Abstract
Posttranslational modifications of histone tails are critical epigenetic marks that regulate diverse cellular processes. Histone lysine methylation activates or represses transcription, depending on the site and degree of these modifications. Two classes of histone lysine demethylases remove histone methylation. Lysine demethylase 1 (KDM1, also known as LSD1) is a flavin adenine dinucleotide (FAD)-containing enzyme that removes mono-/di-methylation. The Jumonji C-terminal domain (JmjC) family of histone demethylases uses Fe2+ and α-ketoglutarate as cofactors to remove all methylation states. Structural studies have provided insights into the overall architecture, the catalytic mechanism, and the substrate specificity of histone demethylases. Here, we review these exciting advances in the structure biology of histone demethylases and discuss the general principles applicable to other histone-modifying enzymes.
Introduction
Numerous epigenetic marks, including acetylation, methylation, phosphorylation, and ubiquitylation, are introduced at particular sites on histone tails [1]. These posttranslational modifications of histones modulate chromatin structure and dynamics, and recruit effectors to specific genomic loci in a combinatorial fashion. In doing so, they regulate diverse, chromatin-based processes, ranging from transcription to DNA replication and repair to chromosome condensation and cohesion [2,3]. For example, differential methylation of lysines in histones H3 and H4 lysine can either activate or repress gene transcription, depending on the location and degree (mono-, di-, or tri-methylation) of these modifications [2,3]. H3-K4 di-/tri-methylation (H3K4me2/3) is often associated with actively transcribed gene promoters, whereas H3-K9 di/-tri-methylation (H3K9me2/3) is associated with heterochromatin and generally represses transcription.
Histone lysine methylation had long been thought to be as an irreversible chromatin mark [2,3]. The recent discovery of histone demethylases changed this view [2,3]. Two classes of histone lysine demethylases were discovered, the amine oxidase-related enzymes and the Jumonji C-terminal domain (JmjC)-containing enzymes. Lysine-specific demethylase 1 (KDM1; also known as LSD1) was the first histone demethylase discovered and belonged to the superfamily of the flavin adenine dinucleotide (FAD)-dependent amine oxidases [4••]. Subsequently, the first JmjC-containing histone demethylase (KDM2A; also known as JHDM1a/FBXL11) was biochemically identified [5••]. Following these initially discoveries, other members of these two classes of histone demethylases were identified through sequence homology [3]. Therefore, like all other histone modifications, histone lysine methylation is dynamic and reversible. Its addition and removal are catalyzed by histone methyltransferases and demethylases, respectively. Histone demethylases contribute to the regulation of the steady-state levels of histone methylation and are thus critical for many chromatin-based cellular processes.
The discovery and biochemical characterization of histone demethylases then spurred rapid structural analyses of these enzymes. During the past five years, high-resolution structures have been determined for both classes of histone demethylases. These structural studies have provided significant insights into the architecture, the mechanism of action, and the substrate specificity of these enzymes. This review will highlight these advances and discuss the general principles learned from these studies that may apply to other histone-modifying enzymes.
Histone lysine demethylase 1 (KDM1)
Discovery and function
Human lysine demethylase 1 (KDM1) was originally identified a component of a transcriptional corepressor complex that also contained the REST corepressor (CoREST) and HDAC1/2 [6-8]. This transcriptional corepressor complex could be recruited to RE1 element-containing gene promoters by REST and repressed the transcription of neuron-specific genes in non-neuronal cells. KDM1 contained a domain that was related to FAD-dependent amine oxidases. Yang Shi and coworkers realized that the chemistry used by amine oxidases could conceivably be used to catalyze lysine demethylation [4••]. In this reaction, FAD oxidized the methyl-lysine to form an imine intermediate, which is then hydrolyzed to yield unmodified lysine and formaldehyde. The reduced FAD was then re-oxidized by oxygen (Figure 1a). They then tested whether KDM1 indeed demethylated lysines in histones. Recombinant KDM1 indeed catalyzed the demethylation of H3K4me1/2 in bulk histones, leading to the discovery of the first histone lysine demethylase [4••]. Because the formation of the obligatory imine intermediate in the demethylation reaction required a lone pair of electrons in the nitrogen atom of the methyllysine, KDM1 only demethylates mono-/di-methylated lysines, not tri-methylated ones. Intriguingly, KDM1 alone demethylates H3K4me1/2 in peptides or bulk histones, but not in nucleosomes [9•,10•]. Only the KDM1-CoREST complex efficiently demethylates H3K4me1/2 in nucleosomal substrates.
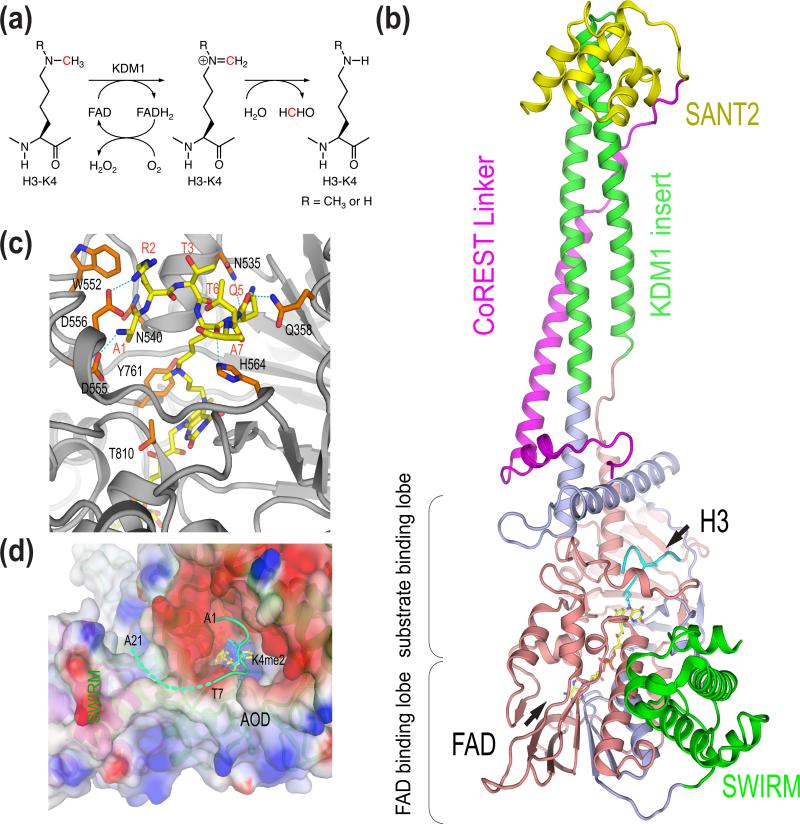
Figure 1.
