Abstract
Overexpression of human AP-endonuclease (APE1/Ref-1), a key enzyme in the DNA base excision repair (BER) pathway, is often associated with tumor cell resistance to various anticancer drugs. In this study, we examined the molecular basis of transcriptional regulatory (non repair) function of APE1 in promoting resistance to certain types of drugs. We have recently shown that APE1 stably interacts with Y-box-binding protein 1 (YB-1), and acts as its coactivator for the expression of multidrug resistance gene MDR1, thereby causing drug-resistance. Here we show for the first time that APE1 is stably associated with the basic transcription factor RNA polymerase II (RNA pol II) and the coactivator p300 on the endogenous MDR1 promoter. APE1’s depletion significantly reduces YB-1/p300 recruitment to the promoter, resulting in reduced RNA pol II loading. Drug-induced APE1 acetylation which is mediated by p300 enhances formation of acetylated APE1 (AcAPE1)/YB-1/p300 complex on the MDR1 promoter. Enhanced recruitment of this complex increases MDR1 promoter dependent luciferase activity and its endogenous expression. Using APE1 downregulated cells and cells overexpressing wild type APE1 or its nonacetylable mutant we have demonstrated that the loss of APE1’s acetylation impaired MDR1 activation and sensitizes the cells to cisplatin or etoposide. We have thus established the basis for APE1’s acetylation-dependent regulatory function in inducing MDR1-mediated drug resistance.
Keywords: APE1, acetylation, YB-1/p300, RNA pol II, MDR1
Introduction
The multifunctional mammalian AP-endonuclease (APE1) plays a central role in the DNA base excision repair (BER) pathway for repairing damaged bases, abasic (AP) sites and their oxidation products generated in the genome either spontaneously or after excision of oxidized and alkylated bases by DNA glycosylases (Demple and Harrison, 1994; Doetsch and Cunningham, 1990; Fan and Wilson, 2005; Izumi et al., 2003; Matsumoto and Kim, 1995; Mitra et al., 2002; Singhal et al., 1995). Unrepaired AP sites and DNA strand breaks induce apoptosis and cytotoxicity (Loeb and Preston, 1986). Besides its repair function, the mammalian APE1 possesses two unique and apparently distinct transcriptional regulatory activities that require its nonconserved N-terminal domain (Bhakat et al., 2009; Evans et al., 2000; Izumi et al., 2005; Tell et al., 2009). APE1 was identified as a reductive activator of c-Jun invitro and named redox effector factor-1 (Ref-1; (Xanthoudakis and Curran, 1992) and was subsequently shown to activate several other transcription factors e.g., NF-κB, HIF1-α, p53, Pax5, Pax8, and c-Myb, presumably via its redox activity (Evans et al., 2000; Jayaraman et al., 1997; Xanthoudakis et al., 1992). APE1 could also act as a trans-acting factor that was discovered in the trans-acting complex that binds to the negative Ca2+ response elements (nCaRE-A and B) during Ca2+-dependent downregulation of the parathyroid hormone (PTH) gene (Okazaki et al., 1994). The presence of the nCaRE-B element and binding of APE1 to this element was also shown in the human renin gene promoter (Fuchs et al., 2003). Thus APE1’s role as a regulatory factor in diverse transacting complexes involved in the activation or repression of various genes were documented (Bhakat et al., 2009). We discovered acetylation of human APE1 at Lys6 and Lys7 by the histone acetyltransferase (HAT) activity of p300 (Bhakat et al., 2003a), and that a significant fraction of APE1 is normally present in the acetylated form (AcAPE1) in many cell lines (Bhakat et al., 2009; Bhattacharyya et al., 2009; Chattopadhyay et al., 2008). APE1’s acetylation is regulated by the deacetylase activity of SIRT1 both in cells and invitro (Yamamori et al.,2010) and SIRT1 mediated deacetylation of APE1 enhances its association with XRCC1 which regulates its BER activity (Yamamori et al.,2010). It appears that APE1 is in dynamic equilibrium between its acetylated and unmodified state and the fraction of APE1 that is acetylated varies among different tumor cell lines (Bhakat et al., 2009). We showed that APE1 acetylation stimulates formation of the nCaRE-B complex which contains hnRNP-L and HDAC1 leading to repression of the PTH gene (Bhakat et al., 2003a; Kuninger et al., 2002). Similarly, Crowe’s group in collaboration with us has recently shown that Helicobacter pylori infection induces acetylation of APE1 in gastric epithelial cells; AcAPE1 suppresses Bax expression, and modulates p53-dependent apoptosis of H. Pyroli-infected cells (Bhattacharyya et al., 2009). Furthermore, we have shown that early growth response 1 (Egr-1)-mediated activation of phosphoinositol phosphatase and tensin homologue (PTEN) gene is dependent on APE1 acetylation (Fantini et al., 2008). Thus we have established a novel role of acetylation-mediated transcriptional regulatory function of APE1 in the regulation of diverse genes. APE1 was also found to be ubiquitinated at multiple lysine residues in the N terminal region (Busso et al., 2009).
APE1 is often overexpressed in tumor cells, and its altered level or intracellular distribution has been found in various cancer tissues including ovarian, cervical, prostate, glioma, head and neck, and non-small-cell lung carcinomas (Evans et al., 2000; Kelley et al., 2001; Robertson et al., 2001; Xu et al., 1997). APE1 overexpression is invariably associated with resistance to various anticancer drugs; its downregulation or functional impairment sensitizes cells to diverse genotoxic agents including MMS, H2O2, bleomycin, TMZ, BCNU, etoposide, cisplatin and doxorubicin (Bapat et al., 2009; Bobola et al., 2005; Chattopadhyay et al., 2008; Fishel and Kelley, 2007; Jiang et al., 2008; McNeill et al., 2009; Robertson et al., 1997; Wang et al., 2004). While APE1-dependent repair of the cytotoxic lesions induced by some of these agents, eg. MMS or H2O2 could explain enhanced sensitivity to some chemicals, the DNA damage induced by many other drugs such as etoposide or doxorubicin is not repaired via APE1-mediated BER pathway. Hence the higher sensitivity to these drugs cannot be explained by the loss of APE1’s DNA repair function. So, at least in these cases, the loss of APE1’s regulatory function is likely to be responsible for drug sensitivity. In support of this possibility, we have recently shown that APE1 stably interacts with YB-1, a major transcription factor for the activation of multidrug resistance gene MDR1 (Chattopadhyay et al., 2008). The MDR1 gene product, P-glycoprotein, an N-glycosylated plasma membrane protein and a member of ATP-binding cassette (ABC) transporters consists of two highly homologous halves each of which contains a transmembrane domain with several membrane spanning segments and an ATP-binding fold (Germann and Chambers, 1998). Increased level of this P-glycoprotein in mammalian cells has been found to be related to ATP-dependent reduced drug accumulation and this suggests its role as an energydependent drug efflux pump (Germann and Chambers, 1998). Overexpression of MDR1 confers resistance to a variety of structurally and functionally unrelated antitumor drugs such as vincristine, doxorubicin, etoposide and many others (Chaudhary and Roninson, 1993; Goldstein et al., 1989; Gottesman et al., 2002; Gottesman and Pastan, 1993; Kohno et al., 1989). Both transcriptional activation and gene amplification have been implicated in acquisition of high levels of MDR1 expression in intrinsic or acquired multidrug resistance in tumor cells (Kohno et al., 1994; Shen et al., 1986). Furthermore, protein kinase C mediated phosphorylation of this transporter enhances its drug-efflux and ATPase activity (Ahmad and Glazer, 1993; Ahmad et al., 1994; Yu et al., 1991).
We showed that APE1 downregulation in cisplatin-resistant ovarian cancer cell line A2780 and doxorubicin-resistant breast cancer cell line MCF7 decreases their MDR1 levels and sensitizes these cells to cisplatin or doxorubicin (Chattopadhyay et al., 2008). However, the molecular basis for YB-1 mediated activation of MDR1 by APE1 and its acetylation is not clear. In this study, we have shown that drug-induced APE1 acetylation enhances YB-1/p300 complex formation on MDR1 promoter. Furthermore, we have shown for the first time that APE1 is stably associated with RNA polymerase II (RNA pol II) on the MDR1 promoter and plays a key role in both basal and drug-induced recruitment of YB-1/p300 complex and RNA pol II loading. Thus, we have documented a novel mechanism by which APE1 and its acetylation regulate MDR1 expression, and provided a molecular basis for sensitization of cells to many drugs via APE1 downregulation.
