Abstract
PubChem is a public repository of small molecules and their biological properties. Currently, it contains over 25 million unique chemical structures and 90 million bioactivity outcomes associated with several thousand macromolecular targets. To address the potential utility of this public resource for drug discovery, we systematically summarized the protein targets in PubChem by the function, three-dimensional (3D) structure and biological pathway. Moreover, we analyzed the potency, selectivity and promiscuity of the bioactive compounds identified for these biological targets, including the chemical probes generated by the NIH Molecular Libraries Program (MLP). As a public resource, PubChem lowers the barrier for researchers to advance the development of chemical tools for modulating biological processes and drug candidates for disease treatments.
Keywords: PubChem, Drug target, High-throughput screening, Chemical probe, Drug discovery
PubChem [1,2] (http://pubchem.ncbi.nlm.nih.gov) is a public repository for chemical structures and their biological properties. The bioactivity results in PubChem are contributed by over a hundred organizations, with the majority data coming from the screening center network under the NIH Molecular Libraries Program (MLP) [3]. This program aims to expand the use of small molecules as chemical probes, which offer dynamic, reversible and tunable perturbations for biological systems [4], to study the functions of genes and proteins in physiology and pathology. Unlike the pharmaceutical industry and biotechnology companies, which primarily focus on the “druggable genome” [5, 6] to screen the “drug-like” small molecules against relatively limited types of targets, such as kinases, G protein-coupled receptors (GPCRs), enzymes, ion channels and nuclear hormone receptors, an extensive collection of biological targets and chemical compounds are being investigated by the MLP to answer a wide scope of biological questions, from identifying inhibitors of a specific enzyme to looking for small molecules that affect protein-protein interactions or modulate splicing events [3]. With the rapid growth in data capacity, PubChem is becoming a valuable resource for drug development and has attracted significant interest from researchers in both academia and industry.
PubChem consists of three interconnected databases: Substance, BioAssay and Compound. The Substance database contains the descriptions of molecules (primarily small molecules) provided by depositors; the BioAssay database contains the screening results of substances by assay providers; and the Compound database contains unique chemical structures derived by structural standardization of the records in the Substance database. Currently, over 25 million unique chemical structures were in the Compound database, which were derived from a collection of 70 million substances. As of April 2010, the BioAssay database comprised over 2,700 bioassays associated with more than one million compounds tested against several thousand molecular targets. In addition, several bioassays from RNAi screening experiment also have been deposited in the BioAssay database.
A review of this public resource allows the community to better understand the information content and utilize the data in PubChem, which may ultimately help to advance the development of new chemical tools and drug candidates by enabling researchers to study the structure-activity relationship, investigate the interaction mechanisms between small molecules and their targets [7] and gain insights into the chemical and biological space in their research area. In this work, we provide a comprehensive summary of the protein targets in PubChem with respect to the functional classification, the availability of three-dimensional (3D) structure and biological pathway. Meanwhile, the potency, selectivity and promiscuity of the bioactive compounds including the chemical probes developed by the MLP, which are associated with those protein targets, are also investigated as well.
Bioassay targets in PubChem
Target identification is one of the key steps for drug development [8, 9]. Tremendous efforts have been made in the past decades by pharmaceutical industries and biotechnology companies that focus on the druggable genome [5, 6] to identify novel drug targets for drug discovery. However, only a few drug targets are successfully used in current therapies [10]. The human genome project has identified about 20,000 to 25,000 genes and an even larger number of transcripts and proteins, which provides great opportunities for drug target investigation [11]. Currently, PubChem records two major types of molecular targets for research, i.e. protein targets for small molecules and gene targets from RNAi reagents, which represent a great diversity of types of assays, including, for example, enzyme inhibitor identification, protein-protein interactions, tumor cell growth inhibition and even organismal phenotypes. As the protein targets are of particular interest to researchers in drug discovery and the majority of bioassays in PubChem focus on enzymes or other proteins, we will focus on the analysis of protein targets in this study. Hence, a collection of 2,206 protein targets was compiled from PubChem at the time of this work.
