See also Figure S4)
Two-dimensional PAGE WB with an antibody to Vim further supported Vim phosphorylation by AKT1 (Figure S4C). Protein lysate extracted from cells after treatment with LY294002 or A563 exhibited differential migration of Vim on the isoelectric point axis compared to that of untreated cells, suggesting Vim post-translational modification (most likely related to changes in phosphorylation state) induced by AKT inhibition.
To confirm that Vim is potentially a direct AKT substrate an in vitro AKT kinase assay was conducted. GST-VIM was phosphorylated by activated AKT1 but not by AKT kinase-dead control (Figure 5C). To dissect out the exact AKT phosphorylation site(s) in Vim, an in vitro kinase assay was conducted using cold ATP. Vim separated on SDS-PAGE after incubation with or without active AKT1, was identified via Coomassie blue staining. Corresponding bands were isolated, excised, and digested into tryptic peptides, which were in turn subjected to three complementary phosphoproteomics MS-based methods: MALDI TOF/TOF, liquid chromatography (Bodenstine & Welch)/MS/MS, and multiple reaction monitoring (MRM) (Table S2 and Figures 5D&S4D–G). These studies identified Ser39 of Vim as a potential AKT1 phosphorylation target; no other AKT1-induced phosphorylation sites were found.
We next mutated the GST-VIM-FL construct at Ser39 and Ser325 (another Motif Scan (http://scansite.mit.edu/) predicted AKT phosphorylation site, Table S3) to create GST-VIM-S39A and GST-VIM-S325A constructs and utilized corresponding GST fusion proteins in an AKT1 in vitro kinase assay. Constitutively activated AKT1 induced the phosphorylation of GST-VIM and GST-VIM-S325A, but not GST-VIM-S39A (Figure 5E). Similarly, IP using an anti-GFP antibody after transfection of STS cells with either VIM-GFP or VIMS39A-GFP constructs showed that Ser39 mutation abrogates AKT1-induced Vim phosphorylation (Figure 5F).
AKT1-induced STS cell migration and invasion are mediated by vimentin
We sought to evaluate the possible functional biological implications of AKT1-Vim interaction and phosphorylation. MTS assays demonstrated that Vim knockdown resulted in a small (22% ± 5%; Figure 6A) reduction in growth of STS cells within 24hr; markedly more pronounced was the reduction in STS cell migration/invasion (p<0.01). Next, MCF7 cells (lacking endogenous vimentin) were transfected with Vim, AKT1-DD, WTVim, Vim S39A, or both AKT1-DD and WT or S39A Vim (Figure 6B); as previously shown(Hendrix et al., 1997) WT Vim induced an increase in migration/invasion, whereas mutated Vim S39A or AKT1-DD overexpression alone did not significantly affect these processes. Co-transfection of WT Vim and AKTDD resulted in marked vimentin phosphorylation and a significant (p<0.05) increase in migration/invasion, while transfection of the unphosphorylatable mutated Vim S39A together with AKT-DD did not enhance these processes. To further evaluate the importance of AKT1-induced Vim Ser39 phosphorylation, MCF7 cells were transfected with WT or S39D Vim constructs. The phosphomimetic Vim had a more pronounced effect on MCF7 migration/invasion than WTVim (p<0.05; Figure 6C).
FIGURE 6. AKT1-induced STS cell migration and invasion are mediated, at least in part, by vimentin.