Results
Stable association of APE1 with p300 and RNA pol II
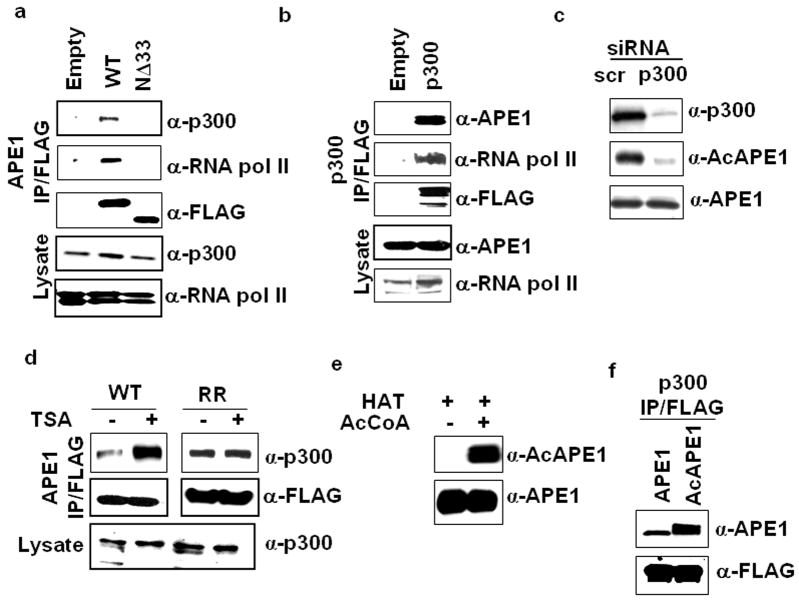
We showed previously that APE1 is acetylated at Lys6/Lys 7 by p300 (Bhakat et al., 2003a) and this acetylation enhances its association with YB-1 (Chattopadhyay et al., 2008). We tested stable association between APE1 and p300 by co-immunoprecipitation (Co-IP) analysis with nuclear extracts of HEK-293T cells ectopically expressing C-terminally FLAGtagged full-length or N terminal 33 amino acid deleted (NΔ33) APE1. Western analysis showed the presence of p300 in the FLAG IP of full length but not of NΔ33 APE1 (Figure 1a) indicating that the N terminal region of APE1 is required for its stable interaction with p300. We further confirmed this interaction by reciprocal analysis and showed the presence of APE1 in the FLAGp300 IP (Figure 1b). To test p300 mediated acetylation of APE1 in cells, we downregulated cellular p300 level with siRNA in HEK-293T cells and checked AcAPE1 level with our AcAPE1 specific antibody (Chattopadhyay et al., 2008). p300 downregulation caused significant decrease in APE1 acetylation without affecting the total APE1 level (Figure 1c). Several studies have shown that acetylation is a common strategy that p300/CBP utilizes to enhance interaction with nonhistone proteins and that the p300 bromodomain is involved in binding to acetylated Lys residues of partner proteins (Mujtaba et al., 2002). We examined whether APE1 acetylation modulates its interaction with p300. We immunoprecipitated FLAG-tagged wild type (WT) APE1 or its nonacetylable K6R/K7R (RR) mutant from the nuclear lysates of Trichostatin A (TSA, a histone deacetylase inhibitor; Yoshida et al., 1990) treated or control cells and examined the level of p300 in the IPs. TSA treatment significantly enhanced the amount of p300 bound to WT but not to RR APE1 (Figure 1d) indicating that acetylation enhances APE1’s association with p300. This was further confirmed by invitro interaction of unmodified or acetylated APE1 (Figure 1e) with FLAG- tagged p300. Higher level of bound AcAPE1 compared to the unmodified APE1 was observed (Figure 1f).
Figure 1.
Association of APE1 with p300 and RNA pol II. (a) Nuclear lysates of HEK-293T cells transfected with empty vector, FLAG-tagged WT APE1 or N terminal 33 aa deleted (NΔ33) APE1 mutant were immunoprecipitated (IP) with FLAG antibody. Western analysis of the IPs with p300, RNA pol II or FLAG antibody; p300 and RNA pol II levels in the lysates (lower panels). (b) Nuclear extracts of HEK-293T cells transfected with empty vector or FLAG tagged p300 were immunoprecipitated with FLAG antibody. Western analysis of the IPs with APE1, RNA pol II or FLAG antibody; APE1 and RNA pol II levels in the lysates (lower panels). (c) HEK-293T cells were transfected with 80 nM of p300–specific or scrambled (scr) duplex siRNAs. Western analysis after 48 hours for p300 (upper panel), AcAPE1 (middle panel) and APE1 (lower panel) levels in cell extracts. (d) HEK-293T cells transfected with FLAG-tagged WT APE1 (WT) or K6R/K7R APE1 (RR) were treated with TSA (100 ng/ml) for 6 hours and then the nuclear extracts were immunoprecipitated with FLAG antibody. Western analysis of the IPs with p300 (upper panel) or FLAG (middle panel) antibody; p300 levels in the lysates (lower panel). (e) Recombinant APE1 (2 μg) was incubated with p300 HAT domain in presence (+) or absence (-) of 1 mM acetyl CoA (AcCoA) for 2 hours at 30°C. Western analysis of the invitro acetylated APE1 with AcAPE1 (upper panel) or APE1 (lower panel) antibody. (f) Immunoprecipitated p300-FLAG beads were incubated with 100 ng of unmodified or invitro acetylated APE1. After washing, the bound proteins were eluted with SDS/Laemmli buffer and immunoblotted with APE1 (upper panel) or FLAG (lower panel) antibody.
Transcriptional activation by a transcription factor involves recruitment of various coactivators to the specific binding site on DNA, which then directly or indirectly interacts with components of the basal transcription machinery including RNA pol II and stabilizes the transcription preinitiation complex (Cho et al., 1998). Because p300 is also known to function as a bridging factor connecting sequence specific transcription factors with the basal transcription machinery (Cho et al., 1998; Neish et al., 1998) including RNA pol II, it is possible that APE1 could be associated with the RNA pol II transcription complex. Western analysis of the FLAG IP of nuclear extract from cells expressing FLAG-APE1 showed the presence of RNA pol II in the immunoprecipitate of full length but not of NΔ33 APE1 (Figure 1a). As expected, we also showed the presence of RNA pol II in immuno-pull down of FLAG-p300 (Figure 1b).
Requirement of APE1 in p300/YB-1complex formation and RNA pol II loading on MDR1 promoter
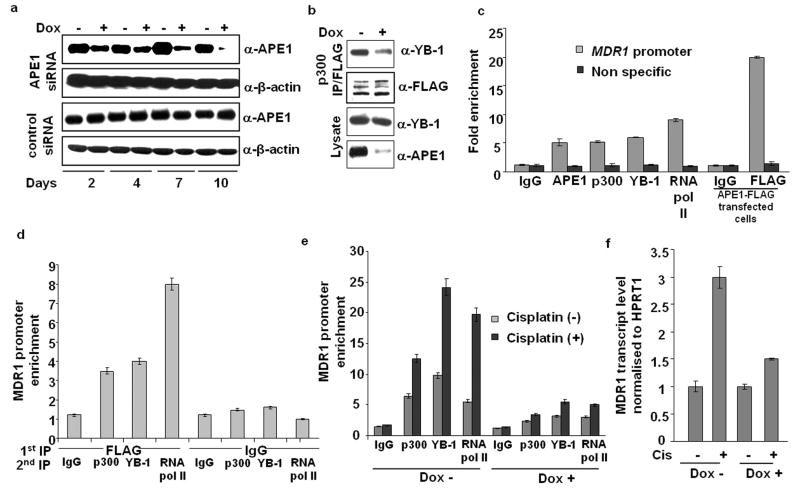
Several studies showed that YB-1 is a major transcription factor for MDR1 gene (Bargou et al., 1997; Kohno et al., 1994). Because p300 interacts with both YB-1 and APE1, we asked whether the interaction between YB-1 and p300 is mediated by APE1. We generated a HEK- 293T cell line stably expressing APE1 siRNA (APE1siRNAHEK-293T) driven by doxycycline (Dox) inducible promoter as described earlier (Vascotto et al., 2009). After 7-10 days of Dox treatment, endogenous APE1 expression was minimal compared to cells expressing control siRNA (Figure 2a). We transfected APE1 downregulated APE1siRNAHEK-293T cells with p300-FLAG construct and immunoprecipitated the nuclear extracts with FLAG Ab for Western analysis. Significant reduction in the amount of YB-1 bound to p300 was observed in the IP from APE1 downregulated cells compared to control cells (Figure 2b). These results indicate APE1’s requirement in facilitating YB-1/p300 complex formation.
Figure 2.