Functional families
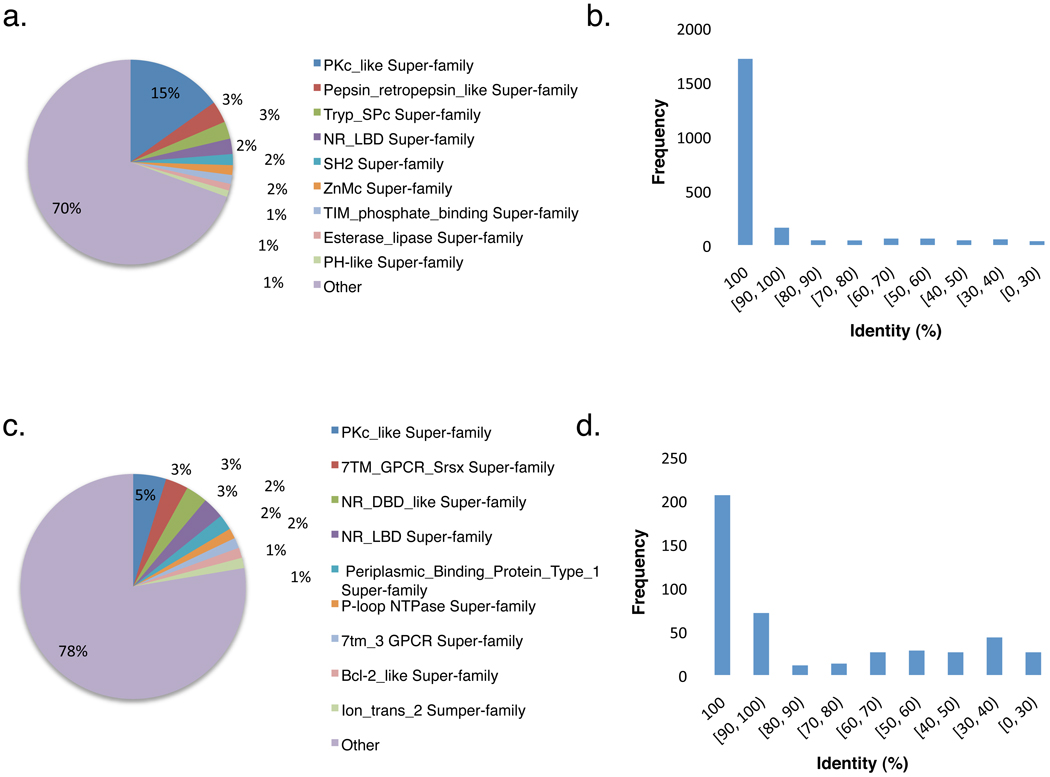
To look into the potential functions of these bioassay targets, we performed sequence similarity search against the annotated functional domains in the NCBI Conserved Domain Database (CDD, http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml) [12] by using the reverse position specific BLAST (RPS-Blast) tool [13]. We found that the 2,206 protein targets fell into 671 unique protein super-families (Fig. 1a). About 15% of them belonged to the protein kinase super-family. Other super-families such as nuclear receptor, trypsin-like serine protease, src homology protein and zinc-dependent metalloprotease comprised about 2–3% of the bioassay targets, respectively. The rest of the super-families (67%, 450/671) contained only one or two bioassay targets for each member. In particular, the high-throughput screening (HTS) assays under the MLP contributed 450 protein targets, scattering into 312 protein super-families (Fig. 1c). Although the protein kinase super-family still dominated this subset, it accounted for 5% of the MLP target set. The other super-families, such as seven transmembrane (7TM) GPCRs and DNA-binding domain of nuclear receptors (NR-DBD) accounted for 2–3% on average. These results suggest that the bioassay targets in PubChem represent a relatively broader functional diversity compared to the known druggable targets. Thus, it allows researchers to study the mechanisms of protein-ligand interactions on a wider scope and to identify novel molecular targets from PubChem for potential treatments.
Figure 1.
Linking protein super-families and three-dimensional structures to PubChem bioassay targets. a & c represent super-family annotations; b & d indicate the availability of related 3D structures derived from homologous analysis at each sequence similarity level. a & b denote the entire set of protein targets in PubChem; c & d represent the protein targets that are involved in high-throughput screening (HTS) assays from the NIH Molecular Libraries Program (MLP).