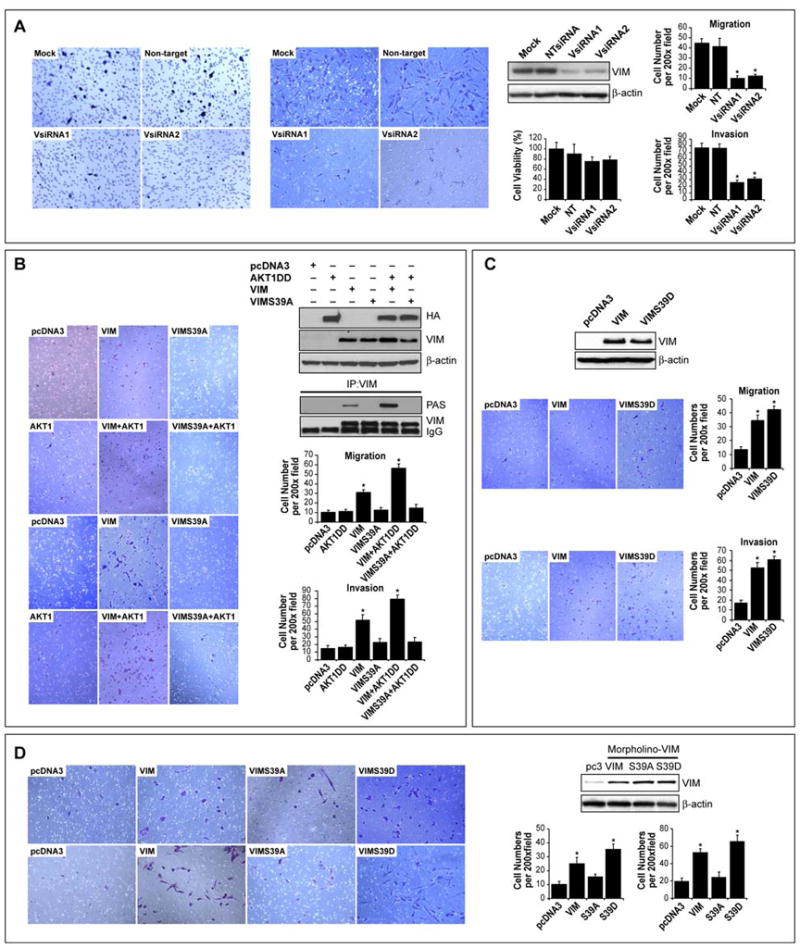
A. Vim knockdown resulted in a small (15% ± 5%) reduction in SKLMS1 growth but a marked decrease in migration/invasion (p<0.01); B. MCF7 cells were transfected with AKT1 DD, Vim, VimS39A individually or in combination. Over-expression of vimentin and to a higher degree vim in conjuncture with AKTDD resulted in the detection of phosphorylated vimentin using Vim IP/PAS WB. WT Vim overexpression in MCF7 cells increased migration/invasion, whereas AKT1-DD overexpression alone did not significantly affect these processes. Moreover, co-transfection of AKT1-DD and WT Vim resulted in the most pronounced effect (p<0.05). In contrast transfection of mutated Vim S39A did not enhance migration/invasion and these processes were not significantly impacted by the co-transfection of AKT-DD and Vim S39A; C. Forced expression of Vim-S39D enhanced the migration/invasion of MCF7 cells as compared to WT Vim (p<0.05); D. Endogenous Vim was knocked down in STS cells (using anti-Vim morpholino) and cells were then transfected with either pcDNA, WT-Vim, Vim-S39A, or Vim-S39D (WB). WT-Vim and Vim-S39D were capable of at least partially rescuing the migratory and invasive phenotype of the Vim knockdown cells (p<0.005), whereas Vim-S39A had only minimal effects (p>0.05). [Graphs represent the average of three repeated experiments ±SEM; * depict statistically significant effects (p<0.05); pAKT WB in all panels refer to pS473]
Next, a rescue experiment in STS cells was conducted where endogenous Vim was first knocked-down using anti-Vim antisense morpholino oligomers (Figure 6D); cells were transfected with either WTVim, Vim-S39A, or Vim-S39D. WTVim and Vim-S39D were capable of, at least partially, rescuing the migratory/invasive phenotype of Vim knocked-down cells (p<0.005), whereas Vim-S39A had only minimal effects (p>0.05). Lastly, SKLMS1 cells stably transfected to overexpress Vim S39D/GFP exhibited significantly larger tumors and a higher experimental metastasis rate when injected to SCID mice as compared to Vim S39A/GFP transfected cells (Table S4).