Requirement of APE1 in YB-1/p300 recruitment and RNA pol II loading on MDR1 promoter. (a) HEK-293T cells expressing APE1 siRNA (APE1siRNAHEK-293T cells) or control duplex siRNA (ControlsiRNAHEK-293T cells) under a Doxycycline (Dox)-inducible promoter were established following the procedure described in Materials & Methods. These cells were treated with Dox (1ug/ml) for the indicated times and APE1 levels were measured by Western analysis; β-actin antibody was used as a loading control. (b) APE1siRNAHEK-293T cells were treated with (+) or without (-) Dox for 8 days and then the cells were transfected with p300-FLAG expression plasmid. Nuclear extracts were immunoprecipitated with FLAG antibody and Western analysis of the IPs was performed with YB-1 or FLAG antibody; YB-1 and APE1 levels in the lysates (lower panels). (c) CHIP assay in HEK-293T cells or APE1-FLAG transfected HEK-293T cells with the indicated antibodies. Immunoprecipitated Y-box element containing MDR1 promoter sequence or a non specific coding sequence in the MDR1 gene distant to Y-box element was amplified and quantified by SYBR GREEN based Real Time PCR analysis. Values in the bar diagram are relative to PCR from input chromatin. (d) re-ChIP assay of immunoprecipitated APE1-FLAG bound chromatin from APE1 FLAG transfected HEK-293T cells with p300, YB-1 or RNA pol II Ab or control IgG. The first IP was carried out with FLAG antibody or control IgG and the second IP was carried out with p300, YB-1 or RNA pol II Antibody or control IgG. (e) APE1 levels in APE1siRNAHEK-293T cells were downregulated with Dox treatment. Then the cells were treated with (+) or without (-) cisplatin for 1 hour, and 5 hours later, ChIP assay was carried out with p300, YB-1 or RNA pol II Antibody or control IgG. (f) Real Time RT-PCR analysis of MDR1 transcript. APE1 levels in APE1siRNAHEK-293T cells were downregulated with Dox treatment. The cells were then treated with cisplatin for 1 hour and 16 hours later total RNA was isolated and subjected to cDNA synthesis and Real Time PCR. Values in the bar diagram (normalized with respect to HPRT1 transcript) are relative to cisplatin untreated control. All the results represent the mean ± standard deviations of 3 independent experiments performed in duplicates.
In order to examine whether APE1-dependent YB-1/p300 complex formation is essential for MDR1 expression, we showed their occupancy on the MDR1 promoter Y-box element by ChIP assay in HEK-293T cells with antibodies to APE1, YB-1, p300 or RNA pol II. The amount of immunoprecipitated MDR1 promoter sequence or a coding sequence of this gene distant to the Y-box element was quantified by Real Time PCR analysis. Significant enrichment of the MDR1 promoter Y-box element and not of the coding sequence was observed with APE1, p300, YB-1 or RNA pol II antibody as compared to control IgG (Figure 2c), indicating that all of these proteins are stably and specifically associated with MDR1 promoter. To establish their simultaneous promoter occupancy, we performed re-ChIP assays using HEK-293T cells transfected with APE1-FLAG construct. Immunoprecipitated chromatin with FLAG antibody or control IgG was secondarily immunoprecipitated with p300, YB-1 or RNA pol II antibody. Significant enrichment of the MDR1 promoter was observed in the FLAG IP as compared to control IgG (Figure 2c) confirming association of FLAG-APE1 with MDR1 promoter. Figure 2d provides strong evidence for simultaneous association of p300, YB-1 and RNA pol II with APE1 on the Y-box element on MDR1 promoter because APE1-bound chromatin (FLAG IP) could be immunoprecipitated with the corresponding antibodies as compared to control IgG.
We next examined whether APE1 is necessary for the recruitment of p300, YB-1 and RNA pol II on MDR1 promoter. ChIP analysis was performed in cisplatin treated or control APE1 downregulated APE1siRNAHEK-293T cells. Figure 2e shows that APE1 downregulation significantly reduced p300, YB-1 and RNA pol II occupancy on the MDR1 promoter. MDR1 promoter sequence was enriched in the IPs with p300, YB-1 or RNA pol II antibody after cisplatin treatment which was inhibited by APE1 downregulation (Figure 2e). These results suggest that APE1 is required for both basal and drug-induced enhanced binding/recruitment of p300 and YB-1 to the Y-box element which enhances RNA pol II loading on the MDR1 promoter. The functional consequence of reduced binding of YB-1/p300 and RNA pol II loading on MDR1 promoter in APE1 downregulated cells was directly tested by measuring the endogenous MDR1 transcript levels by Real Time RT-PCR. As shown in Figure 2f, MDR1 level was increased by 3-fold following cisplatin treatment relative to untreated control which was significantly inhibited in APE1 downregulated cells.
Drug induced oxidative stress enhances APE1’s acetylation and its association with YB-1/p300 on MDR1 promoter
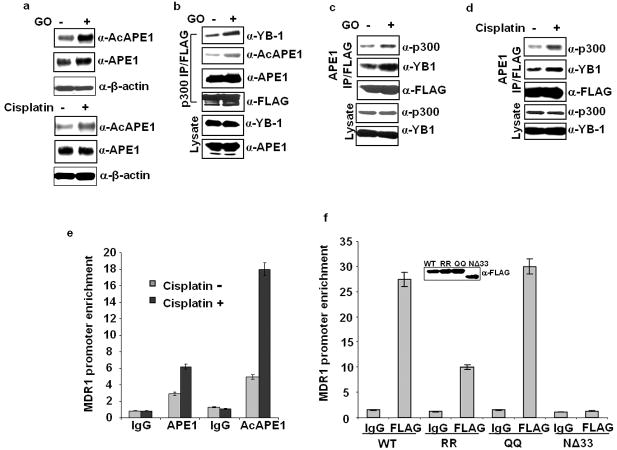
Many DNA damaging agents including chemotherapeutic drugs induce oxidative stress in cells (Kovacic, 2007; Kovacic and Osuna, 2000). To examine whether oxidative stress enhances the AcAPE1 level, we measured its level in HEK-293T cells after treatment with glucose oxidase (GO; 100 ng/ml) or cisplatin (40 ng/ml) for 1 hour. While no significant change in the APE1 polypeptide level was observed, the AcAPE1 level increased significantly after treatment (Figure 3a). Enhancement of APE1 acetylation after drug treatment may be likely to be due to oxidative stress-mediated enhancement of HAT activity of p300 (Rahman, 2002). Because genotoxic agents elevate the AcAPE1 level, we examined the effect of oxidative stress on AcAPE1/YB- 1/p300 complex formation. We transfected HEK-293T cells with p300-FLAG and treated the cells with GO. Western analysis of FLAG IP showed enhanced association of YB-1 with p300 (Figure 3b). While the amount of APE1 associated with p300 under oxidative stress remained unaltered, increased AcAPE1 level in the p300 complex indicates that oxidative stress enhanced APE1 acetylation, which in turn increased the amount of YB-1 in the p300 bound complex (Figure 3b). We also observed enhanced association of p300 and YB-1 in FLAG APE1 IP after treatment with GO (Figure 3c) or cisplatin (Figure 3d). Thus drug-induced oxidative stress raised the level of AcAPE1 and its association with YB-1/p300. This enhanced the assembly/recruitment of AcAPE1/YB-1/p300 complex to the MDR1 promoter and consequently could help RNA pol II loading.
Figure 3.
Oxidative stress enhances APE1’s acetylation and its association with YB-1/p300 on MDR1 promoter. (a) HEK 293T cells were treated with 100 ng/ml of Glucose oxidase (GO) or 40 μg/ml of cisplatin for 1 hour, and 5 hours later the AcAPE1, APE1 or β-actin levels in the whole cell extracts were measured by Western analysis. (b) HEK-293T cells transfected with FLAG-tagged p300 were treated with GO (100 ng/ml) for 1 hour, and 5 hours later, nuclear lysates were prepared and immunoprecipitated with FLAG antibody. Western analysis of the IPs with YB-1, AcAPE1, APE1 or FLAG antibody; YB-1 and APE1 levels in the lysates (lower panels). (c & d) HEK-293T cells transfected with FLAG-tagged APE1 were treated with GO (100 ng/ml) or cisplatin (40 μg/ml) as above, nuclear lysates were prepared and immunoprecipitated with FLAG antibody. Western analysis of the IPs with p300, YB-1 or FLAG antibody; p300 and YB-1 levels in the lysates (lower panels). (e) ChIP assay for the association of AcAPE1 on MDR1 promoter. HEK-293T cells were treated with cisplatin as above and ChIP assay was performed with APE1 or AcAPE1 antibody or control IgG. (f) HEK-293T cells were transfected with WT APE1, RR, QQ or NΔ33 APE1 and ChIP assay was carried out with FLAG antibody or control IgG. Inset panel shows the level of FLAG tagged WT or mutant APE1 proteins in the cell extracts determined by Western analysis with FLAG antibody. Values in the bar diagram in all the cases are relative to PCR from input chromatin and results represent mean ± standard deviations of 3 independent ChIP experiments performed in duplicates.