Three-dimensional structures
The 3D structures of macromolecular targets are important to the study of the mechanisms of protein-protein and protein-ligand interactions. To link the protein targets to relevant 3D structures, we used the BLAST tool [14, 15] to search against the protein sequences derived from the Protein Data Bank (PDB, http://www.pdb.org) [16]. We found that 78% of these targets have corresponding 3D structures with 100% sequence identity in PDB database (Fig. 1b). When looking into the possibility of inferring related structures from the similarity search, another 8% of these targets found related structures in PDB database with sequence identity over 90%. Given the fact that protein structures tend to be highly conserved at this level of sequence identity, this analysis suggests that over 86% of the molecular targets in PubChem have related structural information in PDB. On the other hand, less than 2% of these targets could not be linked to any relevant 3D structures, or were only able to be linked to the related protein structures with the sequence identity below 30%. As to the 450 protein targets from the MLP, over 60% either have corresponding 3D structures with sequence identity of 100% or can be linked to related structures with sequence identity of 90% or above (Fig. 1d).
Related pathways
Most diseases occur due to the misregulation of multiple genes that are involved in mutual interactions, including genes, transcripts and proteins, in a dynamic network. During the past decade, high-throughput technologies have been widely used in biological research and generated a tremendous amount of experimental data, which make it possible to the study the functions of genes or proteins at biological system level. Drug development is inherently a complicated process that drugs and their targets are engaged in a complex system, which is far from being thoroughly understood. Moreover, approximately 35% of the known drugs or drug candidates are active against more than one target [17], which makes the interactions more sophisticated. Therefore, it is essential to investigate the connections of the drug, drug target and disease in the context of biological system.
In this study, we mapped 507 (23%) out of the 2,206 protein targets from PubChem to 287 pathways in the KEGG database (http://www.genome.jp/kegg/) [17–20]. We observed that some pathways, such as the mitogen-activated protein kinase (MAPK) signaling pathway, were related to multiple protein targets in PubChem. On the other hand, some bioassay targets were involved in multiple KEGG pathways. A list of top 20 pathways that contain multiple bioassay targets and top 20 targets that are involved in multiple pathways are provided in Table S1 and S2, respectively, as a supplementary. Targets involved in the same pathway are likely to play similar roles in regulating a specific biological process. Thus, selectively inhibiting or activating a target in the same pathway might effectively modulate a specific biological process or restore the function from a disease state back to a normal one. Thus, the wealth of bioactivity data in PubChem may facilitate the researches in chemical biology and drug development at the system level.
Bioactive compounds in PubChem
The characteristics of small molecules make them not only as drugs that modulate physiological functions, but also as chemical tools that interrogate the functions of novel genes, pathways and cells [3]. The purpose of the NIH MLP is to develop chemical probes for modulating biological process and facilitate the development of new drugs by offering the capacity of the HTS to the public sector [3]. Currently, more than one million compounds were tested against several thousand targets and deposited in PubChem. About two hundred thousands of them were reported active, among which there were 116 chemical probes generated by the MLP projects at the time of this work.
Potency
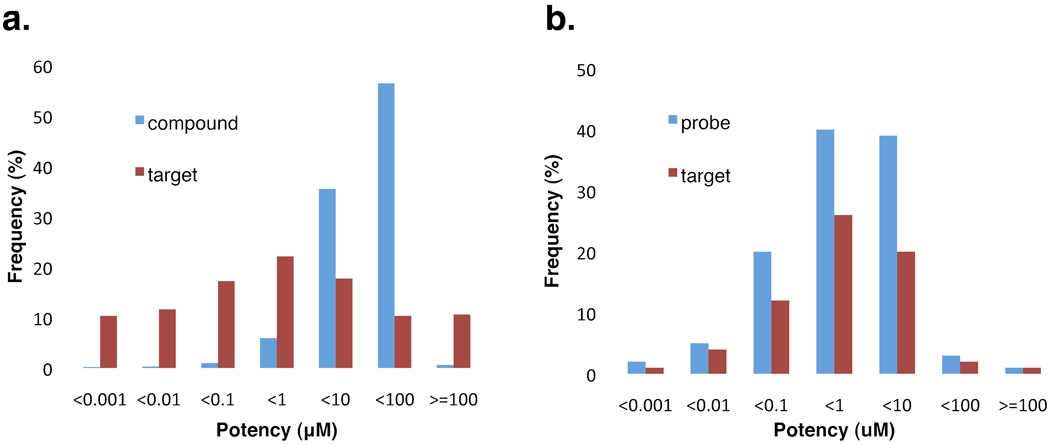
A large fraction of the bioactive compounds (91,022) in PubChem were assayed with a confirmed potency measurement, which were associated with 1,771 out of the 2,206 protein targets in total. The distribution of bioactivity potency was analyzed, with the results showing that nearly 10% of the compounds have a potency of • 1 µM (Fig. 2a). These compounds were associated with over 60% of the1771 targets, i.e. each of these targets had at least one bioactive compound with a potency of • 1 µM. On the other hand, we found that about 40% of the targets had no active compound with the potency better than 10 µM (Fig. 2a), which indicates that there are great chances to develop highly potent compounds for these targets through further study by medicinal chemistry approaches. When focusing on the 116 MPL chemical probes, we found that most of them demonstrated much higher potency in the range of 0.001 ~ 1 µM (Fig. 2b). The MLP probes will be discussed in detail in the following section of “Chemical probes”.
