SUMMARY
Knowledge of both the global chromatin structure and the gene expression programs of human embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs) should provide a robust means to assess whether the genomes of these cells have similar pluripotent states. Recent studies have suggested that ESCs and iPSCs represent different pluripotent states with substantially different gene expression profiles. We describe here a comparison of global chromatin structure and gene expression data for a panel of human ESCs and iPSCs. Genome-wide maps of nucleosomes with histone H3K4me3 and H3K27me3 modifications indicate that there is little difference between ESCs and iPSCs with respect to these marks. Gene expression profiles confirm that the transcriptional programs of ESCs and iPSCs show very few consistent differences. Although some variation in chromatin structure and gene expression was observed in these cell lines, these variations did not serve to distinguish ESCs from iPSCs.
INTRODUCTION
Mammalian cells can be directly reprogrammed into induced pluripotent stem cells (iPSCs) by introduction of defined sets of transcriptional regulators (Takahashi and Yamanaka, 2006; Maherali et al., 2007; Nakagawa et al., 2008; Okita et al., 2007; Takahashi et al., 2007; Wernig et al., 2007; Yu et al., 2007; Aoi et al., 2008). These iPSCs hold great potential for regenerative medicine because they are similar to pluripotent embryonic stem cells (ESCs) and can be derived in a patient-specific manner from adult somatic cells (Yamanaka, 2007; Saha and Jaenisch, 2009). ESCs and iPSCs are highly similar in a broad range of phenotypic behaviors, including cell morphology, expression of pluripotency markers, teratoma formation, ability to differentiate into germ layers, and tetraploid complementation (Okita et al., 2007; Wernig et al., 2007; Boland et al., 2009; Kang et al., 2009; Smith et al., 2009; Zhao et al., 2009). However, recent studies comparing the gene expression profiles of ESCs and iPSCs have suggested that iPSCs are a unique cellular subtype distinct from ESCs (Chin et al., 2009; Marchetto et al., 2009) and that iPSCs retain some of the expression program of their cell of origin (Ghosh et al., 2010). It is important to understand whether there are genuine differences in the global chromatin structure and the gene expression programs of human ESCs and iPSCs, given that such differences may impact the potential therapeutic use of iPSCs.
Trithorax group (TrxG) and Polycomb group (PcG) protein complexes are key regulators of chromatin structures that are required for segmental identity in the developing embryo and contribute to maintenance of the pluripotent ESC state (Ringrose and Paro, 2004; Schuettengruber et al., 2007; Pietersen and van Lohuizen, 2008). TrxG complexes catalyze histone H3 lysine 4 trimethylation (H23K4me3) at promoters of protein-coding genes (Bernstein et al., 2002; Santos-Rosa et al., 2002; Ng et al., 2003; Schneider et al., 2004; Guenther et al., 2007; Mikkelsen et al., 2007), miRNA loci (Marson et al., 2008; Ozsolak et al., 2008), and noncoding lincRNA loci (Guttman et al., 2009). The PcG protein complex PRC2 catalyzes histone H3 lysine 27 trimethylation (H3K27me3), which contributes to repression of developmental genes (Schuettengruber et al., 2007; Schwartz and Pirrotta, 2008; Simon and Kingston, 2009). Histone H3K4me3 and H3K27me3 histone modifications are generally associated with transcriptionally active and repressed domains of the genome, respectively, although both modifications can occur at silent genes encoding developmental regulators that are poised for future activation (Bernstein et al., 2006; Lee et al., 2006; Guenther et al., 2007). Genome-wide maps of these histone modifications, which produce robust signals in ChIP-Seq experiments, can be especially useful for comparing transcriptional and developmental states of cells, particularly when coupled with gene expression profiling.
Microarray-based gene expression profiling has proven to be a powerful approach to characterize the transcriptional state of cells and to identify differences between cells of different types or states (Ebert and Golub, 2004; Ivanova et al., 2006). Comparing the gene expression profiles of ESCs and iPSCs could permit identification of any unique and consistent differences between these two cell types. However, comparative analysis of expression data can be challenging because ofdifferences in the homogeneity of cell populations, cell handling, reagents, and analytical techniques. In comparing the expression profiles of ESCs and iPSCs, it is therefore important to use analytical methods that account for the noise in the data and require reproducible results across multiple experiments (Bammler et al., 2005).
We have investigated whether a panel of human ESCs differs consistently from a panel of human iPSCs using both genome-wide maps of histone H3K4me3 and H3K27me3 modifications and gene expression analysis. We have also reanalyzed a large collection of previously published gene expression data by using different analysis methods. Our results reveal that small variations in the chromatin structure or gene expression occur among different ESC and iPSC lines, but we do not observe a consistent signature that distinguishes iPSC lines from ESC lines when examined after extended culture.