We then directly tested the presence of AcAPE1 on MDR1 promoter by ChIP analysis. Significant enrichment of MDR1 promoter with AcAPE1 Ab over control IgG (Figure 3e) indicates AcAPE1’s stable association with MDR1 promoter. Furthermore, significantly increased enrichment of the MDR1 promoter in the IP with AcAPE1 antibody relative to APE1 antibody after cisplatin treatment indicates that drug-induced APE1 acetylation enhances its promoter recruitment (Figure 3e). ChIP assay (Figure 3f) in HEK-293T cells transfected with FLAG-tagged WT APE1, RR, acetylation mimic K6Q/K7Q (QQ) or NΔ33 APE1 constructs with FLAG antibody shows that replacing lysine to acetylation mimic glutamine did not affect APE1’s association with MDR1 promoter; but changing lysine residues to nonacetylable arginine significantly reduced its promoter association. Furthermore, NΔ33 abolished APE1’s promoter association. Together, these data suggest that APE1 acetylation enhances its recruitment on the MDR1 promoter.
p300-mediated APE1 acetylation enhances Y-box-dependent MDR1 promoter activity
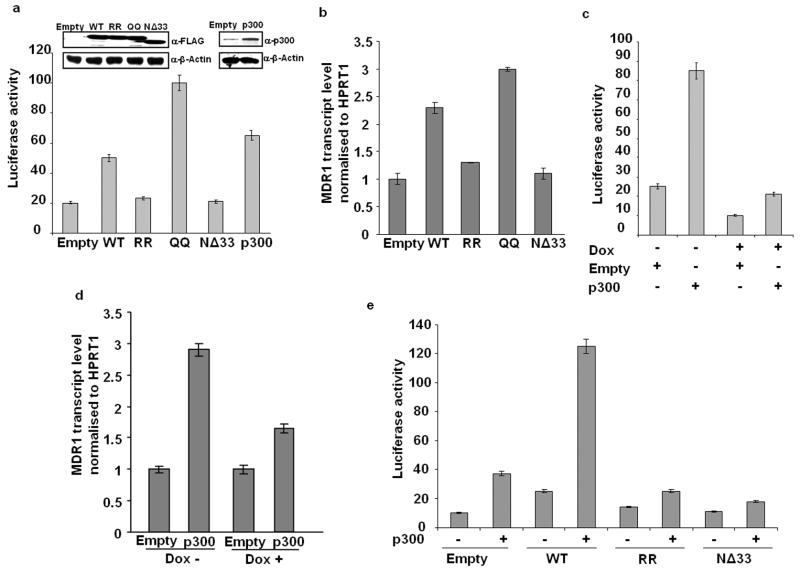
To evaluate the functional consequence of APE1 acetylation and its enhanced binding with p300 and YB-1 on its transcriptional coactivator function, we performed MDR1 promoter-dependent luciferase assay and Real Time RT-PCR assay to measure endogenous MDR1 transcript level. We examined the effect of overexpression of WT APE1, acetylation mimic QQ, nonacetylable RR or NΔ33 APE1 mutants. Figure 4a shows that overexpression of WT or QQ APE1 but not RR or NΔ33 APE1 mutant significantly enhanced MDR1 promoter activity indicating that APE1 acetylation is required for this activity. Figure 4b shows that while ectopic expression of WT or QQ APE1 increased endogenous MDR1 expression by more than 2- fold relative to empty vector, the RR or NΔ33 mutant had no significant effect. To further evaluate the effect of APE1 acetylation, we examined the effect of p300 level on MDR1 promoter activity. Figure 4a shows that p300 overexpression activated the MDR1 promoter. To confirm that p300-mediated activation of the MDR1 promoter is mediated through APE1, it was downregulated in APE1siRNAHEK-293T cells before cotransfection with MDR1 promoter-reporter and p300 expression plasmids. As predicted, APE1 downregulation significantly reduced MDR1 promoter activity compared to the control cells (Figure 4c). p300 overexpression had less stimulatory effect in APE1-downregulated cells, confirming that p300-mediated activation of MDR1 promoter is dependent on the APE1 level (Figure 4c). Similarly p300 overexpression significantly increased MDR1 transcript level in Dox-untreated cells relative to empty vector, but had less effect in APE1 downregulated cells (Figure 4d). Moreover, while overexpression of p300 or WT APE1 individually activated the MDR1 promoter, simultaneous overexpression of p300 and WT but not RR had a synergistic effect (Figure 4e). These results confirm the acetylation dependent regulatory role of APE1 in enhancing MDR1 promoter activity and its expression.
Figure 4.
Enhancement of MDR1 promoter activity and expression by APE1 acetylation. (a) HEK-293T cells were cotransfected with MDR1 promoter-luciferase reporter plasmid and empty vector, WT, RR, QQ, NΔ33 APE1 or p300 expression plasmids. 48 hours later, luciferase activity was measured and normalized with total protein content in the lysates. Inset panel shows the level of FLAG tagged WT/ mutant APE1 proteins or p300 protein in the cell extracts determined by Western analysis. (b) Real Time RT-PCR assay for MDR1 transcript levels in HEK-293T cells ectopically expressing WT, RR, QQ or NΔ33 APE1 mutant proteins. Values in the bar diagram are relative to empty vector transfected cells. (c) APE1 levels in APE1siRNAHEK-293T cells were downregulated with Dox treatment for 8 days. Then the cells were cotransfected with MDR1 promoter reporter plasmid and expression plasmids for p300 or its empty vector. Luciferase activity was measured as mentioned above. (d) MDR1 transcript levels in Dox-treated (APE1 downregulated) or untreated APE1siRNAHEK-293T cells ectopically expressing p300. Values in the bar diagram are relative to empty vector transfected cells. (e) HEK-293T cells were cotransfected with MDR1 promoter reporter plasmid along with WT, RR or NΔ33 APE1 constructs and p300 expression plasmid. Luciferase activity was measured as mentioned above. Results in all the cases represent the mean ± standard deviations of 3 independent experiments performed in duplicates.
Acetylation-dependent transcriptional regulatory function of APE1 is critical for resistance to cisplatin or etoposide
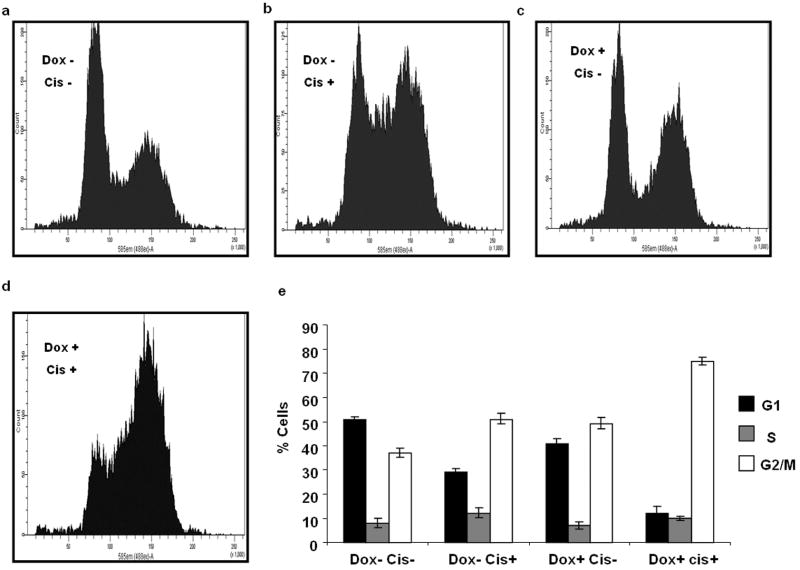
APE1 knockdown sensitizes cells to different chemotherapeutic drugs and induces cell death (Bapat et al., 2009; Bobola et al., 2005; Chattopadhyay et al., 2008; Fishel and Kelley, 2007; Vascotto et al., 2009). To understand APE1’s role in drug resistance we examined the effect of cisplatin treatment on cell cycle progression of APE1 downregulated and control cells by FACS analysis. Cisplatin treatment of control APE1siRNAHEK-293T cells results in an increase in cells at G2/M stage with a parallel decrease of G1 population (Figure 5a, 5b and 5e). When the endogenous APE1 level was downregulated by Dox treatment, significantly more cells were arrested at G2/M stage following cisplatin treatment (Figure 5c, 5d and 5e). Next, clonogeneic survival assays were performed in control and APE1 knockdown cells to measure the cytotoxicity to cisplatin and etoposide. Figures 6a and 6c show that downregulation of APE1 significantly enhanced sensitivity to cisplatin and etoposide as compared to untreated control which support our earlier observation (Chattopadhyay MCB 2008). Dox treatment itself did not affect the survival of the cells with control siRNA (data not shown). Additionally, while ectopic overexpression of WT APE1 or QQ mutant increased resistance to drug-induced cytotoxicity, the RR mutant did not confer resistance to these drugs (Figures 6b and 6d). These results indicate that acetylation-mediated transcriptional regulatory function of APE1 plays a critical role in protecting the cells from cisplatin or etoposide-induced cytotoxicity.