AKT inhibition induces vimentin proteolysis
The effect of AKT inhibition on Vim in STS cells was next evaluated. A decrease in the total level of full length Vim was observed secondary to both the AKT inhibitor A563 and the PI3K inhibitor LY294002 and was accompanied by an increase in cleaved Vim products and increased activated caspase-3 levels (Figure 7A–B). In contrast, neither the MEK inhibitor UO126 nor the mTORC1 inhibitor rapamycin affected Vim expression or induced caspase activation in STS cells (Figure 7B). Previous data suggest that the initial step in Vim proteolysis is its cleavage by caspase 3(Byun et al., 2001). Caspase activity blockade using Z-VAD but not the proteosome inhibitor MG132 significantly decreased LY294002-induced Vim proteolysis (Figure 7C). Use of frozen tissues from our previously published animal experiments (Zhu et al., 2008), in which HT1080 xenografts were treated with A563, demonstrated that AKT inhibition–induced Vim proteolysis and caspase 3 activation occurs in vivo as well (Figure 7D).
FIGURE 7. AKT inhibition induces caspase dependent vimentin proteolysis in STS cells.
A. Treatment with A563 resulted in a dose dependent decrease in total Vim and an increase in Vim cleavage products; B. Similarly, a dose dependent decrease in full length Vim and its increased proteolysis as well as caspase 3 activation was seen after LY294002 treatment but not with UO126 or rapamycin; C. Caspase activity blockade using Z-VAD markedly decreased LY294002-induced Vim proteolysis; no such effect was observed using the proteosome inhibitor MG132; D. Increase in Vim proteolysis and caspase 3 activation was found in protein extracts from HT1080 xenografts treated with A563; E. SKLMS1 cells after Vim knockdown (using morpholino oligos) were transfected with pcDNA (control), WT or mutated (S39A or S39D) Vim. Treatment with LY294002 resulted in marked proteolysis of WT Vim or S39A VIM, whereas no cleavage was noted in cells expressing the S39D VIM; F. LY294002 treatment resulted in the development of granular GFP-expressing aggregates in SKLMS1 cells transfected with GFP-tagged Vim or GFP-tagged S39A-Vim, but not in cells transfected with GFP-tagged S39D-Vim. [pAKT WB in all panels refer to pS473] (See also Figure S5)
The possible role of Ser39 phosphorylation in protecting Vim from proteolysis was evaluated. Anti-Vim morpholino oligos were used to knockdown native Vim and WT or mutated (S39A or S39D) Vim were forcefully re-expressed. Treatment with LY294002 resulted in marked proteolysis of WTVim or S39A-Vim, whereas no cleavage was noted in cells expressing the S39D-Vim (Figure 7E). Similarly, LY294002 treatment resulted in the development of granular GFP-expressing aggregates in cells transfected with GFP-tagged WTVim or GFP-tagged S39A-Vim, but to a markedly lesser degree in cells transfected with GFP-tagged S39D-Vim (Figure 7F).
Discussion
Our studies identified that AKT1 is constitutively active in STS cells. Moreover, cytokine stimulation induces AKT1 and AKT3 phosphorylation to varying degrees, but not AKT2. The specific mechanisms resulting in this observed differential isoform activation are not known; results here suggest that this may be the consequence of low AKT2 expression levels in STS cells. Other factors such as subcellular localization, differential compartmentalization, selectivity of upstream kinases or phosphatases, and perhaps association with unique scaffold proteins or binding partners affecting their accessibility to their upstream effectors might also play be in effect. While not extensively studied in cells of mesenchymal origin, there is some evidence to support the contention that AKT1 enhances migration and invasion. For example, AKT1 was found to promote migration and invasion of fibroblasts and endothelial cells(Ackah et al., 2005; Kanda et al., 2004). One study evaluated the effect of AKT1 activation on sarcoma cells; overexpression of constitutively active membrane-targeted AKT1 or kinase-dead AKT1 mutants in human fibrosarcoma cells demonstrated that AKT1 enhances motility and invasion, possibly by regulating the expression of MMP9(Kim et al., 2009). Our results using a knockdown approach substantiate and expand this initial observation; taken together, it is possible to conclude that activation of AKT1 (but not AKT2) enhances STS tumor cell motility and invasion. AKT inhibitors are in various development stages; several pan AKT blockers already being evaluated in human clinical trials. Efforts are under way to develop isoform-specific inhibitors to decrease off-target effects and hopefully reduce drug-related toxicity. Based on the results presented here, AKT1 is a particularly attractive candidate for STS therapy.