Figure 2.
The distribution of the potency of the bioactive compounds and protein targets in PubChem. a. blue bars show the distribution of the potency for entire bioactive compounds in PubChem in seven potency groups; red bars denote the frequency of the protein targets with most potent compounds falling in respective potency group. b. blue bars represent the distribution of potency for chemical probes indentified by the NIH Molecular Libraries Program (MLP); red bars show the frequency of the protein targets with most potent chemical probes falling in respective potency group.
Selectivity and promiscuity
It is essential to understand the selectivity and promiscuity of small molecules, when fully exploiting the therapeutic potential and minimizing toxic effects of drugs or drug candidates [17, 21, 22]. To evaluate these properties of a compound, a straightforward approach is to investigate the bioactivity profile by screening this compound across a broad panel of targets, however, which could be expensive when applying to a large compound library. On the other hand, as more data is available in PubChem, it will be possible to derive such kind of bioactivity profiles for a particular chemical compound, as well as to investigate the selectivity and promiscuity against a specific target by combining the assay results contributed by many organizations. In particular, the projects under the MLP, which share a common library of over 340,000 compounds, make it feasible to systematically derive target profiling information for many bioactive compounds.
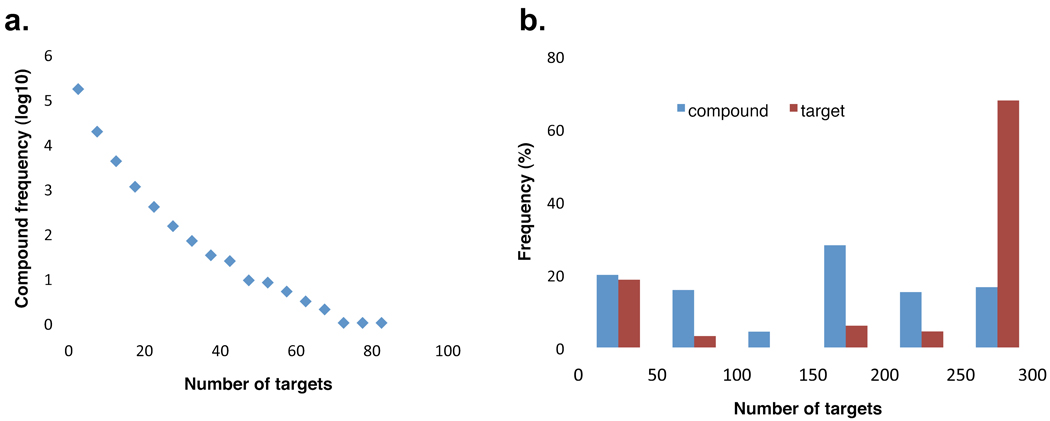
We performed an across-target activity analysis for all the 189,807 active compounds in PubChem to identify the selective as well as promiscuous compounds following the procedure described previously [23]. As a result, 38% (71,627) of those compounds were observed as potentially selective with bioactivity outcome reported active against a single target, while the rest of them (62%) demonstrated active against multiple targets with a portion of them hitting multiple but otherwise related targets (Fig. 3a). Many bioassay targets in PubChem are biologically related as revealed by sequence homology analysis [1]. In particular, the MLP projects usually take a secondary screening against related targets in the search for compounds with higher specificity. Thus, it is not surprising to often observe common hits for related targets. On the other hand, there are many other causes of the promiscuity of a compound [24]. To address this issue, the MLP has developed several profiling bioassays for evaluating aggregation effects, filtering chemical reactivity and identifying interference molecules including screenings for luciferase inhibitors by multiple laboratories. In summary, all the information has made PubChem a valuable resource for studying the promiscuity of chemical compounds and investigating the polypharmacology properties of chemical compounds in system-based drug discovery [25, 26].