Figure 5.
APE1 depletion induces cisplatin-mediated G2/M arrest. (a-d) Cell cycle distribution pattern of APE1 downregulated and control APE1siRNAHEK-293T cells treated with cisplatin. After knockdown of endogenous APE1 level with Dox treatment for 8 days, the cells were treated with 100 μM cisplatin and after 24 hours the cells were analysed for FACS as described in Materials and Methods. (e) The percentage of cells at G1, S and G2/M stages were determined by FACSDiva Version 6.1.2 software programme and plotted and the results in each case represent the mean ± standard deviations of 3 independent experiments performed in duplicates.
Figure 6.
Involvement of APE1’s acetylation-mediated regulatory function in resistance to cisplatin or etoposide. (a & c) Clonogenic survival assay of APE1siRNAHEK-293T cells treated with Dox. APE1 level in these cells was downregulated with Dox treatment for 8 days and seeded in 35 mm dishes (~ 300 cells/dish). Eighteen hours later, the cells were treated with increasing concentrations of cisplatin (0 to 125 μM) or etoposide (0 to 20 μM). Cells were maintained for 10 -14 days and the colonies were fixed and stained with giemsa stain. (b & d) After Dox treatment for 8 days, APE1siRNAHEK-293T cells were transfected with empty vector, WT APE1, RR APE1 or QQ APE1 expression plasmids. Forty eight hours later, the cells were seeded in 35 mm dishes (~ 300 cells/dish) and proceeded as above. The number of visible colonies was counted and one hundred percent corresponds to the number of colonies in the absence of drugs. The graph shows the mean ± standard deviations of 3 independent experiments performed in duplicates
Discussion
Although various mechanisms of drug resistance including alterations of glutathione mediated detoxification and enhanced DNA repair (Kaina and Christmann, 2002; Kaina et al., 2001) have been identified in different cancer cell lines, resistance to many common anticancer drugs often occurs due to enhanced expression of MDR1 and other ATP-binding cassette (ABC) transporter proteins in intrinsically drug-resistant cancer cells, as well as in many tumors that acquire drug resistance after chemotherapy (Ambudkar et al., 2005; Gottesman et al., 2002; Gottesman and Pastan, 1993). These transporters reduce intracellular drug concentration by acting as efflux pumps. Several studies have shown that MDR1 expression is often upregulated due to promoter activation by YB-1 following drug treatment (Chaudhary and Roninson, 1993; Kohno et al., 1989; Kohno et al., 1994). Our earlier study showed that APE1 stably interacts with YB-1 and enhances YB-1’s binding to Y-box element on the MDR1 promoter leading to its activation (Chattopadhyay et al., 2008). In this study, we have shown that APE1 is associated with RNA pol II on the endogenous MDR1 promoter and it is required for both basal and druginduced recruitment of YB-1/p300 complex to the promoter for facilitating RNA pol II loading. Furthermore, our data suggest that drug-induced p300-mediated APE1 acetylation enhances assembly/recruitment of AcAPE1/YB-1/p300 complex on the promoter and plays a critical role in enhancing MDR1 expression. Thus, our study for the first time has provided the basis for non repair function of APE1 for facilitating MDR1 expression.
Some earlier studies showed that APE1 stimulates the DNA-binding of several transcription factors through its redox regulatory function. Initial studies suggested Cys 65 of human APE1/Ref-1 (Cys 64 of mouse APE1/Ref-1) to be critical for redox regulation (Walker et al., 1993), however, subsequent analyses showed that neither Cys 65 nor other cysteine residues are involved in redox regulation (Ando et al., 2008; Ordway et al., 2003). We observed that the C65S and C138S mutants behaved like WT APE1 in modulating YB-1-dependent MDR1 promoter activity (Chattopadhyay et al., 2008), suggesting that APE1’s redox activity may not be involved in MDR1 activation. YB-1, a transcriptional regulator (Kohno et al., 2003), binds to the cognate Y-box element and recruits p300/CBP (Higashi et al., 2003). We showed earlier that APE1 stably interacts with YB-1 and its acetylation enhances its binding to Y box sequence. The observation that deletion of the N-terminal 33 aa which is essential for YB-1 binding (Chattopadhyay et al., 2008) abolished APE1’s promoter association, indicates that this interaction is essential for promoter recruitment. Our ChIP results provided strong evidence for simultaneous MDR1 promoter occupancy of APE1, YB-1, p300 and RNA pol II. We have shown that APE1 downregulation reduced recruitment of the YB-1/p300 complex and of RNA pol II loading on the promoter. It appears likely that the transcriptional activator YB-1 recruits coactivator p300 and APE1 to the promoter which enhances YB-1/p300 binding to the promoter. This induces unfolding of the chromatin structure presumably via p300’s intrinsic HAT activity and thereby facilitates RNA pol II complex loading. Furthermore, drug induced enhanced APE1 acetylation and recruitment of AcAPE1/YB-1/p300 complex to the promoter may cause p300 to communicate with the basal transcription machineries near the transcription start site and thereby stabilize the RNA pol II preinitiation complex. Because the Y-box element (-82 to -73) on the MDR1 promoter is quite close to the transcription start site and p300 may function as a bridging factor between the trans-acting complexes and the basal transcription machinery (Blobel, 2000; Neish et al., 1998), it is likely that APE1 is stably associated with RNA pol II preinitiation complex. Specifically, p300 interacts with the unphosphorylated form of RNA pol II which is competent in forming the transcription preinitiation complex (Cho et al., 1998). One recent study also showed that APE1 interacts with estrogen alpha receptor and is associated with its endogenous promoter for activation of estrogen-responsive gene expression (Curtis et al., 2009). In any case, identification of APE1 as a RNA pol II-associated protein and its requirement in facilitating the RNA pol II loading on the promoter in the context of MDR1 expression is a significant finding of this study.
It appears that overexpression of both YB-1 and APE1, often observed in tumor cells, is associated with resistance to various drugs by activating MDR1. Our earlier study showed that APE1 downregulation in cisplatin resistant A2780/100 and doxorubicin-resistant MCF7MDR1 cell lines decreased their MDR1 levels and enhanced drug sensitivity (Chattopadhyay et al., 2008). Consistent with this, one recent study has shown that upregulation of MDR1 levels in breast cancer tissues in mice is associated with their sensitivity to doxorubicin (Pajic et al., 2009). Although we do not have direct evidence that APE1 or its acetylation mediated regulatory function affect drug transport, activation of MDR1 expression by WT APE1 but not its acetylation mutant support its indirect involvement in enhancing export of the drugs. In the event of drug exposure, an immediate increase in the MDR1 level is warranted for enhancing export of the drugs which could be affected by enhancing APE1 acetylation. However, it is obvious that acetylation of the APE1 is not the sole mechanism for drug resistance; it is a very complex issue considering the interplay of multiple factors. Moreover, the balance between acetyl transferases and deacetylases that control the acetylation of APE1 and other proteins could indirectly regulate drug resistance. Nevertheless, our data show that specific knockdown of endogenous APE1 by conditional expression of siRNA, sensitized cells to cisplatin or etoposide which could be overcome by WT APE1 or the acetylation mimic QQ mutant but not by the acetylation deficient RR mutant. Together, these results confirm that resistance to these drugs is primarily dependent on APE1’s acetylation dependent coregulatory function by modulating MDR1 level.
Finally, we would like to stress that this study provides the first evidence for the involvement of APE1’s transcriptional regulatory function in recruitment of the transcription factors/coactivators and facilitating RNA pol II loading on the promoter and thus documents a novel mechanism by which APE1 and its acetylation controls YB-1-mediated activation of MDR1 gene. Although other factors such as NF-Y, AP-1 may also modulate MDR1 expression (Jin and Scotto, 1998; Okamura et al., 2004), we have provided strong evidence for the key role of APE1 and its acetylation in MDR1 activation, suggesting that the transactivation function of APE1 could be used as a novel therapeutic target.
