Abstract
Epidermolysis bullosa (EB) is a class of intractable, rare, genetic disorders characterized by fragile skin and blister formation as a result of dermal-epidermal mechanical instability. EB presents with considerable clinical and molecular heterogeneity. Viable animal models of junctional epidermolysis bullosa (JEB), that both mimic the human disease and survive beyond the neonatal period, are needed. We identified a spontaneous, autosomal recessive mutation (Lamc2 jeb) due to a Murine Leukemia Virus long terminal repeat insertion in Lamc2 that results in a hypomorphic allele with reduced levels of LAMC2 protein. These mutant mice develop a progressive blistering disease validated at the gross and microscopic levels to closely resemble generalized non-Herlitz JEB. The Lamc2 jeb mice display additional extracutaneous features such as loss of bone mineralization and abnormal teeth, as well as a respiratory phenotype that is recognized but not as well characterized in humans. This model faithfully recapitulates human JEB and provides an important preclinical tool to test novel therapeutic approaches.
INTRODUCTION
The skin and mucus membranes represent the front-line physical barrier and immune defense for the protection of visceral tissues and organs. Skin is composed of an epidermis and dermis which are held together at the basement membrane by numerous adhesion molecules. Epidermolysis bullosa (EB) is a name applied to a heterogeneous group of inherited skin disorders in which minor trauma leads to blistering of the skin and mucous membranes, due to defects in the structure and expression any one of these basement membrane proteins as well as intra-epidermal proteins, desmoplakin and plakophilin 1 (PKP1) (Fine and Hintner, 2009; Fine et al., 2008a; Uitto et al., 1997).
Depending upon where the tissue separation occurs, EB can be subdivided into four main groups: EB simplex, in which blister formation is caused by keratin 5 or 14 mutations, leading to the disruption of the basal keratinocytes; junctional EB (JEB) in which tissue cleavage arises in the lamina lucida; and dystrophic EB, in which cleavage occurs beneath the lamina densa of the dermo-epidermal basement membrane zone (Fine et al., 2008a; Petronius et al., 2003). A fourth type, Kindler syndrome, is characterized by cleavage within and beneath the basement membrane zone, and results from mutations in the Kindlin-1 gene (possibly mouse Fermt1) (Fine et al., 2008b; Ussar et al., 2006).
JEB results from mutations within collagen 17a1 (COL17A1) or any of the genes encoding for the three subunits of laminin-332 (Aumailley et al., 2005), a heterotrimeric macromolecule composed of laminin α3 (LAMA3), laminin β3 (LAMB3), and laminin γ2 (LAMC2). Generalized JEB is subclassified into two major subtypes, based on clinical and molecular criteria. The more severe Herlitz subtype, associated commonly with death during infancy or early childhood, is usually characterized by the presence of compound heterozygote mutations leading to premature termination codons within any of the three laminin-332 subunit genes. Non-Herlitz JEB is generally less severe and is associated with better prognosis. The majority of cases of non-Herlitz JEB result from less severe mutations (missense; in-frame splicing) within the laminin-332 genes (Fine et al., 2008a; Uitto and Richard, 2004).
Because EB is a heritable disease with potentially deadly sequelae, gene therapy may offer a desirable alternative for its treatment (Ferrari et al., 2005). Such therapy, however, is in its infancy and must be developed and validated in animal models before it would be justifiable to apply to humans.
Animal models of skin disease are proving invaluable in the study of human genodermatoses. JEB-like phenotypes have been previously described in dogs (Capt et al., 2005; Dunstan et al., 1988; Nagata et al., 1997; Nagata et al., 1995) and horses (Frame et al., 1988; Spirito et al., 2002). A viable mouse model of JEB would be invaluable, since it can manipulated experimentally and genetically. Targeted disruption of laminin alpha 3, Lama3 (Lama3tm1Crt) (Ryan et al., 1999), or laminin gamma 2, Lamc2 (Lamc2tm1Uit) (Meng et al., 2003), as well as the spontaneous intragenic insertion of an intracisternal A particle (IAP) element into laminin beta 3, Lamb3 (Lamb3IAP) (Kuster et al., 1997), results in a JEB-like phenotype in mice, but all lead to perinatal lethality, thus reducing their experimental utility. A recently produced targeted mouse mutant in collagen XVII (Col17a1tm1Shzu) develops a JEB phenotype. However, most of these mice die within 2 weeks of birth (Nishie et al., 2007). A hypomorphic allele of collagen VII (Col7A1tm1Lbt) has been shown to result in a dystrophic EB phenotype; most of these mice die before 28 days (Fritsch et al., 2008). The only long-term viable EB mouse model currently available is an inducible model for the Dowling-Meara variant of generalized EB simplex (Cao et al., 2001). Here we describe a previously unreported model for JEB in which a hypomorphic allele of the gene Lamc2 results in viable mice that progressively develop a syndrome remarkably similar to that observed in humans.
RESULTS
Histopathology of Lamc2jeb mutant mice
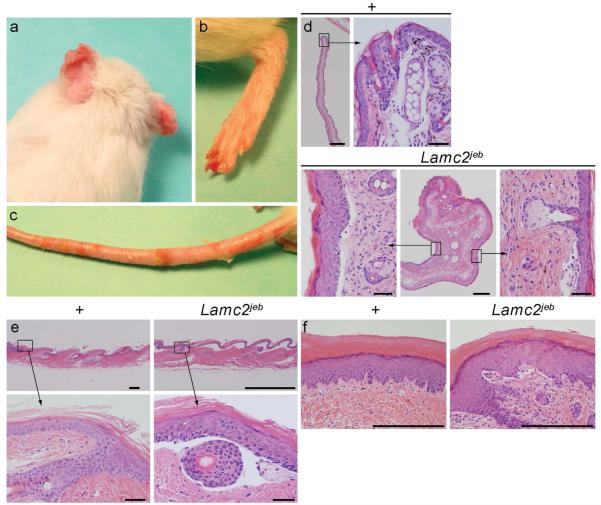
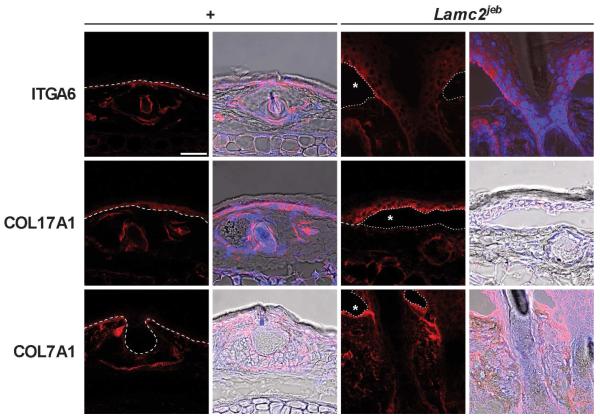
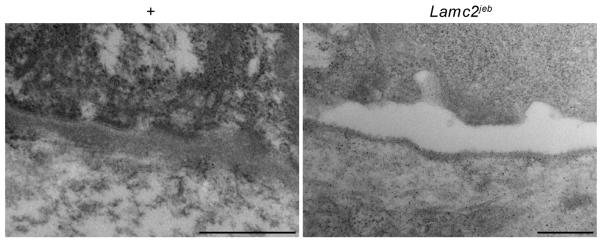
Abnormal mice were first noted in an inbred stock of 129X1/SvJ (129) mice. They developed conspicuously hyperemic, ulcerated pinnae (ears) with similar changes on their tails. These traits were apparent with 100% penetrance in this inbred stock with a mean onset of 154 +/− 4.3 days (n=23) but were never detected in other 129 stocks. Complete necropsies (Relyea et al., 2000; Seymour et al., 2004), were initially performed on two affected and two clinically normal, six month old male and female littermates. Lesions were limited to the skin on several parts of the body (Fig. 1A-C). Histologically, the ears were severely ulcerated with granulation tissue forming at the base of the ulcers (Fig. 1E). Dermal-epidermal separation at the level of the basement membrane was present (Fig. 1E). The most diagnostic areas were the tail (Fig. 1D) and footpads (Fig. 1F), where the skin was not ulcerated. Subepidermal separation, with little to no dermal inflammation, was present. Large areas of epidermis lifted off of the tail (Fig. 1C), confirming the presence of marked mechanical