Figure 3.
An overview of the selectivity property of bioactive compounds in PubChem. a. the distribution of compounds’ across-target activity. X-axis represents the number of distinct active protein targets associated with a compound, while Y-axis represents the frequency of compounds at each across-activity level. It shows that majority compounds are associated with one or a few protein targets, and a small portion of them interact with a large amount of targets; b. the blue bars represent the distribution of the number of tested targets for selective compounds (only active to one protein target in PubChem). Compounds are divided into six selectivity groups. It suggests that the majority of the selective compounds have been tested across over 150 targets. The red bars denote the frequency of the protein targets associated with the compounds in the respective selectivity group.
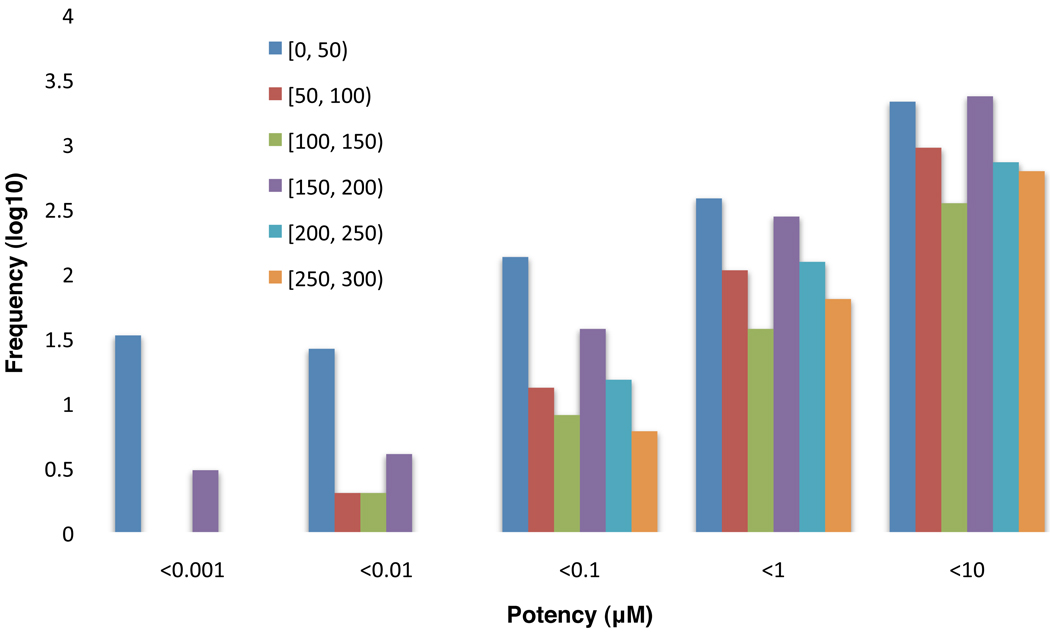
As it would be necessary to assess the selectivity and promiscuity properties in the context of tested targets, we looked into those potentially selective compounds (71,627) and observed that about 80% of them were tested against at least 50 distinct protein targets and a significant portion (60%) was highly selective as tested against more than 150 targets (Fig. 3b). On the other hand, we observed that 14% (316) of the 2,206 targets were associated with at least one of these selective compounds. Among this subset of targets, more than 60% of them were associated with highly selective compounds that were tested broadly across more than 250 distinct protein targets (Fig. 3b). These results indicate that compounds with potentially high selectivity are available for a great portion of protein targets in PubChem. Additionally, we evaluated the potency of these selective compounds by dividing them into several selectivity groups based on the number of targets tested (Fig. 4). This analysis provides further insights into both the selectivity and potency of the bioactive compounds in this subset. It allows one to apply a certain selectivity threshold to identify the compounds with a desired potency and to track down the molecular target associated with the compound as well, which may serve as a starting point for a medicinal chemist to further optimize the bioactive compound towards a chemical probe or a drug candidate.
Figure 4.