Mechanism, structure, and substrate specificity of KDM1. (a) Chemical mechanism of KDM1-catalyzed lysine demethylation. (b) Structure of human KDM1-CoREST-H3 ternary complex (PDB code 2UXN). (c) Binding of H3 at the active site of KDM1. AOD is colored gray. The H3 peptide and residues in AOD involved in H3 binding are shown as sticks. (d) The potential involvement of the AOD-SWIRM interface in substrate binding. Residues 1-7 of H3 interact with the negatively charged active-site cavity of KDM1. The surface of AOD and SWIRM domain is colored by the electrostatic potential (red, negative; blue, positive). Residues 8-21 of H3 (shown as a dashed green line) and might bind at the groove between AOD and SWIRM.
KDM1 homologs exist in organisms from fission yeast (S. pombe) to mammals, with each organism containing at least two KDM1-related genes [3]. In most organisms examined except S. pombe, KDM1 homologs catalyze H3K4me1/2 demethylation [3]. The two S. pombe KDM1 homologs form a heterodimer and demethylate H3K9me1/2, as opposed to H3K4me1/2 [11].
Depletion of KDM1 by RNA interference (RNAi) increased H3K4me2 at the promoters of certain REST-responsive genes and activated their transcription, confirming that KDM1 repressed transcription through histone demethylation [4••]. In addition to repressing REST-responsive genes, KDM1-containing complexes can be recruited by alternative mechanisms to other gene promoters to demethylate H3K4me2 and to repress transcription [12-14].
Mammalian KDM1 has also been reported to be required for estrogen receptor (ER)-dependent or androgen receptor (AR)-dependent gene transcription [15,16]. In both cases, it has been suggested that KDM1 demethylates H3K9me2, an epigenetic mark that represses transcription. Direct demethylation of H3K9me2 by KDM1 alone or in complex with other cellular factors has not yet been demonstrated, however. It remains formally possible that KDM1 plays an indirect role in demethylation of H3K9me2 at these ER- or AR-dependent promoters.
The biochemical and functional studies on KDM1 pose two major questions for structural biology. How does CoREST stimulate the activity of KDM1 toward nucleosomes? How does KDM1 achieve its specificity toward H3K4me1/2? These two questions have been addressed by structural studies on KDM1.
CoREST-dependent nucleosome demethylation by KDM1
Structures of KDM1 alone and in complex with the C-terminal domain of CoREST but without bound histone substrates were determined first [17-19]. KDM1 forms an elongated structure with three major parts: the N-terminal SWARM domain, the amine oxidase domain (AOD) consisting of two lobes, and a long stalk formed by the insert between the two halves of AOD (Figure 1b). The SWIRM and AOD domains pack against each other to form a globular structural core. As expected, the AOD of KDM1 has a fold highly similar to conventional FAD-dependent amine oxidases and contains the substrate-binding lobe and FAD-binding lobe. The active site lies at the interface of the two lobes and forms a large cavity. The isoalloxazine ring of FAD is located at the base of this cavity.
CoREST contains an ELM2 domain and two SANT domains [6]. The KDM1-binding domain of CoREST mainly consists of the linker between the two SANT domains [10]. As revealed by the structure of the KDM1-CoREST complex, the long stalk of KDM1 formed by the two antiparallel helices belonging to the insert between the two AOD lobes provides the binding platform for CoREST [18••]. The CoREST linker folds into a long helix and forms extensive contacts with this stalk. The SANT2 domain of CoREST which is critical for nucleosome demethylation is located at the tip of the stalk and far away from the active site of KDM1. The CoREST SANT2 domain is structurally homologous to the Myb DNA-binding domain and indeed has DNA-binding activity [18••]. Mutations within this domain that disrupt its DNA-binding activity also diminish demethylation of nucleosomes by the KDM1-CoREST complex [18••]. These results indicate that CoREST stimulates the binding and activity of KDM1 toward nucleosomes by binding to nucleosomal DNA.
KDM1, but not CoREST, has recently been identified as a component of the Mi-2/nucleosomes remodeling and deacetylase (NuRD) complex [14•]. In this complex, KDM1 directly binds to metastasis tumor antigen 1-3 (MTA1-3) through its stalk. Similar to CoREST, MTA2 stimulates KDM1-mediated demethylation of nucleosomal substrates [14•]. Interestingly, MTAs also contain SANT domains. It is possible that MTAs stimulate KDM1 activity through nucleosomal DNA binding with their SANT domains. In addition to CoREST and MTAs, there may be other unidentified SANT domain-containing proteins that bind to the stalk of KDM1 and help anchor KDM1 to nucleosomes.
Substrate specificity of KDM1
There are striking similarities between the active sites of KDM1 and maize polyamine oxidase (mPAO) [17-20]. In particular, a critical lysine residue (K661 in KDM1) forms water-bridged hydrogen bonds with the N5 atom of FAD in both enzymes and is essential for their activities. There are, however, important structural differences between the two that can explain their substrate preferences. For example, compare to the active site of mPAO, the opening of the active site of KDM1 is considerably larger, enabling KDM1 to accommodate histone peptides that are relatively bulkier than polyamines.
Although KDM1 cannot demethylate trimethylated lysines, a histone H3 peptide containing tri-methylated K4 blocks the demethylation of H3K4me1/2 and acts as a competitive inhibitor of KDM1 [17]. This finding suggests that KDM1 binds to H3K4me3 with an affinity comparable to or higher than that toward H3K4me1/2. This further supports the notion that the inability of KDM1 to demethylate tri-methylated lysines is due to the limitations of its inherent catalytic mechanism, as opposed to a failure to recognize tri-methylated substrates.