fragility of the skin. The histological features of the footpads in the same mouse ranged from areas of normality, to acute blister formation, or healing by scarring. Dorsal thoracic skin was normal or had areas of ulceration with adjacent dermal-epidermal separation (not shown). Histological, histochemical, and immunohistochemical (IHC) analyses supported the diagnosis of JEB. There was no evidence of any keratinocytes adhering to the base of the cleft, thereby excluding a variant of EB simplex. The most abundant follicular and interfollicular epidermal keratins, most notably keratins 5 and 14, were expressed normally, as would be expected in JEB skin (data not shown). This was also true for mouse specific keratin 6, which is normally expressed in the companion layer of the hair follicle (as it was in these mice) and only in hyperplastic epidermis (data not shown). In particular, immunohistochemical localization of type IV collagen to the base of the blister (not shown), indicative of intralamina lucida cleavage confirms the diagnosis of JEB (Megahed, 2004). Immunofluorescence blister mapping revealed that_integrin alpha 6 (ITGA6) and bullous pemphigoid antigen 180 (COL17A1) expression were located on the roof of the blister while type VII collagen (COL7A1) expression was exclusively present on the blister floor (Fig. 2), thereby localizing the blister to the region where laminin type 5 (laminin-332), which includes laminin gamma 2, is located. These observations closely parallel those seen in skin lesions from patients with JEB. In addition, the presence on transmission electron microscopy (TEM) of intralamina lucida cleavage and reduced numbers of rudimentary-appearing hemidesmosomes further support the diagnosis that this is a model for generalized non-Herlitz type JEB (Fig. 3).
Figure 1. Gross and histological lesions seen in 6 month old 129-Lamc2jeb/Lamc2jeb mutant mice.
A. Ulcers are evident on the tips of the pinnae resulting in distortion. B. Foot pads are hyperemic and ulcerated. C. The tail has multiple ulcers of various size. D. Ear tips in normal +/+ mice. The long, straight auricular cartilage and relatively straight pinna of uniform thickness (A, bar = 500 μm). Higher magnification of the distal end (bar = 100 μm). Deformed ear from a Lamc2jeb/Lamc2jeb mutant mouse. The central figure (bar = 500 μm) is a low magnification of the deformed distal pinna. Note the marked soft tissue thickening due to ulceration, inflammation, and exudation. The auricular cartilage is severely distorted due to contracture from scarring. Boxed areas on either side (bar = 100 μm) are magnified to show marked epidermal hyperplasia (marked acanthosis and mild orthokeratotic hyperkeratosis) with dermal-epidermal separation. The same is seen with separation around the telogen hair follicle. E. Tail skin illustrated the most dramatic lesions. While WT tail skin had a normal moderately thickened epidermis (compared to normal truncal skin) with tight dermal-epidermal adhesion (bar = 200 μm, bar = 100 μm), Lamc2jeb/Lamc2jeb mice at 6 months of age had complete separation of the dermis from the epidermis (bar = 1 mm, bar=100 μm). F. Normal footpads in 6 month old +/+ mice have a thick orthokeratotic hyperkeratotic stratum corneum overlying a thick Malphigian layer that is firmly attached to the underlying dermis. By contrast, Lamc2jeb/Lamc2jeb mutant mice have dermal-epidermal separation in footpads (bar = 200 μm)
Figure 2. Blister mapping confirms separation at level of lamina lucida.
Expression of basement membrane zone proteins integrin alpha 6, collagen type XVII, and collagen type VII. Immunofluorescence of 129X1/SvJ +/+ and Lamc2jeb/Lamc2jeb tail skin using 63x lens on a Leica Confocal microscope at 546nm (red) adjusting threshold and gain to maximize dynamic range. Images on left are at 546nm on right are DAPI and bright field overlay. The blister cavity is indicated by a star. (bar = 50 μm) Dotted line represents dermal epidermal boundary.
Figure 3. Ultrastructural analysis confirms separation at lamina lucida.
Transmission electron micrographs (50,000 x) of +/+ (A) and Lamc2jeb/Lamc2jeb (B) mice footpads. The separation is at the lamina lucida and there are rudimentary or absent hemidesmosomes. (bar = 500 μm)
Mapping Lamc2 jeb
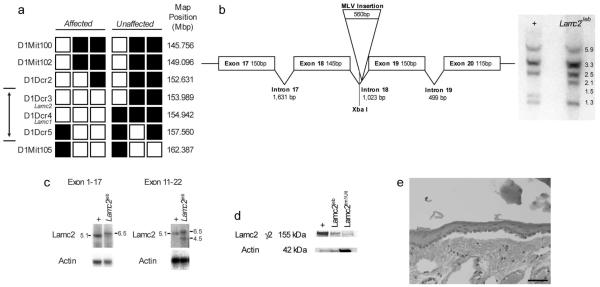
The autosomal recessive junctional epidermolysis bullosa mutation (gene symbol Lamc2 jeb) arose spontaneously. To map the mutated gene, we performed an intraspecific cross between C57BL/6J (B6) wild type (+/+) (females) mice and 129-Lamc2jeb/Lamc2jeb mutant male mice. All F1 progeny were unaffected. These F1 progeny were then backcrossed (129-Lamc2jeb/Lamc2 jeb X B6 +/+)F1 x 129-Lamc2 jeb /Lamc2 jeb mice, with ~50% of their offspring being affected, thus confirming the action of a single autosomal recessive Mendelian trait. To map the mutated gene locus, DNAs from the backcross mice were analyzed using a DNA pooling genome scanning method (Taylor et al., 1994) using allelically discriminant dinucleotide repeat markers mapping to 15-20 cM intervals throughout the genome. Seventeen affected, 11 males/6 females, 18 unaffected littermates, 11 males/7 females, parental 129, B6, and F1 hybrids were used for initial mapping. This low resolution mapping technique revealed that the mutation was linked to a ~16 Mb region on mouse Chromosome (Chr) 1 between 143.810 and 160.466 Mb. Once the preliminary map position for the Lamc2 jeb locus was established, we developed a high resolution map of the region surrounding this locus with the goal of mapping Lamc2 jeb to an interval in which candidate genes could be evaluated for “positional cloning”. DNA from each F2 mouse was allele typed for the most proximal and distal recombinant markers within the ~16 Mb interval. As the mutation arose in 129 mice, the most informative mice were Lamc2 jeb homozygotes with at least one B6 allelic marker in the interval. This mapping narrowed the mutation to a ~5 Mb region between 152.110 and 155.702 (75-82 cM) on Chr 1 to a region including Lamc1 and Lamc2 (Fig. 4A). Lamc1 encodes for the laminin gamma 1 subunit, a component of 10 of the 11 laminin family members. Absence of Lamc1 results in embryonic lethality due to the failure of endoderm differentiation (Bader et al., 2005; Smyth et al., 1999; Willem et al., 2002). However, as summarized earlier, mutations in Lamc2 are associated with forms of JEB (Meng et al., 2003).