The observed dependence of AKT function on cancer type, cell type, and AKT isoform can potentially be attributed to unique downstream substrates expressed in a particular intracellular microenvironment. Here we identified Vim as a potential AKT downstream target in STS. Vim is one of the most widely expressed mammalian intermediate filament proteins. In adults, Vim is present in all mesenchymal cells and tissues(Broers et al., 1989) and is therefore frequently used as a differentiation marker. Insights from genetically engineered Vim knockout mice suggest that the physiological structural role of Vim in quiescent mesenchymal cells is potentially redundant and can be carried out by different cell components; Vim knockout mice are viable and do not exhibit overt phenotypes(Colucci-Guyon et al., 1994). This observation is an important consideration if Vim is to be further investigated as a therapeutic target. However, several lines of investigation suggest that Vim has functions that extend beyond simple mechanical and structural roles, including involvement in adhesion, migration and cell survival(Bhattacharya et al., 2009; Eckes et al., 1998; Hendrix et al., 1997; McInroy & Maatta, 2007; Menet et al., 2001). “Hijacking” of normal physiological processes is a hallmark of cancer; a multitude of potential Vim functions raise the possibility that it could contribute to the pro-tumorigenic, pro-metastatic properties of STS cells. Although the exact mechanisms underlying Vim function and its activation are mostly unknown, a variety of studies suggest that Vim phosphorylation is a key regulator of these effects. Phosphorylation plays a major role in regulating Vim structure, dynamic assembly, and interaction with many intracellular structures (including mitochondria and nuclei) and individual proteins (Pittenger et al., 2008). Vim contains a highly complex phosphorylation pattern involving a multitude of kinase-specific sites(Nelson & Traub, 1983) and is recognized as a substrate for several kinases, including RhoA kinase(Sin et al., 1998), protein kinase C (PKC), cGMP kinase, Yes, Raf-1, PAK, and Aurora B (Goto et al., 1998). For example, PKCε-mediated Vim phosphorylation at N-terminal residues was found to induce cell motility via integrin recycling and trafficking to the cell membrane(Ivaska et al., 2005). To our knowledge, the studies presented here are the first to demonstrate an association between AKT and Vim. Furthermore, direct AKT-induced phosphorylation of Vim at Ser39 was identified; alanine mutation of Ser39 perturbed the ability of Vim to induce migration and invasion, whereas expression of mutant Vim with an acidic residue replacing Ser39 enhanced Vim function both in vitro and in vivo. It is of note that the AKT motif found in vimentin is RXXS/T and not the classical RXRXXS/T AKT consensus site; this motif has been identified in a large panel of potential AKT substrates (See Table S3)(Fang et al., 2007; Gu et al., 2006; Luo et al., 2007; Qi et al., 2006). Being a non-classical AKT phosphorylation motif, we cannot rule out that the observed AKT induced phosphorylation of vimentin in STS cells of functional consequences is not a result of an indirect effect due to AKT activation of AGC kinases that are possibly recruited to vimentin secondary to AKT:vimentin binding and in turn phosphorylate vimentin. Whatever is the case, taken together, our findings identify a novel mechanism of AKT function relevant to STS cells and add an additional downstream target to those with which AKT interacts.