How does KDM1 achieve its site specificity toward H3-K4? The weak binding affinity (Km in the μM range) between KDM1 and histone H3 peptides hindered the structure determination of KDM1 in complex with H3 peptides [21]. Mechanism-based peptide inhibitors provided a solution to this problem [22,23]. Addition of one such inhibitor, N-methylpropargyl-K4 H3, to KDM1 followed by sodium borohydride treatment produced a stable KDM1-H3 peptide complex, in which the substrate analog was covalently linked to the cofactor FAD [24•]. The crystal structure of this KDM1-H3 analog complex was successfully determined [24•]. The structure reveals that residues 1-7 of H3 fit snugly into the active-site cavity of KDM1, making extensive contacts with a large set of conserved, negative-charged residues in KDM1 (Figure 1c). The extreme N terminus of H3 is anchored into an anionic pocket in KDM1. H3 adopts an unusually compacted backbone conformation with three consecutive γ-turns, placing the methyl-lysine above the isoalloxazine ring of FAD for catalysis (Figure 1D). This binding mode places strict steric constrains on the H3 substrate and does not permit more than three residues on the N-terminal side of the methyl-lysine. This structure provides a simple explanation for the observed specificity of KDM1 toward H3K4me.
An H3 peptide with K4 mutated to methionine (referred to as pK4M) bound to KDM1 much more tightly than H3K4me peptides did, allowing the structure determination of the KDM1–pK4M complex [25]. The KDM1-binding modes of N-methylpropargyl-K4 H3 and pK4M are strikingly different. The N-terminal five residues of pK4M form an α-helix, as opposed to the γ-turns in N-methylpropargyl-K4 H3. Replacing M4 in pK4M with a methyl-lysine in the extended configuration produces steric clashes with the isoalloxazine ring, suggesting that the binding mode observed with pK4M is incompatible with catalysis. Nevertheless, the striking differences between the structures of KDM1 bound to two slightly different peptides indicate a high plasticity in substrate recognition by KDM1. As discussed above, KDM1 has been implicated in the demethylation of H3K9me1/2 [15,16]. KDM1 has also been reported to demethylate non-histone substrates, including p53 [26]. It is conceivable that KDM1 can recognize and demethylate different substrates with vastly different binding modes.
H3 peptides shorter than 21 residues are not efficient substrates for KDM1 [21]. Structures of the KDM1-H3 peptide complexes do not include the C-terminal portion of the peptide (residues 10-21), as no interpretable electron density is present for these residues. On the other hand, a conspicuous cleft formed by conserved residues from the SWIRM and AOD domains of KDM1 lies in the vicinity of the active site (Figure 1d). Mutations of residues in this cleft decrease the demethylase activity of KDM1 [17]. This cleft could serve as the binding site for the C-terminal portion of the H3 peptide. The detailed interactions between KDM1 and these residues await further structural investigations, however.
Chemical inhibitors of KDM1
In addition to genetic mutations, cancer cells have an altered epigenetic landscape. Chemical compounds targeting epigenetic modifications can selectively kill cancer cells. For example, HDAC inhibitors have recently been clinically approved to treat human cancers [27,28]. KDM1 and HDAC1/2 are common components of several transcriptional corepresssor complexes. HDAC1/2 cooperate with KDM1 to repress transcription [29]. Chemical inhibitors of KDM1 may also have therapeutic value either as single agents or in combination with HDAC inhibitors.
Because of the high structural and mechanistic similarities between KDM1 and conventional amine oxidases, monoamine oxidase (MAO) inhibitors, such as pargyline and tranylcypromine, can also inhibit KDM1, albeit with low selectivity and potency [16,30]. Modifications of these two MAO inhibitors have led to the development of potent KDM1 inhibitors. First, propargylamine-derived H3 peptides are potent, selective KDM1 inhibitors and have been instrumental in the structural analysis of the KDM1-H3 complex [23]. Second, as predicted from the KDM1-tranylcypromine structures [31,32], chemical substitutions on the phenyl ring of tranylcypromine have greatly improved the potency and selectivity of this class of KDM1 inhibitors [33-36]. High throughput screening of combinatorial chemical libraries constructed with propargylamine or cyclopropylamine moieties may lead to even more potent KDM1 inhibitors.
JmjC domain-containing histone demethylases (JHDMs)
Discovery, catalytic mechanism, and structures
Using conventional biochemical methods, Yi Zhang and coworkers purified a nuclear protein, JmjC domain-containing histone demethylase (KDM2A; also known as JHDM1a/FBXL11), from HeLa cells as the H3K36me1/2 demethylase [5••]. KDM2A is the founding member of a large family of JmjC domain-containing histone demethylases (JHDMs) [37]. The JmjC domain is conserved from yeast to man and belongs to the superfamily of Fe2+-dependent dioxygenases. Using Fe2+ and α-ketoglutarate (αKG) as cofactors and in the presence of oxygen, JHDMs convert the methyl group in the methyllysine to a hydroxymethyl group, which is then released as formaldehyde (Figure 2a). In contrast to KDM1, which can only remove mono- and di-methylated lysines due to inherent chemistry limitations, JHDMs do not such limitations in their catalytic mechanism and can remove all three degrees of methylation, including tri-methylation. JHDMs have been shown to regulate a diverse array of cellular processes [3,37].
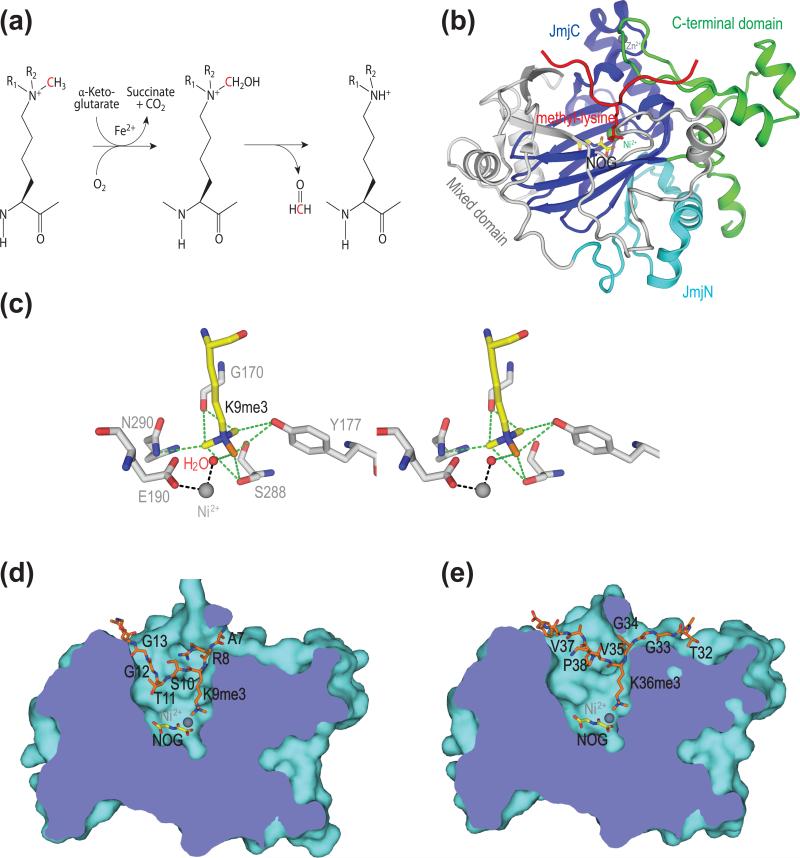
Figure 2.