Figure 4. JEB is a hypomorphic allele of Lamc2.
A. High-resolution haplotype matrix representing the region of interest on mouse Chromosome 1 (the double-headed arrow denotes the maximum non-recombinant interval). Black squares indicate C57BL/6J and 129X1/SvJ heterozygotes; white squares indicate 129X1/SvJ homozygotes. Map positions are based upon public NCBI mouse genome build 37. B. Schematic representation of a portion of Lamc2 (left). Southern blot of Xba I digestion of WT (+/+) and (Lamc2jeb/Lamc2jeb) genomic DNA probed with a cDNA fragment from exons 11-21 (right). An XbaI fragment shift from 1.5 to 2.1 is consistent with the results of Lamc2jeb sequencing, which showed the insertion of murine leukemia virus sequence (MLV) within intron 18 of the Lamc2 gene. C. Northern blot of poly A enriched RNA from skin of mutant (Lamc2jeb/Lamc2jeb) and WT (+/+) control mice hybridized with a probe from exons 1-17 (left) and the second blot an exons 11-21 probe (right). Data are representative of two experiments comparing multiple +/+ and Lamc2jeb/Lamc2jeb mice. D. Western blot indicates altered levels of (155kDa) LAMC2 in Lamc2jeb/Lamc2jeb mice. Blots of total protein from keratinocytes separated on a denaturing 4-12% SDS-PAGE gel were probed with anti-LAMC2 sc-28330 against amino acids 1011-1193 (exons 20-23). When normalized to actin there is substantial reduction in the amount of the LAMC2 protein product in Lamc2jeb/Lamc2jeb mice compared to the WT control. Lamc2tm1Uit targeted mutation was overloaded 10X and shown as a negative control. E. H&E stained section showing dermal epidermal separation observed in the Lamc2jeb/Lamc2tm1Ui product of the complementation test. (bar= 100 μm)
The Lamc2jeb mutation is caused by a retroviral insertion
Because both Lamc1 and Lamc2 were considered candidate genes for explaining the Lamc2jeb mutation, we first evaluated gene expression differences using quantitative real-time PCR. Five primers sets specific to Lamc1, as well as five specific to Lamc2. Neither Lamc1 nor Lamc2 showed a significant change in expression level between Lamc2jeb/Lamc2jeb and + controls when three mutants and three biological control replicates were compared (data not shown). Northern blot analysis of skin poly A RNA probed with a Lamc1 fragment revealed the presence of only the expected 7.6 kb band (data not shown). Lamc2 is reported to encode for two transcripts (5.1 and 2.4 kb), the smaller of which is not expected in adult skin because it exhibits very specific tissue and developmental expression (Airenne et al., 1996; Airenne et al., 2000; Lee et al., 2001). Skin of wild type 129 mice yielded the expected dominant 5.1 kb transcript when probed with a DNA fragment that spanned exons 1-17. By contrast, 129-Lamc2 jeb homozygous mice yielded a major transcript of 6.5 kb and potentially a weak band consistent with the 5.1 kb wild-type transcript (Fig. 4C, left panel). When probed with a more 3′ fragment spanning exons 11-21, two major mutant bands were observed, one again consistent with the 6.5 kb band and a second band of approximately 4.5 kb (Fig. 4C, right panel). This lower size band, detected with the exon 11-21 probe but not with the exon 1-17 probe, suggests that the mutation can also result in a second alternative transcript that initiates 3′ of exon 17.
Sequencing of all exons failed to reveal a change between the jeb and 129 wild type Lamc2; however, sequencing of all introns revealed the presence of a single Murine Leukemia Virus Long Terminal Repeat (MLVLTR) insertion of 560 bp within the 18th intron (between exons 18 and 19, 900 bp from the end of 18 and 130 bp before the beginning of exon 19) of Lamc2 (Fig. 4B). Restriction fragment length polymorphism (RFLP) analysis of genomic DNA by Southern blotting was consistent with this single LTR insertion being the only detected genetic change in the mutant Lamc2 locus (Fig. 4B). Given the increased size of the major Lamc2 jeb transcript observed by Northern blotting, we hypothesized that this increase is because of the incorporation of the LTR as_part of the transcript. Indeed, by use of oligonucleotide primer pairs nested in the LTR and either 3′ or 5′ exonic sequence for RT-PCR, we detected and sequence verified cDNA products containing the intron and LTR (Supp. Figs. 1 and 2). This transcript includes 5′ exons, does not splice out intron 18 and the LTR, and introduces a TAG translational stop codon in intron 18 (Supp. Figs. 1 and 2). However, a correctly spliced WT transcript is also produced at low abundance as it has been detected repeatedly by RT-PCR and sequencing (Supp. Figs. 1 and 3). Consistent with this low abundance transcript, a low abundance LAMC2 protein of the correct WT size is found in skin extract of Lamc2 jeb mice by Western blot but negligibly in skin of mice carrying the null Lamc2tm1Utt allele (Meng et al., 2003) (Fig.4D). Immunoreactive LAMC2 protein is also detected in situ_with antibodies reactive to two different regions of the protein (Fig. 5).
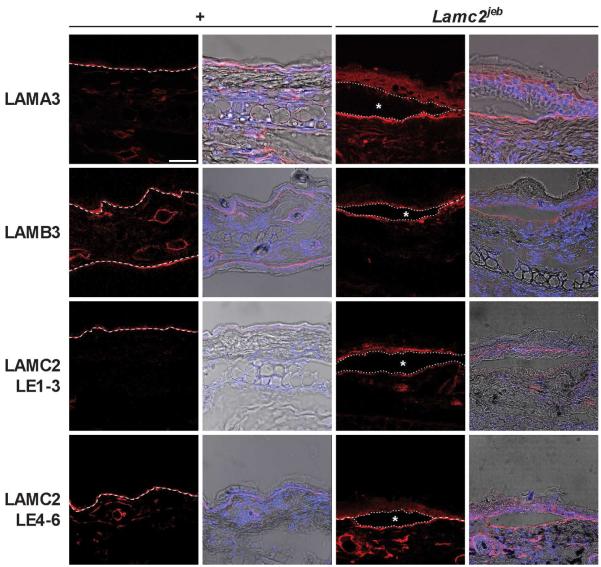
Figure 5. Expression of laminin-332 protein components by immunofluorescence.
Laminin-332 components LAMA3, LAMB3 and LAMC2 were all detected in Lamc2jeb mutant mice, in areas of blistering as well as areas unaffected. Immunofluorescence of 129X1/SvJ +/+ and Lamc2jeb/Lamc2jeb ear skin using 63x lens on a Leica Confocal microscope at 546nm (red) adjusting threshold and gain to maximize dynamic range. Images on left are at 546nm on right are DAPI and bright field overlay. Blister cavity is indicated by a star. (bar = 50 μm) Dotted line represents dermal epidermal boundary.