The distribution of the selective compounds in PubChem. X-axis represents the potency in micromole; Y-axis represents the frequency in the scale of log10. The colour of the bars denotes the ranges of how many targets against which the selective compounds were tested. For example, the range of [100,150) means that this group of compounds were tested against 100 to 150 targets and selectively against one of them.
Chemical probes
At the time of this work, the MLP project has generated 116 chemical probes. The detailed descriptions about the characterizations of the probes are publically available for the community to review (http://mli.nih.gov/mli/mlp-probes/). These MLP chemical probes were associated with 67 individual protein targets, which fell into 89 CDD super-families (some targets belonged to more than one super-family) according to the CDD functional domain annotations. Among them, 36 protein targets had corresponding 3D structures with sequence identity of 100% in PDB database and 41 were mapped to 155 relevant conserved pathways in KEGG database. The distribution of the bioactivity potency of these MLP chemical probes with their corresponding targets is shown in Fig. 2b. The chemical probes with potency in the range of 0.001 ~ 1 μM have been found for over 60% of the protein targets (43/67), which indicates varying quality of the probes with respect to potency. Compared to other bioactive compounds in PubChem, the MLP probes demonstrate relatively higher potency and significantly better selectivity for the respective targets in general. As several literature-based bioactivity databases become publicly available [27–29], it is also possible to gain insights into the novelty of the MLP probes by comparing them with the prior art. The detailed information of the MLP chemical probes, including bioactivity potency, biological pathways and related 3D structures of their targets is provided in Table S3 as a supplementary.
Recently, there have been intensive discussions on the criteria/principles of defining a chemical probe and some contradictory opinions have been raised [21, 30]. Though, only a portion of the MLP chemical probes seem to have medium or high quality based on a crowdsourcing evaluation [31] andmost of them have low citation rates by bibliometric method [30, 32], it would probably take more time to find out their merits in future studies. On the other hand, researchers in both academia and industry can help and are highly encouraged to assess and improve the MLP chemical probes through their own research. To this end, the efforts undertaken by the MLP to further characterize the probes and make the data publicly accessible through PubChem would help make it happen.
Concluding remarks
PubChem is growing rapidly with new data being deposited on a daily basis, which makes it feasible and imperative to evaluate the properties of a particular bioactive compound, a drug candidate or even a known drug on a large scale to identify potentially new functions or off-target effects. It starts to emerge as a valuable resource to explore the functions of genes and proteins in physiology and pathology. A summary of the public services and tools are listed in Table S4 as a supplementary to facility the utilization of the data in PubChem.
As a public molecular information resource at NIH, the free availability of PubChem will undoubtedly lower the barrier for researchers from chemical biology, medicinal chemistry and drug discovery to advance the development of new chemical tools for interrogating the biological functions and potential drug candidates for disease treatments. It also provides great opportunities for researchers in bioinformatics and cheminformatics to tackle the problems in those research fields with computational approaches.
Supplementary Material
Acknowledgments
We thank the National Institutes of Health Fellows Editorial Board for providing editorial assistance. This work is supported by Intramural Research Program of the National Institutes of Health, National Library of Medicine.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Additional information
Supplementary information is available online.
Teaser: PubChem, as a public resource for drug discovery, lowers the barrier for researchers to advance the development of new chemical tools for modulating biological processes and drug candidates for disease treatments.