Mechanism, structure, methylation degree specificity, and site specificity of KDM4A. (a) Chemical mechanism of JmjC-catalyzed lysine demethylation. (b) Structure of human KDM4A in complex with an H3K36me3 peptide (PDB code 2OS2). Instead of αKG and Fe2+, NOG and Ni2+ were used as cofactors to create an inactive enzyme-substrate complex. (c) Stereoview of the methylammonium-binding site in KDM4A (PDB code 2OQ6). The methyl group undergoing catalysis is colored orange. Dashed green lines indicate C-H...O hydrogen bonds. (d) Binding of the H3K9me3 peptide at the active site of KDM4A (PDB code 2OQ6). (e) Binding of the H3K36me3 peptide at the active site of KDM4A (PDB code 2OS2).
The structures of the core catalytic domains of four JHDMs have been determined so far, including KDM2A, KDM4A, PHF8, and both human and C. elegans KDM7A (also known as JHDM1D/KIAA1718) [38•-44]. As expected from sequence similarity, the JmjC domain adopts a jellyroll-like all β fold, similar to other members of the cupin superfamily of metalloenzymes (Figure 2b). The active site is buried in the interior of this jellyroll structural motif, with Fe2+ being coordinated by αKG and three invariable residues (H188, E190, and H276 in human KDM4A). The jellyroll motif is surrounded by other structural elements, which help to maintain the structural integrity of the catalytic core and contribute to substrate recognition. In particular, the JmjN domain is present in all four structurally characterized JHDMs and forms extensive contacts with JmjC. In the case of KDM4A, removal of JmjN impairs the stability and activity of JmjC [38•].
Substrate degree specificity
By using N-oxalylglycine (NOG; an analog and competitive inhibitor of αKG) to replace αKG and Ni2+ to replace Fe2+, several groups have managed to create catalytically inert KDM4A, PHF8, and KDM7A and to determine the structures of these inactive enzymes in complex with their substrates and, in some cases, products [39••-41,43••,44••]. These structures have revealed that JHDMs use distinct strategies to achieve degree and site specificity in the demethylation reactions. These structures have also guided the design of additional αKG analogs that are selective inhibitors of JHDMs [45,46]. All three enzymes, KDM4A, PHF8, and KDM7A, exhibit dual substrate site specificity. KDM4A demethylates H3K9me2/3 and H3K36me2/3 [47,48]. Both PHF8 and KDM7A demethylate H3K9me2 and H3K27me2 [43••]. We discuss the degree and site specificities of these enzymes in this and next sections, respectively.
The structures of these enzymes bound to substrates reveal a highly conserved methyl-lysine-binding pocket in the active site that is distinct from classical methyl-lysine-binding domains such as the PHD finger [39••-41,43••,44••]. The classical methyl-lysine-binding motifs, such as chromo domains, bind methyl-lysine through cation-π interactions between the methylammonium ion and a cage formed by multiple aromatic residues [49]. By contrast, JHDMs bind to methyl-lysine through the formation of a network of unusual C-H...O hydrogen bonds between the methyl groups and the oxygen atoms from the backbone and side chain of active-site residues [39••,43••]. In KDM4A, this hydrogen-bonding network places one of the three methyl groups of the tri-methylated lysine close to the Fe2+ and in an ideal position for catalysis (Figure 2c). When di- or mono-methylated lysines bind at the active site, the methyl groups are sequestered away from the metal ion by the C-H...O hydrogen bonds. Therefore, the catalysis-competent methyl position is energetically disfavored. In the case of a di-methylated lysine, a rotational movement might allow one of the methyl groups to gain access to Fe2+ for catalysis, albeit with less efficiency than a tri-methylated lysine [39••,40•]. A mono-methylated lysine is completely sequestered and cannot reach the proper positioning for catalysis. This model nicely explains the degree specificity of KDM4A, which prefers tri-methylated lysines to di-methylated lysines and does not demethylate mono-methylated lysines. Consistent with this model, mutations of S288 (whose hydroxyl group forms C-H...O hydrogen bonds with methyl groups) in KDM4A increase its activity towards di- or even mono-methylated lysines [19,39].
PHF8 and KDM7A prefer di-methylated lysines to tri- or mono-methylated ones [43••,44••]. Their inability to demethylate tri-methylated lysines is due to steric hindrance. There simply is not enough space in their active sites to accommodate a third methyl group [43••,44••]. The inability of these two enzymes to demethylate mono-methylated lysines could also be due to the sequestration of this methyl group by the C-H...O hydrogen bonds, as in the case of KDM4A.
Substrate site specificity
PHF8 and KDM7A demethylate H3K9me2 and H3K27me2 [43••,44••]. The residues neighboring K9 and K27 are similar, including an “ARKS” motif in both cases. Structures of the C. elegans KDM7A bound to H3K9me2 or H3K27me2 peptides indicate that identical sets of interactions mediate the binding of these two peptides to the active site of the enzyme [44••]. Although only the structure of PHF8 bound to H3K9me2 is available, PHF8 likely recognizes H3K27me2 with the same binding mode. The local sequence similarity at the H3-K9 and H3-K27 sites is thus an important determinant of the dual specificity of PHF8 and KDM7A towards these two sites.
KDM4A demethylates both H3K9me2/3 and H3K36me2/3. Residues neighboring these two sites share no obvious sequence similarity. How does KDM4A achieve dual specificity towards these two sites? Structures of KDM4A bound to H3K9me3 or H3K36me3 indicate that, excluding the methyl-lysine, there are few interactions between KDM4A and the side chains of either peptide (Figure 2D and 2E) [39••-41]. Most interactions involve the peptide backbone, explaining the ability of KDM4A to demethylate two sites that are unrelated in sequence. Why does KDM4A not demethylate other sites? A likely explanation is that certain residues surrounding other sites produce destabilizing binding energies and are selected against by KDM4A. For example, mutation of G12 in H3 to proline, a residue found near K27, drastically reduces H3K9me3 demethylation by KDM4A [39••]. Therefore, KDM4A achieves its dual specificity through recognizing specific backbone conformations of the ligand peptides (positive selection) and minimizing the destabilizing interactions with the ligand sidechains (negative selection).