Finally, to confirm that the mutation was within Lamc2, a genetic complementation test was performed by crossing Lamc2 jeb /Lamc2 jeb mice to mice heterozygous for the Lamc2tm1Ui gene inactivation allele of Lamc2 (Meng et al., 2003). All resulting Lamc2 jeb /Lamc2_tm1Utt heterozygotes but not Lamc2jeb /+ or Lamc2tm1Utt/+ littermates developed the characteristic blisters by two months of age confirming that the mutations were allelic (Fig. 4E). This experiment provided formal genetic proof that Lamc2jeb is the mutant allele. The combined results support the model in which the Lamc2jeb mutation is caused by the insertion of a retroviral LTR. This insertion results in abnormal transcripts, but also permits an apparently intact but low abundance Lamc2 WT transcript that results in a greatly reduced level of LAMC2 protein as detected by Western blot (Fig. 4D). Lamc2jeb is therefore a hypomorphic Lamc2 allele with the reduced amount of LAMC2 protein being sufficient to permit long term viability.
Effect on Bone Mineralization
In order to evaluate if the Lamc2 hypomorphic mouse displayed any of the bone mineralization features observed in humans (Fewtrell et al., 2006), mice were analyzed by dual energy x-ray absorptiometry. As summarized in Table 1, when mice are asymptomatic early on, as measured by gross ear blistering, there were no statistically significant differences in bone mineral composition (BMC) and bone mass density (BMD). However, as mice age and develop blisters, Lamc2jeb/Lamc2 jeb mice display significant decreases in both of these parameters.
Table 1.
Lamc2jeb mutation results in late onset reduction of bone mineral content
| BMD (g/cm2) ± SEM | BMC (g) ± SEM | |
|---|---|---|
| 9 Month 129X1 (n=10) | 0.0575± 0.0036 | 0.6162±0.0465 |
| 9 Month 129X1 Lamc2jeb/Lamc2jeb (n=5) | 0.0580±0.0024 | 0.6160±0.0742 |
| p-value | 0.669 | 0.987 |
|
| ||
| 12 Month 129X1 (n=5) | 0.0591± 0.0021 | 0.6472±0.0488 |
| 12 Month 129X1 Lamc2jeb/Lamc2jeb (n=5) | 0.0510±0.0021 | 0.532±0.0197 |
| p-value* | 0.0003 | 0.0039 |
Two-tailed T-test
Tooth Involvement
Generalized enamel hypoplasia is a principal feature of both Herlitz and non-Herlitz JEB (Wright et al., 1993). As seen in Fig. 6A, the incisors of Lamc2jeb/Lamc2 jeb mice had substantial pitting, as detected by scanning electron microscopy.
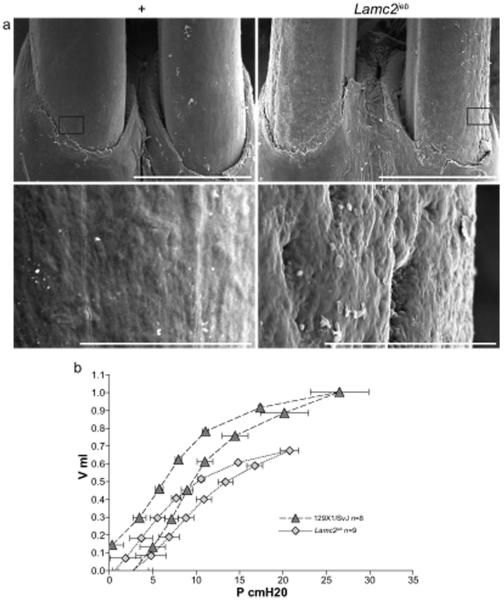
Figure 6. Non-cutaneous skin phenotypes of Lamc2 jebmice.
A. SEM of WT and Lamc2jeb teeth at 35x (bar = 1 mm) and 500x (bar = 100 μm) reveals the surface pitting of the enamel of the teeth in the mutant mice. B. Average lung pressure-volume loops post challenge. The average (±SE) of 8 129X1/SvJ and 9 Lamc2jeb/Lamc2jeb analyzed by forced oscillation technique (at PEEP 3) show clear differences in resistance and/or elastance between WT and Lamc2 jeb/Lamc2 jeb mice. X-axis represents pressure in cm H2O and y-axis represents volume in mL. p < 0.001 by ANOVA.
Respiratory Involvement
Lamc2 is highly expressed in lung ((http://symatlas.gnf.org/SymAtlas/). In addition to the skin manifestations, Lamc2jeb homozygous mutant mice were dyspneic, suggesting pulmonary function abnormalities. No tracheal abnormalities were observed by H&E staining or TEM (data not shown), although abnormal tracheal hemidesmosomes were reported in the neonatal homozygous Lamc2 tm1Utt mice (Nguyen et al., 2006). In order to assess pulmonary mechanics, Lamc2jeb/Lamc2 jeb mice were analyzed by the forced oscillation technique. This process allows for the evaluation of lung function by evaluation of the inflation and deflation phases of each breath and the construction of a classic pressure-volume (PV) curve (Salazar and Knowles, 1964). As shown in Fig. 6B, for a positive end-expiratory pressure (PEEP) of 3 the Lamc2jeb/Lamc2 jeb mice have functional lung defects, resulting in decreased resistance and/or elastance and overall_lung restriction that are similar to those reported in the cystic fibrosis transmembrane conductance regulator (Cftr) deficient mouse (B6.129P2-Cftrtm1Unc/J) (Cohen et al., 2004).
DISCUSSION
We describe a previously unreported, spontaneous mutation in Lamc2 that results in a phenotype of cutaneous blister formation with subsequent ulceration that is remarkable to the extent to which it mimics and is instructive toward understanding human JEB. Affected mice develop blisters as they age but many heal by forming scars under the re-epithelialized surface such that they do not succumb to fluid loss and electrolyte imbalance that would occur with extensive loss of an intact skin barrier. Blister formation above the plane of COL4A1 and COL7A1 and below the plane of ITGA6 and COL17A1 expression support the diagnosis of JEB.
Retroviral insertion within introns is a common mutagenic mechanism in mice (Helms et al., 2005; Seperack et al., 1995; Stoye et al., 1988). Cloning and sequence analysis of Lamc2 revealed that our mouse with the generalized non-Herlitz JEB phenotype had a single MLV LTR insertion within intron 18 of the Lamc2 gene. This insertion gives rise to an abnormally large (6.5 kb) major transcript abrupts the normal excision of intron 18 and introduces a TAG translational stop codon (Supp. Fig. 1). While transcripts with a translational stop codon introduced distantly (>55 bases) from the terminal intron/exon junctions often experience nonsense-mediated decay, there are exceptions (Frankel et al., 2009; Zhao et al., 2005). The apparent stability of the transcript caused by the MLV LTR insertion may have overridden this survey mechanism.
The MLV insertion also leads to the initiation of a smaller (4.5 kb) alternative transcript initiating between exons 18 and 21, perhaps using the known alternate promoter located within exon 19 (Lee et al., 2001). Any protein produced from this truncated transcript is unlikely to have a biological effect as it would lack amino acids including the necessary N-terminal signal sequence. However, a correctly spliced but weakly expressed Lamc2 transcript is detected by Northern and Western blots, and is confirmed by less quantitative in situ immunofluorescence analyses of the dermal-epidermal junction. The fact that the skin lesions are observed only in Lamc2jeb/Lamc2jeb mice is consistent with a recessive mutation, and argues against dominant effects that could arise by the above-described aberrant Lamc2jeb transcripts. This is further supported by the complementation test showing that Lamc2tm1Uit/Lamc2jeb compound heterozygotes develop blisters similar to the Lamc2jeb homozygote. The low abundance, apparently intact transcript is the most parsimonious explanation for this hypomorphic allele._Hypomorphic alleles arising from alternate splicing or exon skipping of LAMC2 and LAMB3 resulting in a reduced WT transcript have also been reported for JEB in humans (Castiglia et al., 2001; Nakano et al., 2002; Pulkkinen et al., 1994).