AKT is known for its pro-survival effects; AKT blockade results in significant apoptosis in a multitude of cancer models, including STS(Zhu et al., 2008). Data presented here offer a potential novel mechanism for AKT blockade induced apoptosis affecting Vim-expressing cells. A role for Vim in apoptosis was recently elucidated(Byun et al., 2001). Vim undergoes rapid proteolysis upon diverse pro-apoptotic stimuli, including ionizing radiation, Fas, TRAIL, TNFα, and tamoxifen. This Vim proteolysis occurs secondary to caspase pathway activation, resulting in cleavage of Vim by multiple caspases at several sites (Asp85, Asp259, and Asp429;(Byun et al., 2001). It is possible that Vim proteolysis and collapse contribute to many of the morphological manifestations of apoptosis, including cellular rounding, nuclear condensation, and packaging of the debris of dying cells into apoptotic bodies. Furthermore, Vim proteolysis releases potential pro-apoptotic proteolytic fragments that can markedly enhance apoptosis. For example, the generation of a short N-terminal cleavage product (amino acids 1-85) was shown to play an active pro-apoptotic role (Morishima, 1999). A positive feedback loop is suggested whereby activated caspases induce the cleavage of Vim and these cleavage products in turn activate caspases to amplify apoptosis. To our knowledge, AKT blockade-induced Vim proteolysis has not been previously described. AKT inhibition is known to induce the activation of caspases; our results elucidate the mechanism by demonstrating that AKT blockade induces caspase-dependent Vim proteolysis in vitro and in vivo. Our results also suggest that AKT-induced Vim phosphorylation protects Vim (to some extent) from caspase proteolysis, suggesting a potential pro-survival effect of AKT. Future clinical trials evaluating the effect of AKT inhibitors on STS could assess the utility of measuring Vim cleavage as a marker of apoptosis/therapeutic efficacy.
In summary, many key regulatory roles played by AKT in STS cells could be at least partially mediated through its interaction with and consequential phosphorylation of Vim. Vim could therefore potentially be selectively targeted for STS therapy. This previously unexplored biology has important therapeutic implications and merits further investigation.
Materials and Methods
Cell Lines and Reagents
A list of cell lines, human cell strains (including isolation and characterization), and culture conditions is provided in the Supplemental Experimental Procedures. A comprehensive list of all commercial antibodies, inhibitors, cytokines, constructs and transfection procedures are described in detail in Supplementary data.
Cell Growth
Cell growth assays were done with a CellTiter96 Aqueous Non-Radioactive Cell Proliferation Assay kit (Promega), per manufacturer instructions. Absorbance was measured at 490 nm; absorbance of treated cells is graphically presented as a percentage of control cell absorbance.
Migration and Invasion Assays
Migration and invasion assays were conducted as described previously(Liu et al., 2006); further detail is provided in supplementary data.
Western Blotting and Coimmunoprecipitation
WB analysis, coimmunoprecipitation and two-dimensional PAGE western blot analysis were performed by standard methods. Brief details are provided in the supplementary data.
Mass Spectrometry
In-Gel Trypsin Digestion methodology is described in supplementary data. MALDI-TOF MS was used for AKT1 binding partners protein identification (detailes are provided in supplementary data). Three complementary MS-based methods were used for phosphoproteomics: MALDI TOF/TOF, liquid chromatography (Bodenstine & Welch)/MS/MS, and multiple reaction monitoring (MRM). Further details can be found in supplementray data.
GST Fusion Protein Pull-Down and In Vitro Protein Binding Assays
Expression of GST and GST-tagged full-length Vim, Vim fragments, full-length AKT1, and AKT1 fragments was induced in E. coli BL21, and proteins were purified by immobilization on glutathione-Sepharose-4B beads (GE Healthcare). Further detail is provided in the Supplemental data. For GST fusion protein pull-down assays, 50 μl of beads (5 μg of immobilized protein) were incubated with 500–1,000 μg of whole-cell lysates in 1% NP-40 buffer for 4 h at 4°C. For in vitro protein binding assays, 10 μl of beads (1 μg of immobilized protein) were incubated with commercially available recombinant human Vim (1 μg) or AKT1 (1 μg) in a 1% NP-40 buffer for 2 h at 4°C. Beads were spun down and washed with cold 1% NP-40 in 1x PBS (1 ml each), boiled in 10 μl of 4x SDS gel loading buffer, and subjected to SDS–PAGE followed by western blotting.
Nonradioactive AKT Kinase Assay and In Vitro AKT Kinase Assay
Nonradioactive Akt Kinase Assay Kit providing all the reagents necessary to measure AKT 1 or 2 kinase activity was purchased from Cell Signaling. Immobilized AKT antibodies were used to immunoprecipitate AKT 1 or w from cell extracts. Then, a kinase assay was performed using GSK-3 Fusion Protein as a substrate in the presence of 200 μM of cold ATP. Phosphorylation of GSK-3 was measured by WB, using pGSK-3 Ab.