Functions of auxiliary domains
In addition to the JmjN and JmjC domains, most JHDMs contain auxiliary functional domains, including PHD, Tudor, F-box, Bright/Arid, Zn2+ finger, and TPR [37]. Among them, PHD and Tudor domains recognize unmodified or methylated histone tails [3]. These additional domains may promote the recruitment of JHDMs to specific genomic loci, contribute to their substrate specificity, and enable mutual regulation between two or more epigenetic marks. Two recent structural studies on PHF8 and KDM7A have beautifully illustrated these principles [43••,44••].
PHF8 and KDM7A belong to the same subfamily of JHDMs. Both contain an N-terminal PHD domain that binds to H3K4me3. Despite high sequence similarity, the orientations the PHD domains of PHF8 and KDM7A relative to their catalytic core domains (containing JmjN, JmjC, and the C-terminal domain) are strikingly different (Figure 3a). When to a single peptide containing both H3K4me3 and H3K9me2, PHF8 adopts a closed, bent conformation, with the PHD and JmjC domains forming a bivalent interaction with H3K4me3 and H3K9me2 (cis binding mode) [43••]. Consistently, PHF8 demethylates peptides containing both modifications much more efficiently than peptides containing only H3K9me2.
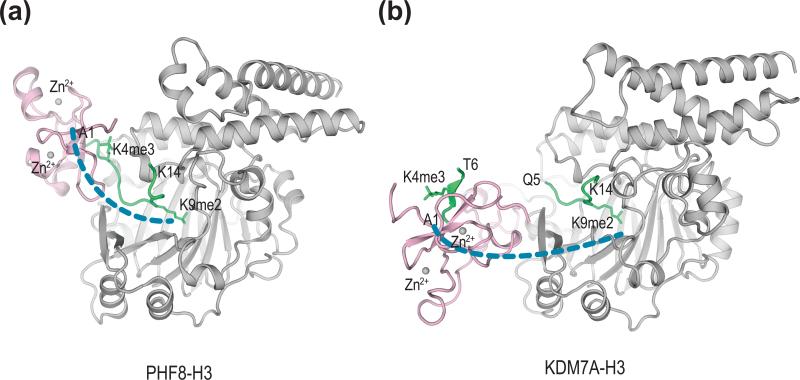
Figure 3.
Different relative orientations of the PHD and JmjC domains in PHF8 and KDM7A. (a) Structure of the PHD and core catalytic domains of human PHF8 bound to an H3 peptide containing both H3K4me3 and H3K9me2 (PDB code 3KV4). The PHD and the catalytic core domains of PHF8 are colored pink and gray, respectively. The H3 peptide is green. (b) Structure of C. elegans KDM7A bound to two different H3 molecules each containing both H3K4me3 and H3K9me2 (PDB code 3N9N). The PHD and the catalytic core domains of PHF8 are colored pink and gray, respectively. The H3 peptide is green. The position of the catalytic core of KDM7A is shown in the same orientation as that of PHF8 in (a).
By contrast, both human and C. elegans KDM7A adopt an open, extended conformation, because of a rigid linker connecting PHD and JmjC (Figure 3b) [43••,44••]. The methyl-lysine-binding sites of PHD and JmjC are separated by about 40Å, a distance far greater than the maximum span of the peptide segment linking K4 and K9 in H3. As a result, KDM7A does not engage both H3K4me3 and H3K9me2 in a single peptide. Instead, its PHD domain binds to H3K4me3 in one peptide while its JmjC domain independently binds to H3K9me3 in another (trans binding mode). It remains to be determined whether KDM7A and PHF8 can recognize both H3K4me3 and H3K27me2 in a single peptide.
Regardless, the PHD domain of C. elegans KDM7A is required for its demethylase activity toward both H3K9me2 and H3K27me2 in vivo [50]. Thus, both cis and trans modes of substrate binding may be used by KDM7A and possibly PHF8 in the context of chromatin (Figure 4a). Interestingly, the Tudor domain in KDM4A binds both H3K4me3 and H4K20me3 [51,52]. It is possible that this domain also contributes to the chromatin recruitment of KDM4A and regulates its demethylase activity in similar ways. Thus, the auxiliary domains in JHDMs fine tune their substrate specificity and enable crosstalk between multiple epigenetic marks.
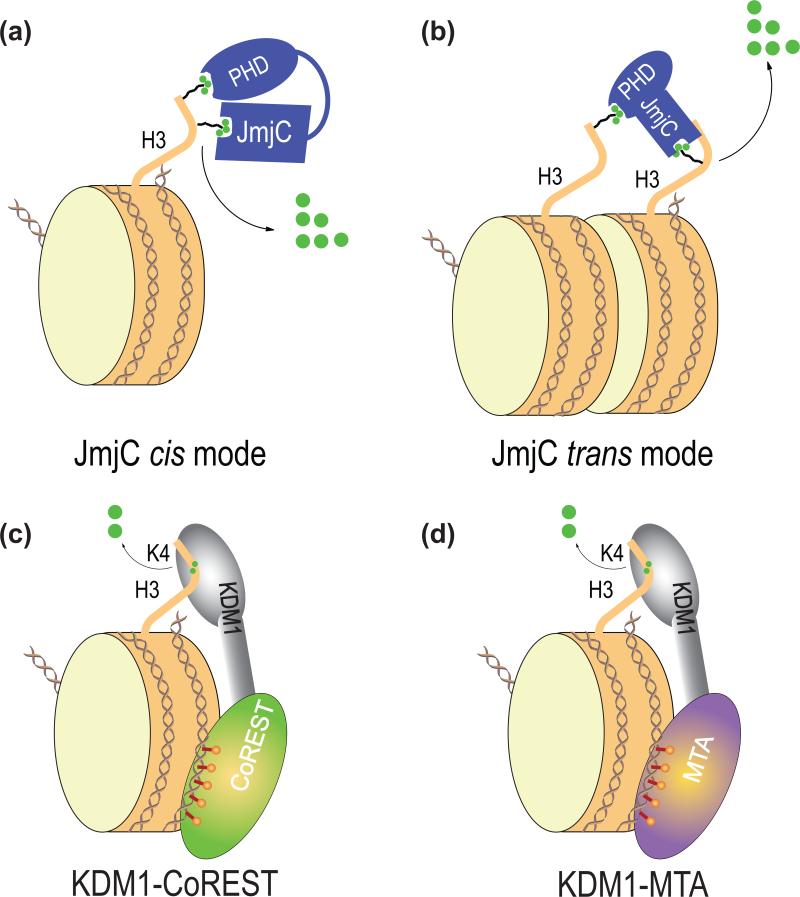
Figure 4.