Classical histological studies evaluated the teeth of JEB patients and concluded that the teeth defects were due to extensive disruption of the ameloblast function and life cycle (Arwill et al., 1965). Unlike humans, rodent incisors are continuously growing. Mice with a targeted knock-out allele of Lamc3 resulted in disrupted ameloblast differentiation and decreased enamel deposition (Ryan et al., 1999). Recent work on the collagen XVII-mutant mouse, Col17a1tm1Shzu, concluded that collagen XVII is also required for tooth enamel formation (Asaka et al., 2008). Laminin-332 is known to have a critical function in the adhesion of the dental epithelial cells with the enamel (Yoshiba et al., 1998). Lamc2jeb mice developed dental abnormalities consistent with that observed in humans (Arwill et al., 1965; Kirkham et al., 2000; Wright et al., 1996; Yoshiba et al., 1998). Our results are consistent with a role for LAMC2 as part of Laminin-332 for proper enamel formation.
They also show the loss of bone mineralization which is also a known characteristic of more severely affected humans with JEB (Fewtrell et al., 2006). Finally, there was a conspicuous respiratory dysfunction in Lamc2jeb mice. Forty percent of JEB patients, regardless of subtype, have moderate to severe tracheolaryngeal involvement (Fine et al., 2007). Of these, a substantial minority, primarily those with Herlitz JEB, die as a direct result of the severity of the upper airway disease while others die of “pneumonia” and other undefined respiratory ailments (Fine et al., 2007). Importantly, respiratory disease in human JEB patients has yet to be described at the level of a quantitative functional assay and this mouse therefore provides a unique opportunity whereby pulmonary function and JEB can be meticulously studied. In addition, since this previously unreported spontaneous mouse model for JEB survives well into adulthood, it provides a useful rodent model for many other levels of investigation, including a mechanistic description of JEB.
Finally, the previously unreported Lamc2 JEB model described here holds promise for evaluating the efficacy and safety of gene therapy. Recent success with ex vivo gene therapy for the treatment of EB appear to be promising (Mavilio et al., 2006) as well as bone marrow transplantation approaches to correct skin defects (Tolar et al., 2009). The Lamc2jeb mice described here provide the biological tool needed to validate these approaches before testing in human patients.
MATERIALS AND METHODS
Mice
Mice were maintained in conventional specific-pathogen free barrier facilities. Routine and skin specific microbiological work done on 129.X1-Lamc2jeb/Lamc2jeb mice with cutaneous ulcers revealed that no known mouse infectious agent was involved in the pathogenesis of JEB. The Jackson Laboratory follows husbandry practices in accordance with the American Association for the Accreditation of Laboratory Animal Care and all work was done with the approval of our Institutional Animal Care and Use Committee. Mice were sex and age matched for each experiment. The mutation arose during the production of 129X1-Fcgrttm1Dcr/Dcr JR#3980 mice and was isolated as 129X1-Lamc2jeb JR#6859 and B6-Lamc2jeb JR#7061 mice. Lamc2tm1Uit (JR#7058) mice were a kind gift from J. Uitto (Thomas Jefferson University).
Gene Mapping
Tail tips were amputated, DNA extracted using a modified hot NaOH protocol (Truett et al., 2000), and DNA pooled from 12 mutant mice. DNA was also collected from 10 clinically normal F2 progeny and 2 parental mice (129-Lamc2jeb/Lamc2jeb) females and B6 +/+ males. Equal amounts of DNA from the 12 mutant mice were mixed to make a pooled sample. A similar pooled sample was assembled from the 12 normal siblings. These two pools were tested for simple sequence length polymorphisms (SSLPs) allele ratios as compared to parental and F1 controls using 87 markers spaced at 7-35cM (mean 14.4 cM spacing) throughout the genome (23). Samples were amplified on a MJ research PTC-1000 Thermal Cycler with initial incubation at 95°C for 5 min, followed by 40 cycles of melting at 95°C for 15s, annealing at 60°C for 20s at 70°C for 30s; followed by holding the samples at 4 °C. Each reaction had a volume of 15 μl containing 3 ng DNA template and 0.025 units/μl Taq DNA Polymerase (Eppendorf, NY). The final concentration of the other reagents were as follows: each primer at 0.4 μM (MWG, High Point NC), 1.5 mM MgCl, 50 mM KCl, 1 mM ß-mercaptoethanol, 25 mM TAPS (N-tris-(hydroxy-methyl)-methyl-3-amino-propanesulfonic acid, sodium salt), 200 μM each dNTP (Promega, Madison WI). All novel dinucleotide repeat based mapping markers are available at MGI (http://www.informatics.jax.org). Markers were electrophoresed in the presence of ethidium bromide and run on a 0.7% agarose with 1.5% Synergel (Diversified BioTech, Boston MA) in Tris-acetate-EDTA. Gels were run at 160 V for 1.5-2 hr and photographed using medium wave UV Syngene Ingenius UV (Frederick, MD) digital camera system.
Histopathology
Organs were fixed overnight in Fekete’s acid-alcohol-formalin fixative, rinsed extensively, held overnight in distilled water, and stored in 70% ethanol until processed. Organs were trimmed, embedded in paraffin, serially sectioned at 6 μm, and then stained with hemotoxylin and eosin (H&E) or periodic acid Schiff.
Immunofluorescence Imaging
Whole ears were cut longitudinally and each half and prepared was rinsed in OCT (Tissue-Tek) until saturated (about 30 seconds to 1 minute). One half of the ear tissue was embedded cut side down into the partially frozen OCT in the base mold. The section was allowed to freeze after which more OCT was added and stored at −80°C. Samples were cut between 10 and 12 μm thick onto super frost plus slides (Fisher). Slides were stored at −20°C until used. For antibody staining the sectioned tissues were fixed in ice cold acetone for 10 minutes, washed three times for 10 minutes in phosphate buffered saline (PBS), and then allowed to dry. Primary antibodies were added with 3% fetal bovine serum for 2 hours at room temperature in a humid chamber. The slides were washed in PBS, incubated in the dark with fluorescence-labeled secondary antibodies for 1 hour, washed and cover slipped using anti-fade gel mounting media (Sigma-Aldrich, St. Louis, MO). Imaging was performed using a SP5 Leica confocal microscope at 63x. The following primary antibodies were used: rabbit anti mouse BP180 and COL7A1 (gifts of Z. Liu), rabbit anti mouse LAMA3, LAMB3, LAMC2 LE1-3 (AA 22-186, exons 2-6), LAMC2 LE4-6 (AA 460-606, exons 11-13, a gift of T. Sasaki) and rat anti-mouse ITGA6 (GeneTex, San Antonio, TX). All secondary antibodies were anti-rabbit or anti-rat Alexa Fluor 546 (Invitrogen/Molecular Probes, Carlsbad, CA).
Electron Microscopy
Samples were prepared according to routine methods previously described (Bechtold, 2000). Skin samples were fixed in 2.5% glutaraldehyde in 0.1M phosphate buffer (PB) pH 7.2 overnight at 4°C. Samples were then washed 2 × 15 minutes in PB and soaked in 2% osmium tetroxide overnight at 4°C followed again by 2 × 15 minute washes in PBS. Samples for TEM were serially dehydrated in ethanol followed by propylene oxide incubation twice for 30 minutes. The samples were then treated with propylene oxide:epon-araldite (1:1) for 24 hrs with rotation followed by 24 hrs of rotation in pure resin. Samples were then cured for 1 day at 70°C, sectioned, and examined by on a JEM-1230 TEM (JEOL, Tokyo, Japan). For scanning electron microscopy, teeth samples were prepared in Karnovsky’s fixative and mounted with double-stick tape on aluminum stubs, sputter-coated with a 4 nm layer of gold, and examined at 20kV at a working distance of approximately 15 mm on a Hitachi S3000N VP Scanning Electron Microscope (Hitachi Science Systems, Japan).