Constitutively activated AKT1 or kinase dead AKT were incubated with 10 μl of GST affinity matrix bead-purified wild-type or mutated Vim proteins (details provided in the Supplemental Experimental Procedures) in the presence of 5 μCi of [γ-32P]ATP, 50 mM cold ATP, and 1.5x AKT1 kinase buffer (75 mM Tris (pH 7.4), 25 mM MgCl2, 2 mM dithiothreitol, 0.1 mM NaVO3, 1.5 μM proteinase inhibitor, and 1.5 mg/ml bovine serum albumin) for 30 min at room temperature. The reaction mixture was spun down, and the beads were washed with PBS. After the addition of 10 μl of 4x SDS gel loading buffer, reaction products were resolved by SDS-PAGE and γ-32P-labeled proteins were visualized by autoradiography. The experiment was repeated excluding [γ-32P]ATP (only cold ATP was added) for submission of Vim to phosphoprotein mass spectrometry analysis. Bands were identified via Coomassie blue staining.
Animal Experiments
All animal procedures and care was approved by the Institutional Animal Care and Usage Committee of UTMDACC and are described in supplementary data.
Statistical Analysis
All experiments were repeated at least three times to ensure reproducibility. Cell proliferation, migration, and invasion assays were repeated three times, and means ± SD were calculated. Two sample t tests were used to assess the differences. Significance was set at p≤0.05. Statistical considerations relating to mass spectrometry studies are described in the supplementary data section. Differences in xenograft growth in vivo were assessed using a two-tailed Student’s t test. Significance was set at p≤0.05. All computations were performed with SAS 9.1 (SAS Institute) and S-PLUS 7.0 (Insightful).
Supplementary Material
Acknowledgments
We thank Dr. Giranda (Abbot Laboratories, Abbott Park, IL) for providing A674563 (A563), Dr. Cryns (Division of Endocrinology, Northwestern University, Chicago, IL) for providing human Vim cDNA, and Dr Fletcher (Brigham and Women’s Hospital, Boston, MA) for the MPNST724 cell line. The UTMB NHLBI Proteomic Center is thanked for the assistance with Mass Spectrometry experiments (The work was supported by the NIH/NHLBI proteomics Initiative NO1-HV-28184 [to K.P.R.]). Ms. Vu is thanked for aid in figure preparation and Ms. Lo for her assistance with scientific editing. This manuscript was supported in part by an NIH RO1 grant CA138345 and a RTOG seed grant (to DL). Dr Rosenblatt is a recipient of a Welch Foundation Endowment in Chemistry and Related Science Fellowship award (Grant # L-AU-0002). All experiments conducted at UT-MDACC Core Facilities were further supported by an NCI Cancer Center Support Grant (CA#16672)
Financial Support: This manuscript was supported by an NIH/NCI RO1 grant CA138345 and a RTOG seed grant (to DL), an NIH/NHLBI proteomics Initiative grant NO1-HV-28184 (to KPR)
Footnotes
Conflict of Interest: The authors declare that they have no conflict of interest
The Supplemental Data include 5 figures, 5 tables, and Supplemental Experimental Procedures
References
- Ackah E, Yu J, Zoellner S, Iwakiri Y, Skurk C, Shibata R, Ouchi N, Easton RM, Galasso G, Birnbaum MJ, Walsh K, Sessa WC. J Clin Invest. 2005;115:2119–27. doi: 10.1172/JCI24726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arboleda MJ, Lyons JF, Kabbinavar FF, Bray MR, Snow BE, Ayala R, Danino M, Karlan BY, Slamon DJ. Cancer Res. 2003;63:196–206. [PubMed] [Google Scholar]