Proposed multivalent interactions between histone demethylases and nucleosomes. (a) Cis binding mode between histone demethylases and nucleosomes, as exemplified by PHF8. In this binding mode, two chromatin-binding modules engage the same histone tail. (b) Trans binding mode between histone demethylases and nucleosomes, as exemplified by KDM7A. In this binding mode, two chromatin-binding modules bind to two different histone tails, in the same nucleosome or two different nucleosomes. (c) The proposed multivalent interaction between KDM1-CoREST and the nucleosome. Histone H3 tail binds to the active site of KDM1 while CoREST SANT domains bind to DNA. (d) The proposed multivalent interaction between KDM1-MTA and the nucleosome. Histone H3 tail binds to the active site of KDM1 while MTA SANT domains bind to DNA.
Conclusions
Histone demethylases control the dynamics and the steady-state levels of histone methylation, thereby regulating diverse chromatin-based biological processes. Structural biology of these enzymes has provided key insights into their catalytic mechanisms, their specificities toward certain sites and degrees of methylation, and their recruitment to chromatin. A general principle that emerges from these studies is the use of multiple, weak interactions by these enzymes to demethylate histones in the context of chromatin (Figure 4). For example, KDM1 is recruited to nucleosomes through an interaction between its active site and H3K4me1/2-containing H3 tail and DNA binding by the SANT domains of CoREST and possibly MTAs. In the cases of PHF8 and KDM7A, both PHD and JmjC domains engage in interactions with H3K4me3 and H3K9me2, either in cis or in trans. The cooperativity between these multiple binding events not only increases chromatin-binding affinity of these enzymes, but also enables the crosstalk between different epigenetic marks.
Most, if not all, histone-modifying enzymes or enzyme complexes contain multiple chromatin-binding functionalities. This multivalency mechanism of chromatin binding is generally applicable to other chromatin-modifying enzymes. Recent advances in chemical biology have allowed the production of nucleosomes with uniform, defined patterns of modifications [53]. Structures of complexes between chromatin-modifying enzymes containing multiple chromatin-binding modules and nucleosomes with defined epigenetic marks will reveal the basis of multivalency binding in atomic details.
Acknowledgements
Research in our laboratory is supported by the National Institutes of Health and the Welch Foundation (I-1441). H.Y. is an Investigator at the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 2.Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- 3.Mosammaparast N, Shi Y. Reversal of histone methylation: biochemical and molecular mechanisms of histone demethylases. Annu. Rev. Biochem. 2010;79:155–179. doi: 10.1146/annurev.biochem.78.070907.103946. [DOI] [PubMed] [Google Scholar]
- 4••.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [This study identified the first histone lysine demethylase and established histone methylation as a reversible and dynamic epigenetic mark.] [DOI] [PubMed] [Google Scholar]
- 5••.Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [This paper describes the biochemical purification and characterization of the founding member of the JmjC family of histone demethylases.] [DOI] [PubMed] [Google Scholar]
- 6.You A, Tong JK, Grozinger CM, Schreiber SL. CoREST is an integral component of the CoREST- human histone deacetylase complex. Proc. Natl. Acad. Sci. U. S. A. 2001;98:1454–1458. doi: 10.1073/pnas.98.4.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hakimi MA, Bochar DA, Chenoweth J, Lane WS, Mandel G, Shiekhattar R. A core-BRAF35 complex containing histone deacetylase mediates repression of neuronal-specific genes. Proc. Natl. Acad. Sci. U. S. A. 2002;99:7420–7425. doi: 10.1073/pnas.112008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hakimi MA, Dong Y, Lane WS, Speicher DW, Shiekhattar R. A candidate X-linked mental retardation gene is a component of a new family of histone deacetylase-containing complexes. J. Biol. Chem. 2003;278:7234–7239. doi: 10.1074/jbc.M208992200. [DOI] [PubMed] [Google Scholar]
- 9•.Lee MG, Wynder C, Cooch N, Shiekhattar R. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature. 2005;437:432–435. doi: 10.1038/nature04021. [DOI] [PubMed] [Google Scholar]
- 10•.Shi YJ, Matson C, Lan F, Iwase S, Baba T, Shi Y. Regulation of LSD1 histone demethylase activity by its associated factors. Mol. Cell. 2005;19:857–864. doi: 10.1016/j.molcel.2005.08.027. [Ref. 9 and 10 demonstrate the requirement of CoREST in LSD1-dependent demethylation of nucleosomal substrates and highlight the importance of CoREST SANT domains in this process.] [DOI] [PubMed] [Google Scholar]
- 11.Lan F, Zaratiegui M, Villen J, Vaughn MW, Verdel A, Huarte M, Shi Y, Gygi SP, Moazed D, Martienssen RA. S. pombe LSD1 homologs regulate heterochromatin propagation and euchromatic gene transcription. Mol. Cell. 2007;26:89–101. doi: 10.1016/j.molcel.2007.02.023. [DOI] [PubMed] [Google Scholar]
- 12.Wang J, Scully K, Zhu X, Cai L, Zhang J, Prefontaine GG, Krones A, Ohgi KA, Zhu P, Garcia-Bassets I, et al. Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature. 2007;446:882–887. doi: 10.1038/nature05671. [DOI] [PubMed] [Google Scholar]
- 13.Saleque S, Kim J, Rooke HM, Orkin SH. Epigenetic regulation of hematopoietic differentiation by Gfi-1 and Gfi-1b is mediated by the cofactors CoREST and LSD1. Mol. Cell. 2007;27:562–572. doi: 10.1016/j.molcel.2007.06.039. [DOI] [PubMed] [Google Scholar]