Transcript Analysis and Sequencing
For Northern blot, total RNA was isolated using the standard Trizol reagent method (Invitrogen). Poly A+ enrichment was performed by passing over oligo(dT) cellulose column (Applied Biosystems/Ambion, Foster City, CA). Two micrograms of RNA per lane was analyzed by Northern blot. 1 ug of total RNA was used to make cDNA using messages Sensor RT (Applied Biosystems/Ambion). The cDNA was used to produce probes for Lamc1 and Lamc2 using primers for Lamc1F, Lamc1R, Lamc2.2F, Lamc2.18R, Lamc2.12F, Lamc2.22R is described in Supplemental Table 1. Additional primers used for quantitative Real Time PCR on cDNA produced from both WT and 129X1/SvJ Lamc2jebmice are listed in Supplemental Table 1. Gene expression differences were determined using the Global Pattern Recognition (GPR) software (Akilesh et al., 2003). For sequencing, total RNA was isolated from spleen using RNAqueous 4-PCR (Ambion). Oligo(dT)18 and random decamer primed cDNA was synthesized using ThermoScript RT (Invitrogen). Sequencing of Lamc2 was done using gene specific primers on an Applied Biosystems 3700 and analyzed using Sequencher 4.8 (Gene Codes Corporation, Ann Arbor, MI).
FlexiVent Measurements
Mice are anesthetized with ketamine/dormitor at doses based on body weight. To prevent the mice from breathing against the respirator, the mice were treated with pancuronium bromide in NaCl as a muscle relaxant at 0.2mg/kg. Once the mouse was properly anesthetized, a small incision was made between the 3rd and 5th tracheal rings whereupon a 1.27 cm long tracheal cannula was inserted. The cannula was secured in place with suture material. The mouse was then fixed to the flexiVent ventilator (SCIREQ, Montreal, CA) and ventilated at 200 breaths/min with a tidal volume of 10 ml/kg body weight. Once the mouse was breathing passively, 2 consecutive sigh breaths were performed to open the airways and lungs. Baseline R, C, and E measures were recorded. Sigh breaths were performed throughout the experiment to keep airways and lungs open.
Bone Density Measurements
Peripheral dual-energy X-ray absorptiometry (pDXA; PIXI-mus, GE-Lunar, Madison WI) was used to assess bone mineral density and bone mineral composition, with head exclusion. This methodology is routinely used in and validated in small animals (Bouxsein et al., 2002). Mice were anesthetized with tribromoethanol at 180 mg/kg intraperitoneal, allowing 5 minutes for sedation before scans.
Western Blot Analysis
Skin lysates were prepared in 20 mM Tris-HCl (pH 7.5), 135 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, 1% Triton X-100, and 10% glycerol supplemented with a complete protease inhibitor cocktail (Gene Technology, Somerville, MA). Total protein was quantified using BCM protein analysis kit (Thermo Scientific/Pierce, Rockford, IL). 10-100 ug was separated on a BioRad Criterion XT 4-12% Bis-Tris gel, electroblotted onto a PVDF membrane (BioRad, Hercules, CA), and probed with a 1:500 dilution of LAMC2 sc-28330 anti-human LAMC2 antibody (Santa Cruz, Santa Cruz, CA). The secondary antibody used (1:1000) was goat anti mouse IgG conjugated to HRP (Southern Biotech, Birmingham, AL) with detection using Pierce ECL western blot substrate. Images were collected on a Fuji LAS1000 CCD luminescent imaging system, with band intensities quanified using ImageJ (NIH). As a loading control the blot was stripped and re-probed with rabbit anti-mouse actin (Sigma) and goat anti-rabbit conjugated HRP.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by Dystrophic Epidermolysis Research Association (DebRA) - International and The National Institutes of Health (DK56597, DCR; AR49288, JPS). The authors thank L. E. King, Jr. for discussions on the comparative aspects of this mouse model with the human disease, and A. Brown and especially W. Frankel for advice regarding MLVs and genomic analyses. We appreciate Drs. J. Uitto and J. Klement for making the Lamc2tm1Uit mouse available, T. Sasaki for anti- LAMA3, LAMB3, and LAMC2 antibodies, and Z. Liu for the anti-BP180 and anti COL7A1 antibodies. We also thank K.A. Silva and J. Miller for initial sample preparation and immunohistochemical analysis as well as L. Rowe and M. Barter for SSLP mapping services. We thank A. Nicholson and J. Ryan for pulmonary physiology analyses, B. King and T. Sterns for statistical analysis, and L. Bechtold and P. Finger for EM sample preparation.
Abbreviations
- EB
epidermolysis bullosa
- jeb
junctional epidermolysis bullosa
- Lamc2
laminin gamma 2 gene
- Lamc2jeb
mouse laminin gamma 2 allelic mutation
- MLV
Murine Leukemia Virus
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
References
- Airenne T, Haakana H, Sainio K, Kallunki T, Kallunki P, Sariola H, et al. Structure of the human laminin gamma 2 chain gene (LAMC2): alternative splicing with different tissue distribution of two transcripts. Genomics. 1996;32:54–64. doi: 10.1006/geno.1996.0076. [DOI] [PubMed] [Google Scholar]
- Airenne T, Lin Y, Olsson M, Ekblom P, Vainio S, Tryggvason K. Differential expression of mouse laminin gamma2 and gamma2* chain transcripts. Cell Tissue Res. 2000;300:129–37. doi: 10.1007/s004410000182. [DOI] [PubMed] [Google Scholar]
- Akilesh S, Shaffer DJ, Roopenian D. Customized molecular phenotyping by quantitative gene expression and pattern recognition analysis. Genome Res. 2003;13:1719–27. doi: 10.1101/gr.533003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arwill T, Olsson O, Bergenholtz A. Epidermolysis Bullosa Hereditaria. 3. A Histologic Study of Changes in Teeth in the Polydysplastic Dystrophic and Lethal Forms. Oral Surg Oral Med Oral Pathol. 1965;19:723–44. doi: 10.1016/0030-4220(65)90342-7. [DOI] [PubMed] [Google Scholar]
- Asaka T, Akiyama M, Domon T, Nishie W, Natsuga K, Fujita Y, et al. Type XVII Collagen is a Key Player in Tooth Enamel Formation. Am J Pathol. 2008 doi: 10.2353/ajpath.2009.080573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aumailley M, Bruckner-Tuderman L, Carter WG, Deutzmann R, Edgar D, Ekblom P, et al. A simplified laminin nomenclature. Matrix Biol. 2005;24:326–32. doi: 10.1016/j.matbio.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Bader BL, Smyth N, Nedbal S, Miosge N, Baranowsky A, Mokkapati S, et al. Compound genetic ablation of nidogen 1 and 2 causes basement membrane defects and perinatal lethality in mice. Mol Cell Biol. 2005;25:6846–56. doi: 10.1128/MCB.25.15.6846-6856.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold LS. Ultrastructural evaluation of mouse mutations. In: Sundberg JP, Boggess D, editors. Systematic characterization of mouse mutations. CRC Press; Boca Raton: 2000. pp. 121–9. [Google Scholar]