- Bhattacharya R, Gonzalez AM, Debiase PJ, Trejo HE, Goldman RD, Flitney FW, Jones JC. J Cell Sci. 2009;122:1390–400. doi: 10.1242/jcs.043042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenstine TM, Welch DR. Cancer Microenviron. 2008;1:1–11. doi: 10.1007/s12307-008-0001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broers JL, de Leij L, Rot MK, ter Haar A, Lane EB, Leigh IM, Wagenaar SS, Vooijs GP, Ramaekers FC. Differentiation. 1989;40:119–28. doi: 10.1111/j.1432-0436.1989.tb00821.x. [DOI] [PubMed] [Google Scholar]
- Byun Y, Chen F, Chang R, Trivedi M, Green KJ, Cryns VL. Cell Death Differ. 2001;8:443–50. doi: 10.1038/sj.cdd.4400840. [DOI] [PubMed] [Google Scholar]
- Clark MA, Fisher C, Judson I, Thomas JM. N Engl J Med. 2005;353:701–11. doi: 10.1056/NEJMra041866. [DOI] [PubMed] [Google Scholar]
- Colucci-Guyon E, Portier MM, Dunia I, Paulin D, Pournin S, Babinet C. Cell. 1994;79:679–94. doi: 10.1016/0092-8674(94)90553-3. [DOI] [PubMed] [Google Scholar]
- Eckes B, Dogic D, Colucci-Guyon E, Wang N, Maniotis A, Ingber D, Merckling A, Langa F, Aumailley M, Delouvee A, Koteliansky V, Babinet C, Krieg T. J Cell Sci. 1998;111 ( Pt 13):1897–907. doi: 10.1242/jcs.111.13.1897. [DOI] [PubMed] [Google Scholar]
- Fang D, Hawke D, Zheng Y, Xia Y, Meisenhelder J, Nika H, Mills GB, Kobayashi R, Hunter T, Lu Z. J Biol Chem. 2007;282:11221–9. doi: 10.1074/jbc.M611871200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto H, Kosako H, Tanabe K, Yanagida M, Sakurai M, Amano M, Kaibuchi K, Inagaki M. J Biol Chem. 1998;273:11728–36. doi: 10.1074/jbc.273.19.11728. [DOI] [PubMed] [Google Scholar]
- Grille SJ, Bellacosa A, Upson J, Klein-Szanto AJ, van Roy F, Lee-Kwon W, Donowitz M, Tsichlis PN, Larue L. Cancer Res. 2003;63:2172–8. [PubMed] [Google Scholar]
- Gu YM, Jin YH, Choi JK, Baek KH, Yeo CY, Lee KY. FEBS Lett. 2006;580:305–10. doi: 10.1016/j.febslet.2005.12.024. [DOI] [PubMed] [Google Scholar]
- Hendrix MJ, Seftor EA, Seftor RE, Trevor KT. Am J Pathol. 1997;150:483–95. [PMC free article] [PubMed] [Google Scholar]
- Hernando E, Charytonowicz E, Dudas ME, Menendez S, Matushansky I, Mills J, Socci ND, Behrendt N, Ma L, Maki RG, Pandolfi PP, Cordon-Cardo C. Nat Med. 2007;13:748–53. doi: 10.1038/nm1560. [DOI] [PubMed] [Google Scholar]
- Hutchinson JN, Jin J, Cardiff RD, Woodgett JR, Muller WJ. Cancer Res. 2004;64:3171–8. doi: 10.1158/0008-5472.can-03-3465. [DOI] [PubMed] [Google Scholar]
- Irie HY, Pearline RV, Grueneberg D, Hsia M, Ravichandran P, Kothari N, Natesan S, Brugge JS. J Cell Biol. 2005;171:1023–34. doi: 10.1083/jcb.200505087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivaska J, Vuoriluoto K, Huovinen T, Izawa I, Inagaki M, Parker PJ. Embo J. 2005;24:3834–45. doi: 10.1038/sj.emboj.7600847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju X, Katiyar S, Wang C, Liu M, Jiao X, Li S, Zhou J, Turner J, Lisanti MP, Russell RG, Mueller SC, Ojeifo J, Chen WS, Hay N, Pestell RG. Proc Natl Acad Sci U S A. 2007;104:7438–43. doi: 10.1073/pnas.0605874104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanda S, Miyata Y, Kanetake H. J Biol Chem. 2004;279:4007–16. doi: 10.1074/jbc.M307569200. [DOI] [PubMed] [Google Scholar]