- 14•.Wang Y, Zhang H, Chen Y, Sun Y, Yang F, Yu W, Liang J, Sun L, Yang X, Shi L, et al. LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell. 2009;138:660–672. doi: 10.1016/j.cell.2009.05.050. [The authors show that another SANT domain-containing protein MTA2 helps to recruit LSD1 to chromatin and promotes LSD1-dependent demethylation of nucleosomal substrates, presumably using a mechanism similar to CoREST.] [DOI] [PubMed] [Google Scholar]
- 15.Garcia-Bassets I, Kwon YS, Telese F, Prefontaine GG, Hutt KR, Cheng CS, Ju BG, Ohgi KA, Wang J, Escoubet-Lozach L, et al. Histone methylation-dependent mechanisms impose ligand dependency for gene activation by nuclear receptors. Cell. 2007;128:505–518. doi: 10.1016/j.cell.2006.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, Gunther T, Buettner R, Schule R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 17.Stavropoulos P, Blobel G, Hoelz A. Crystal structure and mechanism of human lysine-specific demethylase-1. Nat. Struct. Mol. Biol. 2006;13:626–632. doi: 10.1038/nsmb1113. [DOI] [PubMed] [Google Scholar]
- 18••.Yang M, Gocke CB, Luo X, Borek D, Tomchick DR, Machius M, Otwinowski Z, Yu H. Structural basis for CoREST-dependent demethylation of nucleosomes by the human LSD1 histone demethylase. Mol. Cell. 2006;23:377–387. doi: 10.1016/j.molcel.2006.07.012. [The authors determined the structure of LSD1 in complex with CoREST. They further showed that the SANT2 domain of CoREST bound DNA and that the DNA-binding activity of SANT2 was required for the demethlyation of nucleosomes by LSD1-CoREST. This study thus established that LSD1-CoREST recognized nucleosomal substrates through multivalent binding.] [DOI] [PubMed] [Google Scholar]
- 19.Chen Y, Yang Y, Wang F, Wan K, Yamane K, Zhang Y, Lei M. Crystal structure of human histone lysine-specific demethylase 1 (LSD1). Proc. Natl. Acad. Sci. U. S. A. 2006;103:13956–13961. doi: 10.1073/pnas.0606381103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Binda C, Coda A, Angelini R, Federico R, Ascenzi P, Mattevi A. A 30-angstrom-long U-shaped catalytic tunnel in the crystal structure of polyamine oxidase. Structure. 1999;7:265–276. doi: 10.1016/s0969-2126(99)80037-9. [DOI] [PubMed] [Google Scholar]
- 21.Forneris F, Binda C, Vanoni MA, Battaglioli E, Mattevi A. Human histone demethylase LSD1 reads the histone code. J. Biol. Chem. 2005;280:41360–41365. doi: 10.1074/jbc.M509549200. [DOI] [PubMed] [Google Scholar]
- 22.Culhane JC, Szewczuk LM, Liu X, Da G, Marmorstein R, Cole PA. A mechanism-based inactivator for histone demethylase LSD1. J. Am. Chem. Soc. 2006;128:4536–4537. doi: 10.1021/ja0602748. [DOI] [PubMed] [Google Scholar]
- 23.Szewczuk LM, Culhane JC, Yang M, Majumdar A, Yu H, Cole PA. Mechanistic analysis of a suicide inactivator of histone demethylase LSD1. Biochemistry. 2007;46:6892–6902. doi: 10.1021/bi700414b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24•.Yang M, Culhane JC, Szewczuk LM, Gocke CB, Brautigam CA, Tomchick DR, Machius M, Cole PA, Yu H. Structural basis of histone demethylation by LSD1 revealed by suicide inactivation. Nat. Struct. Mol. Biol. 2007;14:535–539. doi: 10.1038/nsmb1255. [This paper described the structure of LSD1-CoREST bound to an H3 peptide inhibitor and revealed the structural basis for the H3-K4 specificity of LSD1.] [DOI] [PubMed] [Google Scholar]
- 25.Forneris F, Binda C, Adamo A, Battaglioli E, Mattevi A. Structural basis of LSD1-CoREST selectivity in histone H3 recognition. J. Biol. Chem. 2007;282:20070–20074. doi: 10.1074/jbc.C700100200. [DOI] [PubMed] [Google Scholar]
- 26.Huang J, Sengupta R, Espejo AB, Lee MG, Dorsey JA, Richter M, Opravil S, Shiekhattar R, Bedford MT, Jenuwein T, et al. p53 is regulated by the lysine demethylase LSD1. Nature. 2007;449:105–108. doi: 10.1038/nature06092. [DOI] [PubMed] [Google Scholar]
- 27.Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nat. Rev. Drug Discov. 2006;5:769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- 28.Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat. Rev. Cancer. 2006;6:38–51. doi: 10.1038/nrc1779. [DOI] [PubMed] [Google Scholar]
- 29.Lee MG, Wynder C, Bochar DA, Hakimi MA, Cooch N, Shiekhattar R. Functional interplay between histone demethylase and deacetylase enzymes. Mol. Cell Biol. 2006;26:6395–6402. doi: 10.1128/MCB.00723-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee MG, Wynder C, Schmidt DM, McCafferty DG, Shiekhattar R. Histone H3 lysine 4 demethylation is a target of nonselective antidepressive medications. Chem. Biol. 2006;13:563–567. doi: 10.1016/j.chembiol.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 31.Yang M, Culhane JC, Szewczuk LM, Jalili P, Ball HL, Machius M, Cole PA, Yu H. Structural basis for the inhibition of the LSD1 histone demethylase by the antidepressant trans-2-phenylcyclopropylamine. Biochemistry. 2007;46:8058–8065. doi: 10.1021/bi700664y. [DOI] [PubMed] [Google Scholar]
- 32.Mimasu S, Sengoku T, Fukuzawa S, Umehara T, Yokoyama S. Crystal structure of histone demethylase LSD1 and tranylcypromine at 2.25 A. Biochem. Biophys. Res. Commun. 2008;366:15–22. doi: 10.1016/j.bbrc.2007.11.066. [DOI] [PubMed] [Google Scholar]
- 33.Ueda R, Suzuki T, Mino K, Tsumoto H, Nakagawa H, Hasegawa M, Sasaki R, Mizukami T, Miyata N. Identification of cell-active lysine specific demethylase 1-selective inhibitors. J. Am. Chem. Soc. 2009;131:17536–17537. doi: 10.1021/ja907055q. [DOI] [PubMed] [Google Scholar]
- 34.Culhane JC, Wang D, Yen PM, Cole PA. Comparative analysis of small molecules and histone substrate analogues as LSD1 lysine demethylase inhibitors. J. Am. Chem. Soc. 2010;132:3164–3176. doi: 10.1021/ja909996p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Binda C, Valente S, Romanenghi M, Pilotto S, Cirilli R, Karytinos A, Ciossani G, Botrugno OA, Forneris F, Tardugno M, et al. Biochemical, structural, and biological evaluation of tranylcypromine derivatives as inhibitors of histone demethylases LSD1 and LSD2. J. Am. Chem. Soc. 2010;132:6827–6833. doi: 10.1021/ja101557k. [DOI] [PubMed] [Google Scholar]