Materials and Methods
Cell lines, plasmids and transfection
HEK-293T cell line was cultured in DMEM-high glucose medium (Gibco-BRL) with 10% fetal calf serum. The HEK-293T cells stably expressing APE1 siRNA or control siRNA under doxycycline inducible promoter were generated as described earlier (Vascotto et al., 2009) by selection with blasticidin (Gibco-BRL; 5 μg/ml) and zeocine (Gibco-BRL; 100 μg/ml). The APE1 siRNA expressing HEK-293T clone (APE1siRNAHEK-293T) that showed maximum APE1 knockdown by treatment with Doxycycline (Dox, Sigma) was used in subsequent experiments. Cells were transfected using lipofectamine 2000 (Invitrogen). PCMV 5.1 FLAG expression plasmids for wild type (WT) APE1, K6R/K7R (RR) APE1, N terminal 33 amino acid deleted (NΔ33) APE1 and p300 were described elsewhere (Bhakat and Mitra, 2000; Chattopadhyay et al., 2008). The PCMV 5.1 FLAG tagged K6Q/K7Q (QQ) APE1 expression plasmid was generated by site-directed mutagenesis kit (Stratagene). The p300 siRNA used was described earlier (Bhakat et al., 2006).
Co-immunoprecipitation (Co-IP) and Western analysis
Immunoprecipitation (IP) was done with mouse anti-FLAG M2 antibody-conjugated agarose beads (Sigma) in nuclear extracts of cells transfected with FLAG tagged constructs as described previously (Bhakat et al., 2006; Chattopadhyay et al., 2008; Das et al., 2007). The immunoprecipitated proteins were resolved in SDS-PAGE and identified by Western analysis with the indicated antibodies; rabbit α-APE1 (Ramana et al., 1998), rabbit α-AcAPE1 (Chattopadhyay et al., 2008), rabbit α-YB-1 (Chattopadhyay et al., 2008; Das et al., 2007), rabbit α-p300 (N15; Santa Cruz), rabbit α-RNA pol II (N20; Santa Cruz), mouse α-FLAG (M2; Sigma) and mouse α-β-actin (AC15; Sigma).
Luciferase assay
Cells were cotransfected with MDR1 promoter reporter constructs (Chattopadhyay et al., 2008) and the expression plasmids for WT, RR, QQ or NΔ33 APE1 and p300. Luciferase activity in the cells extracts was measured in a luminometer (AutoLumant LB 953; Berthold) using the luciferase assay kit (Promega). The luciferase activity was normalized with respect to total protein content of the lysates.
RNA isolation and Real Time RT-PCR assay
Total RNA was isolated from cells with Qiagen RNeasy mini kit followed by DNase 1 (NEB) treatment and cDNA synthesis using Superscript III first-strand synthesis kit (Invitrogen). MDR1 expression in the samples were analysed by SYBR GREEN based Real Time PCR (7000 Fast Real-Time PCR System; Applied Biosystems) using SYBR Premix Ex Taq (TaKaRa) and primers appropriate for MDR1 (forward: 5’-CCCATCATTGCAATAGCAGG-3’ and reverse: 5’-TGTTCAAACTTCTGCTCCTGA-3) or HPRT1 expression (internal control; primer sequences: RealTimePrimers.com).
Invitro acetylation and binding assay
Recombinant APE1 (2 μg) was acetylated with recombinant p300 HAT domain polypeptide in the presence of 1 mM acetyl CoA as described previously (Bhakat et al., 2003b). Acetylated or unmodified APE1 (100 ng) was incubated with p300 FLAG-IP beads and eluted with SDS Laemmli buffer for Western analysis with APE1 or FLAG antibodies.
Chromation immunoprecipitation (ChIP) assay
ChIP assay was performed using Magna chromatin immunoprecipitation assay kit (Upstate) with the following antibodies; α-RNA pol II (N20), α-p300 (N15), α-YB-1, α-APE1 (Novus Biologicals), α-AcAPE1, α-FLAG (M2) or control IgG (Santa Cruz). The immunoprecipitated purified DNA was then subjected to SYBR GREEN based Real Time PCR with primers (forward: 5’-TCTCGAGGAATCAGCATTCA-3’ and reverse: 5’-AAGAGCCGCTACTCGAATGA-3’) for the MDR1 promoter sequence (-138 to +18) containing the Y-box element and a sequence of the MDR1 gene distant to the Y-box element (forward: 5’- AACTGAGGAGATGGGGCATA-3’ and reverse: 5’- TGGGAACAAATCACTGATCG-3’). For re-ChIP assay, after the first IP was performed with FLAG antibody or control IgG, the second IP was performed in the eluents with RNA pol II, p300 or YB-1 antibody or control IgG .
Cell cycle assay
Cell cycle studies were performed by flow cytometry using a BD FACSCanto (Becton Dickinson). About 106 cells washed and resuspended in PBS were fixed in 100% ethanol at -20°C for 15 mins and then rehydrated in PBS at room temperature. The cells were then stained with 3μM propidium iodide in Staining buffer containing 100 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM CaCl2, 0.5 mM MgCl2, 0.1% NP40 and 0.2 mg/ml RNase A, incubated at room temperature for 15 mins in dark and analyzed on a Becton-Dickinson FACSCanto using an Ar laser (excitation 488 nm). For each sample, 10,000 single events were detected and data analysis was performed using FACSDiva version 6.1.2 software programme.
Clonogenic survival assay
Endogenous APE1 downregulated APE1siRNAHEK-293T or control cells were plated on 35 mm dishes (~300 cells/dish). Next day, the cells were treated with cisplatin (0-125 μM) or etoposide (0-20 μM) and allowed to grow for 10-14 days until visible colonies appear. In another set, after APE1 downregulation the cells were transfected with WT, RR or QQ APE1 expression constructs. Forty eight hours later, the cells were plated on 35 mm dishes and proceeded for testing drug sensitivity. The colonies were fixed with 100% methanol, stained with giemsa staining solution (1:50) and counted.
Acknowledgments
We acknowledge the help of Ranajay Chattopadhyay, University of Virginia, Charlottesville in this study. This work was supported by grants: RO1 ESO8457, RO1 CA53791 and P50 ES66076 to S.M.
Footnotes
Conflict of Interest
The authors declare no potential conflict of interest.
References
- Ahmad S, Glazer RI. Expression of the antisense cDNA for protein kinase C alpha attenuates resistance in doxorubicin-resistant MCF-7 breast carcinoma cells. Mol Pharmacol. 1993;43:858–862. [PubMed] [Google Scholar]
- Ahmad S, Safa AR, Glazer RI. Modulation of P-glycoprotein by protein kinase C alpha in a baculovirus expression system. Biochemistry. 1994;33:10313–10318. doi: 10.1021/bi00200a011. [DOI] [PubMed] [Google Scholar]
- Ambudkar SV, Sauna ZE, Gottesman MM, Szakacs G. A novel way to spread drug resistance in tumor cells: functional intercellular transfer of P-glycoprotein (ABCB1) Trends Pharmacol Sci. 2005;26:385–387. doi: 10.1016/j.tips.2005.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando K, Hirao S, Kabe Y, Ogura Y, Sato I, Yamaguchi Y, et al. A new APE1/Ref-1-dependent pathway leading to reduction of NF-kappaB and AP-1, and activation of their DNA-binding activity. Nucleic Acids Res. 2008;36:4327–4336. doi: 10.1093/nar/gkn416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bapat A, Fishel M, Kelley MR. Going Ape as an Approach to Cancer Therapeutics. Antioxid Redox Signal. 2009;11:651–668. doi: 10.1089/ars.2008.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargou RC, Jurchott K, Wagener C, Bergmann S, Metzner S, Bommert K, et al. Nuclear localization and increased levels of transcription factor YB-1 in primary human breast cancers are associated with intrinsic MDR1 gene expression. Nat Med. 1997;3:447–450. doi: 10.1038/nm0497-447. [DOI] [PubMed] [Google Scholar]
- Bhakat KK, Izumi T, Yang SH, Hazra TK, Mitra S. Role of acetylated human AP-endonuclease (APE1/Ref-1) in regulation of the parathyroid hormone gene. Embo J. 2003a;22:6299–6309. doi: 10.1093/emboj/cdg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhakat KK, Mantha AK, Mitra S. Transcriptional Regulatory Functions of Mammalian AP-endonuclease (APE1/Ref-1), an Essential Multifunctional Protein. Antioxid Redox Signal. 2009;11:621–638. doi: 10.1089/ars.2008.2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhakat KK, Mitra S. Regulation of the human O(6)-methylguanine-DNA methyltransferase gene by transcriptional coactivators cAMP response elementbinding protein-binding protein and p300. J Biol Chem. 2000;275:34197–34204. doi: 10.1074/jbc.M005447200. [DOI] [PubMed] [Google Scholar]