- Kim MA, Lee HS, Lee HE, Kim JH, Yang HK, Kim WH. Histopathology. 2009;54:442–51. doi: 10.1111/j.1365-2559.2009.03247.x. [DOI] [PubMed] [Google Scholar]
- Liu H, Radisky DC, Nelson CM, Zhang H, Fata JE, Roth RA, Bissell MJ. Proc Natl Acad Sci U S A. 2006;103:4134–9. doi: 10.1073/pnas.0511342103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Langlais P, Yi Z, Lefort N, De Filippis EA, Hwang H, Christ-Roberts CY, Mandarino LJ. Endocrinology. 2007;148:4895–905. doi: 10.1210/en.2007-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroulakou IG, Oemler W, Naber SP, Tsichlis PN. Cancer Res. 2007;67:167–77. doi: 10.1158/0008-5472.CAN-06-3782. [DOI] [PubMed] [Google Scholar]
- McInroy L, Maatta A. Biochem Biophys Res Commun. 2007;360:109–14. doi: 10.1016/j.bbrc.2007.06.036. [DOI] [PubMed] [Google Scholar]
- Menet V, Gimenez y Ribotta M, Chauvet N, Drian MJ, Lannoy J, Colucci-Guyon E, Privat A. J Neurosci. 2001;21:6147–58. doi: 10.1523/JNEUROSCI.21-16-06147.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki T, Honda K, Ohata H. Am J Physiol Cell Physiol. 2007;293:C1216–25. doi: 10.1152/ajpcell.00083.2007. [DOI] [PubMed] [Google Scholar]
- Morishima N. Genes Cells. 1999;4:401–14. doi: 10.1046/j.1365-2443.1999.00270.x. [DOI] [PubMed] [Google Scholar]
- Nelson WJ, Traub P. Mol Cell Biol. 1983;3:1146–56. doi: 10.1128/mcb.3.6.1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phung TL, Ziv K, Dabydeen D, Eyiah-Mensah G, Riveros M, Perruzzi C, Sun J, Monahan-Earley RA, Shiojima I, Nagy JA, Lin MI, Walsh K, Dvorak AM, Briscoe DM, Neeman M, Sessa WC, Dvorak HF, Benjamin LE. Cancer Cell. 2006;10:159–70. doi: 10.1016/j.ccr.2006.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittenger JT, Hess JF, Budamagunta MS, Voss JC, Fitzgerald PG. Biochemistry. 2008;47:10863–70. doi: 10.1021/bi801137m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi XJ, Wildey GM, Howe PH. J Biol Chem. 2006;281:813–23. doi: 10.1074/jbc.M505546200. [DOI] [PubMed] [Google Scholar]
- Sin WC, Chen XQ, Leung T, Lim L. Mol Cell Biol. 1998;18:6325–39. doi: 10.1128/mcb.18.11.6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulis ML, Parsons R. Trends Cell Biol. 2003;13:478–83. doi: 10.1016/s0962-8924(03)00175-2. [DOI] [PubMed] [Google Scholar]
- Sumitani S, Goya K, Testa JR, Kouhara H, Kasayama S. Endocrinology. 2002;143:820–8. doi: 10.1210/endo.143.3.8687. [DOI] [PubMed] [Google Scholar]
- Tanno S, Tanno S, Mitsuuchi Y, Altomare DA, Xiao GH, Testa JR. Cancer Res. 2001;61:589–93. [PubMed] [Google Scholar]
- Tomita Y, Morooka T, Hoshida Y, Zhang B, Qiu Y, Nakamichi I, Hamada K, Ueda T, Naka N, Kudawara I, Aozasa K. Clin Cancer Res. 2006;12:3070–7. doi: 10.1158/1078-0432.CCR-05-1732. [DOI] [PubMed] [Google Scholar]
- Yang J, Weinberg RA. Dev Cell. 2008;14:818–29. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- Yoeli-Lerner M, Yiu GK, Rabinovitz I, Erhardt P, Jauliac S, Toker A. Mol Cell. 2005;20:539–50. doi: 10.1016/j.molcel.2005.10.033. [DOI] [PubMed] [Google Scholar]
- Zhu QS, Ren W, Korchin B, Lahat G, Dicker A, Lu Y, Mills G, Pollock RE, Lev D. Cancer Res. 2008;68:2895–903. doi: 10.1158/0008-5472.CAN-07-6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.