References
- 1.Wang Y, et al. An overview of the PubChem BioAssay resource. Nucleic Acids Res. 2009;38:D255–D266. doi: 10.1093/nar/gkp965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Y, et al. PubChem: a public information system for analyzing bioactivities of small molecules. Nucleic Acids Res. 2009;37:W623–W633. doi: 10.1093/nar/gkp456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Austin CP, et al. NIH Molecular Libraries Initiative. Science. 2004;306:1138–1139. doi: 10.1126/science.1105511. [DOI] [PubMed] [Google Scholar]
- 4.Editorial. Perfecting probes. Nat. Chem. Biol. 2009;5:435. doi: 10.1038/nchembio0709-435. [DOI] [PubMed] [Google Scholar]
- 5.Hopkins AL, Groom CR. The druggable genome. Nat. Rev. Drug Discov. 2002;1:727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 6.Russ AP, Lampel S. The druggable genome: an update. Drug Discov. Today. 2005;10:1607–1610. doi: 10.1016/S1359-6446(05)03666-4. [DOI] [PubMed] [Google Scholar]
- 7.Drews J. What's in a number? Nat. Rev. Drug Discov. 2006;5:975. doi: 10.1038/nrd2205. [DOI] [PubMed] [Google Scholar]
- 8.Lindsay MA. Target discovery. Nat. Rev. Drug Discov. 2003;2:831–838. doi: 10.1038/nrd1202. [DOI] [PubMed] [Google Scholar]
- 9.Harland L, Gaulton A. Drug target central. Expert. Opin. Drug Discov. 2009;4:857–872. doi: 10.1517/17460440903049290. [DOI] [PubMed] [Google Scholar]
- 10.Overington JP, et al. How many drug targets are there? Nat. Rev. Drug Discov. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 11.Mayr LM, Bojanic D. Novel trends in high-throughput screening. Curr. Opin. Pharmacol. 2009;9:580–588. doi: 10.1016/j.coph.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 12.Marchler-Bauer A, et al. CDD: a Conserved Domain Database for protein classification. Nucleic Acids Res. 2005;33:D192–D196. doi: 10.1093/nar/gki069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marchler-Bauer A, et al. CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res. 2002;30:281–283. doi: 10.1093/nar/30.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Altschul SF, et al. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 15.Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bernstein FC, et al. The Protein Data Bank: a computer-based archival file for macromolecular structures. J. Mol. Biol. 1977;112:535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- 17.Xie L, et al. Drug discovery using chemical systems biology: identification of the protein-ligand binding network to explain the side effects of CETP inhibitors. PLoS Comput. Biol. 2009;5:e1000387. doi: 10.1371/journal.pcbi.1000387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanehisa M, et al. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 38:D355–D360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanehisa M, et al. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frye SV. The art of the chemical probe. Nat. Chem. Biol. 2010;6:159–161. doi: 10.1038/nchembio.296. [DOI] [PubMed] [Google Scholar]
- 22.Rix U, Superti-Furga G. Target profiling of small molecules by chemical proteomics. Nat. Chem. Biol. 2009;5:616–624. doi: 10.1038/nchembio.216. [DOI] [PubMed] [Google Scholar]
- 23.Han L, et al. A survey of across-target bioactivity results of small molecules in PubChem. Bioinformatics. 2009;25:2251–2255. doi: 10.1093/bioinformatics/btp380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng BY, et al. High-throughput assays for promiscuous inhibitors. Nat. Chem. Biol. 2005;1:146–148. doi: 10.1038/nchembio718. [DOI] [PubMed] [Google Scholar]
- 25.Yildirim Ma, et al. Drug-target network. Nat. Biotechnol. 2007;25:1119–1126. doi: 10.1038/nbt1338. [DOI] [PubMed] [Google Scholar]
- 26.Chen B, et al. PubChem as a source of polypharmacology. J. Chem. Inf. Model. 2009;49:2044–2055. doi: 10.1021/ci9001876. [DOI] [PubMed] [Google Scholar]
- 27.Liu T, et al. BindingDB: a web-accessible database of experimentally determined protein-ligand binding affinities. Nucleic Acids Res. 2007;35:D198–D201. doi: 10.1093/nar/gkl999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Overington J. ChEMBL. An interview with John Overington, team leader, chemogenomics at the European Bioinformatics Institute Outstation of the European Molecular Biology Laboratory (EMBL-EBI). Interview by Wendy A. Warr. J. Comput. Aided Mol. Des. 2009;23:195–198. doi: 10.1007/s10822-009-9260-9. [DOI] [PubMed] [Google Scholar]
- 29.Harmar AJ, et al. IUPHAR-DB: the IUPHAR database of G protein-coupled receptors and ion channels. Nucleic Acids Res. 2009;37:D680–D685. doi: 10.1093/nar/gkn728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Workman P, Collins I. Probing the probes: fitness factors for small molecule tools. Chem. Biol. 2010;17:561–577. doi: 10.1016/j.chembiol.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oprea T, et al. A crowdsourcing evaluation of the NIH chemical probes. Nat. Chem. Biol. 2009;5:441–447. doi: 10.1038/nchembio0709-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bologa C. Promiscuity and Pubchem: A Retrospective Analysis. Proceedings of the Society for Biomolecular Screening 16th Annual Conference and Exhibition; 2010. p. 119. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.