- Bhakat KK, Mokkapati SK, Boldogh I, Hazra TK, Mitra S. Acetylation of human 8-oxoguanine-DNA glycosylase by p300 and its role in 8-oxoguanine repair in vivo. Mol Cell Biol. 2006;26:1654–1665. doi: 10.1128/MCB.26.5.1654-1665.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhakat KK, Yang SH, Mitra S. Acetylation of human AP-endonuclease 1, a critical enzyme in DNA repair and transcription regulation. Methods Enzymol. 2003b;371:292–300. doi: 10.1016/S0076-6879(03)71022-2. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya A, Chattopadhyay R, Burnette BR, Cross JV, Mitra S, Ernst PB, et al. Acetylation of apurinic/apyrimidinic endonuclease-1 regulates Helicobacter pylori-mediated gastric epithelial cell apoptosis. Gastroenterology. 2009;136:2258–2269. doi: 10.1053/j.gastro.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel GA. CREB-binding protein and p300: molecular integrators of hematopoietic transcription. Blood. 2000;95:745–755. [PubMed] [Google Scholar]
- Bobola MS, Finn LS, Ellenbogen RG, Geyer JR, Berger MS, Braga JM, et al. Apurinic/apyrimidinic endonuclease activity is associated with response to radiation and chemotherapy in medulloblastoma and primitive neuroectodermal tumors. Clin Cancer Res. 2005;11:7405–7414. doi: 10.1158/1078-0432.CCR-05-1068. [DOI] [PubMed] [Google Scholar]
- Busso CS, Iwakuma T, Izumi T. Ubiquitination of mammalian AP endonuclease (APE1) regulated by the p53-MDM2 signaling pathway. Oncogene. 2009;28:1616–1625. doi: 10.1038/onc.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay R, Das S, Maiti AK, Boldogh I, Xie J, Hazra TK, et al. Regulatory role of human AP-endonuclease (APE1/Ref-1) in YB-1-mediated activation of the multidrug resistance gene MDR1. Mol Cell Biol. 2008;28:7066–7080. doi: 10.1128/MCB.00244-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary PM, Roninson IB. Induction of multidrug resistance in human cells by transient exposure to different chemotherapeutic drugs. J Natl Cancer Inst. 1993;85:632–639. doi: 10.1093/jnci/85.8.632. [DOI] [PubMed] [Google Scholar]
- Cho H, Orphanides G, Sun X, Yang XJ, Ogryzko V, Lees E, et al. A human RNA polymerase II complex containing factors that modify chromatin structure. Mol Cell Biol. 1998;18:5355–5363. doi: 10.1128/mcb.18.9.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis CD, Thorngren DL, Ziegler YS, Sarkeshik A, Yates JR, Nardulli AM. Apurinic/apyrimidinic endonuclease 1 alters estrogen receptor activity and estrogen-responsive gene expression. Mol Endocrinol. 2009;23:1346–1359. doi: 10.1210/me.2009-0093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Chattopadhyay R, Bhakat KK, Boldogh I, Kohno K, Prasad R, et al. Stimulation of NEIL2-mediated oxidized base excision repair via YB-1 interaction during oxidative stress. J Biol Chem. 2007;282:28474–28484. doi: 10.1074/jbc.M704672200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demple B, Harrison L. Repair of oxidative damage to DNA: enzymology and biology. Annu Rev Biochem. 1994;63:915–948. doi: 10.1146/annurev.bi.63.070194.004411. [DOI] [PubMed] [Google Scholar]
- Doetsch PW, Cunningham RP. The enzymology of apurinic/apyrimidinic endonucleases. Mutat Res. 1990;236:173–201. doi: 10.1016/0921-8777(90)90004-o. [DOI] [PubMed] [Google Scholar]
- Evans AR, Limp-Foster M, Kelley MR. Going APE over ref-1. Mutat Res. 2000;461:83–108. doi: 10.1016/s0921-8777(00)00046-x. [DOI] [PubMed] [Google Scholar]
- Fan J, Wilson DM., 3rd Protein-protein interactions and posttranslational modifications in mammalian base excision repair. Free Radic Biol Med. 2005;38:1121–1138. doi: 10.1016/j.freeradbiomed.2005.01.012. [DOI] [PubMed] [Google Scholar]
- Fantini D, Vascotto C, Deganuto M, Bivi N, Gustincich S, Marcon G, et al. APE1/Ref-1 regulates PTEN expression mediated by Egr-1. Free Radic Res. 2008;42:20–9. doi: 10.1080/10715760701765616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishel ML, Kelley MR. The DNA base excision repair protein Ape1/Ref-1 as a therapeutic and chemopreventive target. Mol Aspects Med. 2007;28:375–395. doi: 10.1016/j.mam.2007.04.005. [DOI] [PubMed] [Google Scholar]
- Fuchs S, Philippe J, Corvol P, Pinet F. Implication of Ref-1 in the repression of renin gene transcription by intracellular calcium. J Hypertens. 2003;21:327–335. doi: 10.1097/00004872-200302000-00024. [DOI] [PubMed] [Google Scholar]
- Germann UA, Chambers TC. Molecular analysis of the multidrug transporter, P-glycoprotein. Cytotechnology. 1998;27:31–60. doi: 10.1023/A:1008023629269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein LJ, Galski H, Fojo A, Willingham M, Lai SL, Gazdar A, et al. Expression of a multidrug resistance gene in human cancers. J Natl Cancer Inst. 1989;81:116–124. doi: 10.1093/jnci/81.2.116. [DOI] [PubMed] [Google Scholar]
- Gottesman MM, Fojo T, Bates SE. Multidrug resistance in cancer: role of ATP-dependent transporters. Nat Rev Cancer. 2002;2:48–58. doi: 10.1038/nrc706. [DOI] [PubMed] [Google Scholar]
- Gottesman MM, Pastan I. Biochemistry of multidrug resistance mediated by the multidrug transporter. Annu Rev Biochem. 1993;62:385–427. doi: 10.1146/annurev.bi.62.070193.002125. [DOI] [PubMed] [Google Scholar]
- Higashi K, Inagaki Y, Fujimori K, Nakao A, Kaneko H, Nakatsuka I. Interferon-gamma interferes with transforming growth factor-beta signaling through direct interaction of YB-1 with Smad3. J Biol Chem. 2003;278:43470–43479. doi: 10.1074/jbc.M302339200. [DOI] [PubMed] [Google Scholar]
- Izumi T, Brown DB, Naidu CV, Bhakat KK, Macinnes MA, Saito H, et al. Two essential but distinct functions of the mammalian abasic endonuclease. Proc Natl Acad Sci U S A. 2005;102:5739–5743. doi: 10.1073/pnas.0500986102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi T, Wiederhold LR, Roy G, Roy R, Jaiswal A, Bhakat KK, et al. Mammalian DNA base excision repair proteins: their interactions and role in repair of oxidative DNA damage. Toxicology. 2003;193:43–65. doi: 10.1016/s0300-483x(03)00289-0. [DOI] [PubMed] [Google Scholar]
- Jayaraman L, Murthy KG, Zhu C, Curran T, Xanthoudakis S, Prives C. Identification of redox/repair protein Ref-1 as a potent activator of p53. Genes Dev. 1997;11:558–570. doi: 10.1101/gad.11.5.558. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Guo C, Vasko MR, Kelley MR. Implications of apurinic/apyrimidinic endonuclease in reactive oxygen signaling response after cisplatin treatment of dorsal root ganglion neurons. Cancer Res. 2008;68:6425–6434. doi: 10.1158/0008-5472.CAN-08-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S, Scotto KW. Transcriptional regulation of the MDR1 gene by histone acetyltransferase and deacetylase is mediated by NF-Y. Mol Cell Biol. 1998;18:4377–4384. doi: 10.1128/mcb.18.7.4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaina B, Christmann M. DNA repair in resistance to alkylating anticancer drugs. Int J Clin Pharmacol Ther. 2002;40:354–367. doi: 10.5414/cpp40354. [DOI] [PubMed] [Google Scholar]
- Kaina B, Ochs K, Grosch S, Fritz G, Lips J, Tomicic M, et al. BER, MGMT, and MMR in defense against alkylation-induced genotoxicity and apoptosis. Prog Nucleic Acid Res Mol Biol. 2001;68:41–54. doi: 10.1016/s0079-6603(01)68088-7. [DOI] [PubMed] [Google Scholar]
- Kelley MR, Cheng L, Foster R, Tritt R, Jiang J, Broshears J, et al. Elevated and altered expression of the multifunctional DNA base excision repair and redox enzyme Ape1/ref-1 in prostate cancer. Clin Cancer Res. 2001;7:824–830. [PubMed] [Google Scholar]
- Kohno K, Izumi H, Uchiumi T, Ashizuka M, Kuwano M. The pleiotropic functions of the Y-box-binding protein, YB-1. Bioessays. 2003;25:691–698. doi: 10.1002/bies.10300. [DOI] [PubMed] [Google Scholar]