- 36.Mimasu S, Umezawa N, Sato S, Higuchi T, Umehara T, Yokoyama S. Structurally designed Trans-2-phenylcyclopropylamine derivatives potently inhibit histone demethylase LSD1/KDM1. Biochemistry. 2010 doi: 10.1021/bi100299r. in press. [DOI] [PubMed] [Google Scholar]
- 37.Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat. Rev. Genet. 2006;7:715–727. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- 38•.Chen Z, Zang J, Whetstine J, Hong X, Davrazou F, Kutateladze TG, Simpson M, Mao Q, Pan CH, Dai S, et al. Structural insights into histone demethylation by JMJD2 family members. Cell. 2006;125:691–702. doi: 10.1016/j.cell.2006.04.024. [This paper described the first structure determination of an JmjC-containing histone demethylase and identified active-site residues that were critical for the specificity of JMJD2A toward tri-methylated lysines.] [DOI] [PubMed] [Google Scholar]
- 39••.Couture JF, Collazo E, Ortiz-Tello PA, Brunzelle JS, Trievel RC. Specificity and mechanism of JMJD2A, a trimethyllysine-specific histone demethylase. Nat. Struct. Mol. Biol. 2007;14:689–695. doi: 10.1038/nsmb1273. [The authors provided an elegant explanation for the methylation-state specificity of JMJD2A and identified a key residue in controlling this specificity. They further provided evidence to suggest that negative selection contributes to the site specificity of JMJD2A.] [DOI] [PubMed] [Google Scholar]
- 40•.Ng SS, Kavanagh KL, McDonough MA, Butler D, Pilka ES, Lienard BM, Bray JE, Savitsky P, Gileadi O, von Delft F, et al. Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity. Nature. 2007;448:87–91. doi: 10.1038/nature05971. [This study demonstrated the limited contacts between KDM4A and the sidechains of its peptide substrates and the importance of backbone conformation in the substrate recognition by KDM4A, thus providing a simple explanation of the dual site specificity of KDM4A.] [DOI] [PubMed] [Google Scholar]
- 41.Chen Z, Zang J, Kappler J, Hong X, Crawford F, Wang Q, Lan F, Jiang C, Whetstine J, Dai S, et al. Structural basis of the recognition of a methylated histone tail by JMJD2A. Proc. Natl. Acad. Sci. U. S. A. 2007;104:10818–10823. doi: 10.1073/pnas.0704525104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Han Z, Liu P, Gu L, Zhang Y, Li H, Chen S, Chai J. Structural basis for histone demethylation by JHDM1. Front. Science. 2007;1:52–61. [Google Scholar]
- 43••.Horton JR, Upadhyay AK, Qi HH, Zhang X, Shi Y, Cheng X. Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases. Nat. Struct. Mol. Biol. 2010;17:38–43. doi: 10.1038/nsmb.1753. [This paper, along with Ref. 44, established the distinct mechanisms by which the auxiliary domains of histone demethylases regulate chromatin recruitment of these enzymes and contribute to the crosstalk among multiple epigenetic marks.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44••.Yang Y, Hu L, Wang P, Hou H, Lin Y, Liu Y, Li Z, Gong R, Feng X, Zhou L, et al. Structural insights into a dual-specificity histone demethylase ceKDM7A from Caenorhabditis elegans. Cell Res. 2010 doi: 10.1038/cr.2010.86. [DOI] [PubMed] [Google Scholar]
- 45.Rose NR, Woon EC, Kingham GL, King ON, Mecinovic J, Clifton IJ, Ng SS, Talib-Hardy J, Oppermann U, McDonough MA, et al. Selective inhibitors of the JMJD2 histone demethylases: combined nondenaturing mass spectrometric screening and crystallographic approaches. J. Med. Chem. 2010;53:1810–1818. doi: 10.1021/jm901680b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hamada S, Suzuki T, Mino K, Koseki K, Oehme F, Flamme I, Ozasa H, Itoh Y, Ogasawara D, Komaarashi H, et al. Design, synthesis, enzyme-inhibitory activity, and effect on human cancer cells of a novel series of jumonji domain-containing protein 2 histone demethylase inhibitors. J. Med. Chem. 2010;53:5629–5638. doi: 10.1021/jm1003655. [DOI] [PubMed] [Google Scholar]
- 47.Klose RJ, Yamane K, Bae Y, Zhang D, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y. The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature. 2006 doi: 10.1038/nature04853. [DOI] [PubMed] [Google Scholar]
- 48.Whetstine JR, Nottke A, Lan F, Huarte M, Smolikov S, Chen Z, Spooner E, Li E, Zhang G, Colaiacovo M, et al. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125:467–481. doi: 10.1016/j.cell.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 49.Jacobs SA, Khorasanizadeh S. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science. 2002;295:2080–2083. doi: 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- 50.Lin H, Wang Y, Tian F, Pu P, Yu Y, Mao H, Yang Y, Wang P, Hu L, Lin Y, et al. Coordinated regulation of active and repressive histone methylations by a dual-specificity histone demethylase ceKDM7A from Caenorhabditis elegans. Cell Res. 2010 doi: 10.1038/cr.2010.84. [DOI] [PubMed] [Google Scholar]
- 51.Huang Y, Fang J, Bedford MT, Zhang Y, Xu RM. Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A. Science. 2006;312:748–751. doi: 10.1126/science.1125162. [DOI] [PubMed] [Google Scholar]
- 52.Lee J, Thompson JR, Botuyan MV, Mer G. Distinct binding modes specify the recognition of methylated histones H3K4 and H4K20 by JMJD2A-tudor. Nat. Struct. Mol. Biol. 2008;15:109–111. doi: 10.1038/nsmb1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McGinty RK, Kim J, Chatterjee C, Roeder RG, Muir TW. Chemically ubiquitylated histone H2B stimulates hDot1L-mediated intranucleosomal methylation. Nature. 2008;453:812–816. doi: 10.1038/nature06906. [DOI] [PMC free article] [PubMed] [Google Scholar]