- Bouxsein ML, Rosen CJ, Turner CH, Ackert CL, Shultz KL, Donahue LR, et al. Generation of a new congenic mouse strain to test the relationships among serum insulin-like growth factor I, bone mineral density, and skeletal morphology in vivo. J Bone Miner Res. 2002;17:570–9. doi: 10.1359/jbmr.2002.17.4.570. [DOI] [PubMed] [Google Scholar]
- Cao T, Longley MA, Wang XJ, Roop DR. An inducible mouse model for epidermolysis bullosa simplex: implications for gene therapy. J Cell Biol. 2001;152:651–6. doi: 10.1083/jcb.152.3.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capt A, Spirito F, Guaguere E, Spadafora A, Ortonne JP, Meneguzzi G. Inherited junctional epidermolysis bullosa in the German Pointer: establishment of a large animal model. J Invest Dermatol. 2005;124:530–5. doi: 10.1111/j.0022-202X.2004.23584.x. [DOI] [PubMed] [Google Scholar]
- Castiglia D, Posteraro P, Spirito F, Pinola M, Angelo C, Puddu P, et al. Novel mutations in the LAMC2 gene in non-Herlitz junctional epidermolysis bullosa: effects of laminin-5 assembly, secretion, and deposition. J Invest Dermatol. 2001;117:731–9. doi: 10.1046/j.0022-202x.2001.01453.x. [DOI] [PubMed] [Google Scholar]
- Cohen JC, Lundblad LK, Bates JH, Levitzky M, Larson JE. The “Goldilocks effect” in cystic fibrosis: identification of a lung phenotype in the cftr knockout and heterozygous mouse. BMC genetics. 2004;5:21. doi: 10.1186/1471-2156-5-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunstan RW, Sills RC, Wilkinson JE, Paller AS, Hashimoto KH. A disease resembling junctional epidermolysis bullosa in a toy poodle. Am J Dermatopath. 1988;10:442–7. Dunstan, R. W., R. C. Sills, J. E. Wilkinson, A. S. Paller, and K. H. Hashimoto. 1988. [PubMed] [Google Scholar]
- Ferrari S, Pellegrini G, Mavilio F, De Luca M. Gene therapy approaches for epidermolysis bullosa. Clin Dermatol. 2005;23:430–6. doi: 10.1016/j.clindermatol.2004.07.017. [DOI] [PubMed] [Google Scholar]
- Fewtrell MS, Allgrove J, Gordon I, Brain C, Atherton D, Harper J, et al. Bone mineralization in children with epidermolysis bullosa. The British journal of dermatology. 2006;154:959–62. doi: 10.1111/j.1365-2133.2005.07123.x. [DOI] [PubMed] [Google Scholar]
- Fine J-D, Hintner H. Life with epidermolysis bullosa (EB): etiology, diagnosis, multidisciplinary care, and therapy. xix. Springer: Wien; New York: 2009. p. 338. [Google Scholar]
- Fine JD, Eady RA, Bauer EA, Bauer JW, Bruckner-Tuderman L, Heagerty A, et al. The classification of inherited epidermolysis bullosa (EB): Report of the Third International Consensus Meeting on Diagnosis and Classification of EB. Journal of the American Academy of Dermatology. 2008a;58:931–50. doi: 10.1016/j.jaad.2008.02.004. [DOI] [PubMed] [Google Scholar]
- Fine JD, Johnson LB, Weiner M, Suchindran C. Tracheolaryngeal Complications of Inherited Epidermolysis Bullosa: Cumulative Experience of the National Epidermolysis Bullosa Registry. Laryngoscope. 2007;117:1652–60. doi: 10.1097/MLG.0b013e318093ed8e. [DOI] [PubMed] [Google Scholar]
- Fine JD, Johnson LB, Weiner M, Suchindran C. Cause-specific risks of childhood death in inherited epidermolysis bullosa. The Journal of pediatrics. 2008b;152:276–80. doi: 10.1016/j.jpeds.2007.06.039. [DOI] [PubMed] [Google Scholar]
- Frame SR, Harrington DD, Fessler J, Frame PF. Hereditary junctional mechanobullous disease in a foal. J Am Vet Med Assoc. 1988;193:1420–4. [PubMed] [Google Scholar]
- Frankel WN, Yang Y, Mahaffey CL, Beyer BJ, O’Brien TP. Szt2, a novel gene for seizure threshold in mice. Genes Brain Behav. 2009;8:568–76. doi: 10.1111/j.1601-183X.2009.00509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsch A, Loeckermann S, Kern JS, Braun A, Bosl MR, Bley TA, et al. A hypomorphic mouse model of dystrophic epidermolysis bullosa reveals mechanisms of disease and response to fibroblast therapy. The Journal of clinical investigation. 2008;118:1669–79. doi: 10.1172/JCI34292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms C, Pelsue S, Cao L, Lamb E, Loffredo B, Taillon-Miller P, et al. The Tetratricopeptide repeat domain 7 gene is mutated in flaky skin mice: a model for psoriasis, autoimmunity, and anemia. Experimental biology and medicine (Maywood, NJ. 2005;230:659–67. doi: 10.1177/153537020523000908. [DOI] [PubMed] [Google Scholar]
- Kirkham J, Robinson C, Strafford SM, Shore RC, Bonass WA, Brookes SJ, et al. The chemical composition of tooth enamel in junctional epidermolysis bullosa. Arch Oral Biol. 2000;45:377–86. doi: 10.1016/s0003-9969(00)00003-0. [DOI] [PubMed] [Google Scholar]
- Kuster JE, Guarnieri MH, Ault JG, Flaherty L, Swiatek PJ. IAP insertion in the murine LamB3 gene results in junctional epidermolysis bullosa. Mamm Genome. 1997;8:673–81. doi: 10.1007/s003359900535. [DOI] [PubMed] [Google Scholar]
- Lee G, Kim MG, Yim JB, Hong SH. Alternative transcriptional initiation and splicing of mouse Lamc2 message. Mol Cells. 2001;12:380–90. [PubMed] [Google Scholar]
- Lee JW, Beebe K, Nangle LA, Jang J, Longo-Guess CM, Cook SA, et al. Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature. 2006;443:50–5. doi: 10.1038/nature05096. [DOI] [PubMed] [Google Scholar]
- Mavilio F, Pellegrini G, Ferrari S, Di Nunzio F, Di Iorio E, Recchia A, et al. Correction of junctional epidermolysis bullosa by transplantation of genetically modified epidermal stem cells. Nat Med. 2006;12:1397–402. doi: 10.1038/nm1504. [DOI] [PubMed] [Google Scholar]
- Megahed M. Histopathology of Blistering Diseases. Vol. 405. Springer-Verlag; Beline Heidelberg New York: 2004. [Google Scholar]
- Meng X, Klement JF, Leperi DA, Birk DE, Sasaki T, Timpl R, et al. Targeted inactivation of murine laminin gamma2-chain gene recapitulates human junctional epidermolysis bullosa. J Invest Dermatol. 2003;121:720–31. doi: 10.1046/j.1523-1747.2003.12515.x. [DOI] [PubMed] [Google Scholar]
- Nagata M, Iwasaki T, Masuda H, Shimizu H. Non-lethal junctional epidermolysis bullosa in a dog. The British journal of dermatology. 1997;137:445–9. [PubMed] [Google Scholar]