- Kohno K, Sato S, Takano H, Matsuo K, Kuwano M. The direct activation of human multidrug resistance gene (MDR1) by anticancer agents. Biochem Biophys Res Commun. 1989;165:1415–1421. doi: 10.1016/0006-291x(89)92761-7. [DOI] [PubMed] [Google Scholar]
- Kohno K, Tanimura H, Sato S, Nakayama Y, Makino Y, Wada M, et al. Cellular control of human multidrug resistance 1 (mdr-1) gene expression in absence and presence of gene amplification in human cancer cells. J Biol Chem. 1994;269:20503–20508. [PubMed] [Google Scholar]
- Kovacic P. Unifying mechanism for anticancer agents involving electron transfer and oxidative stress: clinical implications. Med Hypotheses. 2007;69:510–516. doi: 10.1016/j.mehy.2006.08.046. [DOI] [PubMed] [Google Scholar]
- Kovacic P, Osuna JA., Jr Mechanisms of anti-cancer agents: emphasis on oxidative stress and electron transfer. Curr Pharm Des. 2000;6:277–309. doi: 10.2174/1381612003401046. [DOI] [PubMed] [Google Scholar]
- Kuninger DT, Izumi T, Papaconstantinou J, Mitra S. Human APendonuclease 1 and hnRNP-L interact with a nCaRE-like repressor element in the AP-endonuclease 1 promoter. Nucleic Acids Res. 2002;30:823–829. doi: 10.1093/nar/30.3.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb LA, Preston BD. Mutagenesis by apurinic/apyrimidinic sites. Annu Rev Genet. 1986;20:201–230. doi: 10.1146/annurev.ge.20.120186.001221. [DOI] [PubMed] [Google Scholar]
- Matsumoto Y, Kim K. Excision of deoxyribose phosphate residues by DNA polymerase beta during DNA repair. Science. 1995;269:699–702. doi: 10.1126/science.7624801. [DOI] [PubMed] [Google Scholar]
- McNeill DR, Lam W, DeWeese TL, Cheng YC, Wilson DM., 3rd Impairment of APE1 function enhances cellular sensitivity to clinically relevant alkylators and antimetabolites. Mol Cancer Res. 2009;7:897–906. doi: 10.1158/1541-7786.MCR-08-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra S, Izumi T, Boldogh I, Bhakat KK, Hill JW, Hazra TK. Choreography of oxidative damage repair in mammalian genomes. Free Radic Biol Med. 2002;33:15–28. doi: 10.1016/s0891-5849(02)00819-5. [DOI] [PubMed] [Google Scholar]
- Mujtaba S, He Y, Zeng L, Farooq A, Carlson JE, Ott M, et al. Structural basis of lysine-acetylated HIV-1 Tat recognition by PCAF bromodomain. Mol Cell. 2002;9:575–586. doi: 10.1016/s1097-2765(02)00483-5. [DOI] [PubMed] [Google Scholar]
- Neish AS, Anderson SF, Schlegel BP, Wei W, Parvin JD. Factors associated with the mammalian RNA polymerase II holoenzyme. Nucleic Acids Res. 1998;26:847–853. doi: 10.1093/nar/26.3.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura H, Yoshida K, Sasaki E, Morimoto H, Haneji T. Transcription factor NF-Y regulates mdr1 expression through binding to inverted CCAAT sequence in drug-resistant human squamous carcinoma cells. Int J Oncol. 2004;25:1031–1037. [PubMed] [Google Scholar]
- Okazaki T, Chung U, Nishishita T, Ebisu S, Usuda S, Mishiro S, et al. A redox factor protein, ref1, is involved in negative gene regulation by extracellular calcium. J Biol Chem. 1994;269:27855–27862. [PubMed] [Google Scholar]
- Ordway JM, Eberhart D, Curran T. Cysteine 64 of Ref-1 is not essential for redox regulation of AP-1 DNA binding. Mol Cell Biol. 2003;23:4257–4266. doi: 10.1128/MCB.23.12.4257-4266.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajic M, Iyer JK, Kersbergen A, van der Burg E, Nygren AO, Jonkers J, et al. Moderate increase in Mdr1a/1b expression causes in vivo resistance to doxorubicin in a mouse model for hereditary breast cancer. Cancer Res. 2009;69:6396–6404. doi: 10.1158/0008-5472.CAN-09-0041. [DOI] [PubMed] [Google Scholar]
- Rahman I. Oxidative stress, transcription factors and chromatin remodelling in lung inflammation. Biochem Pharmacol. 2002;64:935–942. doi: 10.1016/s0006-2952(02)01153-x. [DOI] [PubMed] [Google Scholar]
- Ramana CV, Boldogh I, Izumi T, Mitra S. Activation of apurinic/apyrimidinic endonuclease in human cells by reactive oxygen species and its correlation with their adaptive response to genotoxicity of free radicals. Proc Natl Acad Sci U S A. 1998;95:5061–5066. doi: 10.1073/pnas.95.9.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson KA, Bullock HA, Xu Y, Tritt R, Zimmerman E, Ulbright TM, et al. Altered expression of Ape1/ref-1 in germ cell tumors and overexpression in NT2 cells confers resistance to bleomycin and radiation. Cancer Res. 2001;61:2220–2225. [PubMed] [Google Scholar]
- Robertson KA, Hill DP, Xu Y, Liu L, Van Epps S, Hockenbery DM, et al. Down-regulation of apurinic/apyrimidinic endonuclease expression is associated with the induction of apoptosis in differentiating myeloid leukemia cells. Cell Growth Differ. 1997;8:443–449. [PubMed] [Google Scholar]
- Shen DW, Fojo A, Chin JE, Roninson IB, Richert N, Pastan I, et al. Human multidrug-resistant cell lines: increased mdr1 expression can precede gene amplification. Science. 1986;232:643–645. doi: 10.1126/science.3457471. [DOI] [PubMed] [Google Scholar]
- Singhal RK, Prasad R, Wilson SH. DNA polymerase beta conducts the gapfilling step in uracil-initiated base excision repair in a bovine testis nuclear extract. J Biol Chem. 1995;270:949–957. doi: 10.1074/jbc.270.2.949. [DOI] [PubMed] [Google Scholar]
- Tell G, Quadrifoglio F, Tiribelli C, Kelley MR. The Many Functions of APE1/Ref-1: Not Only a DNA Repair Enzyme. Antioxid Redox Signal. 2009;11:601–620. doi: 10.1089/ars.2008.2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vascotto C, Cesaratto L, Zeef LA, Deganuto M, D’Ambrosio C, Scaloni A, et al. Genome-wide analysis and proteomic studies reveal APE1/Ref-1 multifunctional role in mammalian cells. Proteomics. 2009;9:1058–1074. doi: 10.1002/pmic.200800638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LJ, Robson CN, Black E, Gillespie D, Hickson ID. Identification of residues in the human DNA repair enzyme HAP1 (Ref-1) that are essential for redox regulation of Jun DNA binding. Mol Cell Biol. 1993;13:5370–5376. doi: 10.1128/mcb.13.9.5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Luo M, Kelley MR. Human apurinic endonuclease 1 (APE1) expression and prognostic significance in osteosarcoma: enhanced sensitivity of osteosarcoma to DNA damaging agents using silencing RNA APE1 expression inhibition. Mol Cancer Ther. 2004;3:679–686. [PubMed] [Google Scholar]
- Xanthoudakis S, Curran T. Identification and characterization of Ref-1, a nuclear protein that facilitates AP-1 DNA-binding activity. Embo J. 1992;11:653–665. doi: 10.1002/j.1460-2075.1992.tb05097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xanthoudakis S, Miao G, Wang F, Pan YC, Curran T. Redox activation of Fos-Jun DNA binding activity is mediated by a DNA repair enzyme. Embo J. 1992;11:3323–3335. doi: 10.1002/j.1460-2075.1992.tb05411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Moore DH, Broshears J, Liu L, Wilson TM, Kelley MR. The apurinic/apyrimidinic endonuclease (APE/ref-1) DNA repair enzyme is elevated in premalignant and malignant cervical cancer. Anticancer Res. 1997;17:3713–3719. [PubMed] [Google Scholar]
- Yamamori T, DeRicco J, Naqvi A, Hoffman TA, Mattagajasingh I, Kasuno K, et al. SIRT1 deacetylates APE1 and regulates cellular base excision repair. Nucleic Acids Res. 2010;38:832–845. doi: 10.1093/nar/gkp1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M, Kijima M, Akita M, Beppu T. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J Biol Chem. 1990;265:17174–17179. [PubMed] [Google Scholar]
- Yu G, Ahmad S, Aquino A, Fairchild CR, Trepel JB, Ohno S, et al. Transfection with protein kinase C alpha confers increased multidrug resistance to MCF-7 cells expressing P-glycoprotein. Cancer Commun. 1991;3:181–189. doi: 10.3727/095535491820873263. [DOI] [PubMed] [Google Scholar]