- Nagata M, Shimizu H, Masunaga T, Nishikawa T, Nanko H, Kariya K, et al. Dystrophic form of inherited epidermolysis bullosa in a dog (Akita Inu) The British journal of dermatology. 1995;133:1000–3. doi: 10.1111/j.1365-2133.1995.tb06942.x. [DOI] [PubMed] [Google Scholar]
- Nakano A, Lestringant GG, Paperna T, Bergman R, Gershoni R, Frossard P, et al. Junctional epidermolysis bullosa in the Middle East: clinical and genetic studies in a series of consanguineous families. Journal of the American Academy of Dermatology. 2002;46:510–6. doi: 10.1067/mjd.2002.119673. [DOI] [PubMed] [Google Scholar]
- Nguyen NM, Pulkkinen L, Schlueter JA, Meneguzzi G, Uitto J, Senior RM. Lung development in laminin gamma2 deficiency: abnormal tracheal hemidesmosomes with normal branching morphogenesis and epithelial differentiation. Respir Res. 2006;7:28. doi: 10.1186/1465-9921-7-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishie W, Sawamura D, Goto M, Ito K, Shibaki A, McMillan JR, et al. Humanization of autoantigen. Nature Medicine. 2007;13:378–83. doi: 10.1038/nm1496. [DOI] [PubMed] [Google Scholar]
- Petronius D, Bergman R, Ben Izhak O, Leiba R, Sprecher E. A comparative study of immunohistochemistry and electron microscopy used in the diagnosis of epidermolysis bullosa. Am J Dermatopathol. 2003;25:198–203. doi: 10.1097/00000372-200306000-00003. [DOI] [PubMed] [Google Scholar]
- Pulkkinen L, Christiano AM, Airenne T, Haakana H, Tryggvason K, Uitto J. Mutations in the gamma 2 chain gene (LAMC2) of kalinin/laminin 5 in the junctional forms of epidermolysis bullosa. Nature genetics. 1994;6:293–7. doi: 10.1038/ng0394-293. [DOI] [PubMed] [Google Scholar]
- Relyea MJ, Sundberg JP, Ward JM. Immunohistochemical and immunofluorescence methods. In: Sundberg JP, Boggess D, editors. Systematic approach to evaluation of mouse mutations. CRC Press; Boca Raton, FL: 2000. pp. 131–44. [Google Scholar]
- Ryan MC, Lee K, Miyashita Y, Carter WG. Targeted disruption of the LAMA3 gene in mice reveals abnormalities in survival and late stage differentiation of epithelial cells. J Cell Biol. 1999;145:1309–23. doi: 10.1083/jcb.145.6.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar E, Knowles JH. An Analysis of Pressure-Volume Characteristics of the Lungs. Journal of applied physiology. 1964;19:97–104. doi: 10.1152/jappl.1964.19.1.97. [DOI] [PubMed] [Google Scholar]
- Seperack PK, Mercer JA, Strobel MC, Copeland NG, Jenkins NA. Retroviral sequences located within an intron of the dilute gene alter dilute expression in a tissue-specific manner. The EMBO journal. 1995;14:2326–32. doi: 10.1002/j.1460-2075.1995.tb07227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seymour R, Ichiki T, Mikaelian I, Boggess D, Silva KA, Sundberg JP. Necropsy methods. In: Hedrich HJ, editor. Laboratory mouse. Academic Press; London: 2004. pp. 495–516. [Google Scholar]
- Smyth N, Vatansever HS, Murray P, Meyer M, Frie C, Paulsson M, et al. Absence of basement membranes after targeting the LAMC1 gene results in embryonic lethality due to failure of endoderm differentiation. J Cell Biol. 1999;144:151–60. doi: 10.1083/jcb.144.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spirito F, Charlesworth A, Linder K, Ortonne JP, Baird J, Meneguzzi G. Animal models for skin blistering conditions: absence of laminin 5 causes hereditary junctional mechanobullous disease in the Belgian horse. J Invest Dermatol. 2002;119:684–91. doi: 10.1046/j.1523-1747.2002.01852.x. [DOI] [PubMed] [Google Scholar]
- Stoye JP, Fenner S, Greenoak GE, Moran C, Coffin JM. Role of endogenous retroviruses as mutagens: the hairless mutation of mice. Cell. 1988;54:383–91. doi: 10.1016/0092-8674(88)90201-2. [DOI] [PubMed] [Google Scholar]
- Taylor BA, Navin A, Phillips SJ. PCR-amplification of simple sequence repeat variants from pooled DNA samples for rapidly mapping new mutations of the mouse. Genomics. 1994;21:626–32. doi: 10.1006/geno.1994.1323. [DOI] [PubMed] [Google Scholar]
- Tolar J, Ishida-Yamamoto A, Riddle M, McElmurry RT, Osborn M, Xia L, et al. Amelioration of epidermolysis bullosa by transfer of wild-type bone marrow cells. Blood. 2009;113:1167–74. doi: 10.1182/blood-2008-06-161299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truett GE, Heeger P, Mynatt RL, Truett AA, Walker JA, Warman ML. Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT) BioTechniques. 2000;29:52, 4. doi: 10.2144/00291bm09. [DOI] [PubMed] [Google Scholar]
- Uitto J, Pulkkinen L, McLean WH. Epidermolysis bullosa: a spectrum of clinical phenotypes explained by molecular heterogeneity. Mol Med Today. 1997;3:457–65. doi: 10.1016/s1357-4310(97)01112-x. [DOI] [PubMed] [Google Scholar]
- Uitto J, Richard G. Progress in epidermolysis bullosa: genetic classification and clinical implications. Am J Med Genet C Semin Med Genet. 2004;131C:61–74. doi: 10.1002/ajmg.c.30035. [DOI] [PubMed] [Google Scholar]
- Ussar S, Wang HV, Linder S, Fassler R, Moser M. The Kindlins: subcellular localization and expression during murine development. Experimental cell research. 2006;312:3142–51. doi: 10.1016/j.yexcr.2006.06.030. [DOI] [PubMed] [Google Scholar]
- Willem M, Miosge N, Halfter W, Smyth N, Jannetti I, Burghart E, et al. Specific ablation of the nidogen-binding site in the laminin gamma1 chain interferes with kidney and lung development. Development. 2002;129:2711–22. doi: 10.1242/dev.129.11.2711. [DOI] [PubMed] [Google Scholar]
- Wright JT, Hall KI, Deaton TG, Fine JD. Structural and compositional alteration of tooth enamel in hereditary epidermolysis bullosa. Connect Tissue Res. 1996;34:271–9. doi: 10.3109/03008209609005271. [DOI] [PubMed] [Google Scholar]
- Wright JT, Johnson LB, Fine JD. Development defects of enamel in humans with hereditary epidermolysis bullosa. Arch Oral Biol. 1993;38:945–55. doi: 10.1016/0003-9969(93)90107-w. [DOI] [PubMed] [Google Scholar]
- Yoshiba N, Yoshiba K, Aberdam D, Meneguzzi G, Perrin-Schmitt F, Stoetzel C, et al. Expression and localization of laminin-5 subunits in the mouse incisor. Cell Tissue Res. 1998;292:143–9. doi: 10.1007/s004410051044. [DOI] [PubMed] [Google Scholar]
- Zhao L, Longo-Guess C, Harris BS, Lee JW, Ackerman SL. Protein accumulation and neurodegeneration in the woozy mutant mouse is caused by disruption of SIL1, a cochaperone of BiP. Nature genetics. 2005;37:974–9. doi: 10.1038/ng1620. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.