RESULTS AND DISCUSSION
Genome-wide Maps of Chromatin Modifications Show that Human iPSCs Share Key Features with ESCs
We used ChIP-Seq to map H3K4me3 and H3K27me3 occupancy genome-wide in six independent ESC lines and six independent iPSC lines grown under identical conditions (Figure 1). The ESCs included two male lines (BG01 and WIBR1) and four female lines (BG03, WIBR2, WIBR3, and WIBR7), each derived from a different donor (Lengner et al., 2010). The iPSC lines were generated from human fibroblasts with a doxycycline-inducible reprogramming system with OCT4, SOX2, and KLF4 genes (Hockemeyer et al., 2008; Soldner et al., 2009). Four of the iPSCs were derived from a female donor (iPS A1, iPS C1, iPS4, and iPS A6; described and characterized in Hockemeyer et al., 2008) and two from a male donor (iPS PDB2lox-17 and iPS PDB2lox-21; described and characterized in Soldner et al., 2009). All iPSC lines contained integrated transgenes, but were doxycycline independent for growth, indicating that transgene expression was not required for propagation of these pluripotent cells.
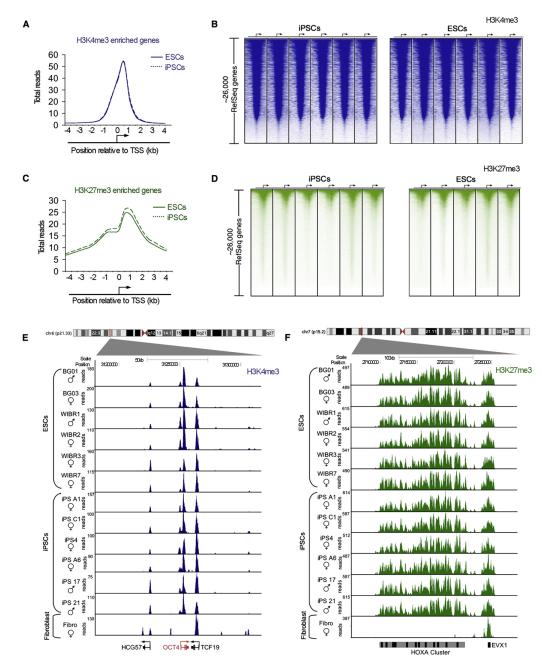
Figure 1. Genome-wide Maps of Chromatin Modifications Show Human iPSCs Share Key Features with ESCs.
(A) Composite H3K4me3 enrichment profile for all RefSeq genes in ESCs (solid blue) and iPSCs (dashed blue). The transcription start site (TSS) and direction of transcription of the average gene is noted by an arrow.
(B) ChIP-Seq density heat map of histone H3K4me3 (blue) for all RefSeq genes. Gene order was determined by highest average ChIP-Seq density in ESCs and arranged from highest to lowest density. The TSS and direction of transcription of genes is indicated by an arrow and the genomic region from −4.5kb to +4.5kb relative to the TSS shown.
(C) Composite H3K27me3 enrichment profile for all RefSeq genes enriched for H3K27me3 in ESCs (solid green) and iPSCs (dashed green). The TSS and direction of transcription of the average gene is noted by an arrow.
(D) ChIP-Seq density heat map of histone H3K27me3 (green) for all RefSeq genes. Gene order was determined by highest average ChIP-Seq density in ESCs and arranged from highest to lowest density. The TSS and direction of transcription of genes is indicated by an arrow and the genomic region from −4.5kb to +4.5kb relative to the TSS shown.
(E) ChIP-Seq density for H3K4me3 (blue) at the OCT4 locus in human ESC, iPSC, and fibroblast cell lines. The position of the loci within chromosome 6 and the scale is shown above the gene tracks.
(F) ChIP-Seq density for H3K27me3 (green) in the HOXA cluster in human ESC, iPSC, and fibroblast cell lines. The position of the cluster within chromosome 7 and the scale is shown above the gene tracks. See also Tables S1–S4.
The maps of histone H3K4me3 and H3K27me3 were highly similar at protein-coding and noncoding genes in all 12 ESC and iPSC lines when examined by enrichment profiles (Figures 1A and 1C), heat maps (Figures 1B and 1D), or inspection of gene tracks (Figures 1E and 1F). Nucleosomes with H3K4me3 occurred at the vast majority of protein-coding genes in both ESCs and iPSCs (~85%), with maximal enrichment occurring ~200 bp downstream of transcriptional start sites (Figures 1A, 1B, and 1E; Table S1 available online). H3K4me3-modified nucleosomes also occupied the start sites of known and predicted noncoding RNAs in both cell types, which include ESC-specific RNAs associated with pluripotency (Marson et al., 2008) (Table S1). H3K27me3-modified nucleosomes occurred primarily in the promoters of ~2000 repressed genes, many of which encode key regulators of development (Figures 1C, 1D, and 1F; Table S1). H3K27me3 marked small domains (1–5 kb) within certain gene promoter regions and large domains extending across >100 kb of the HOX gene clusters. These results indicate that the genomes of human iPSCs possess the general features of Trx- and PcG-mediated histone modifications previously described in ESCs (Bernstein et al., 2006; Boyer et al., 2006; Bracken et al., 2006; Lee et al., 2006; Guenther et al., 2007; Pan et al., 2007; Zhao et al., 2007; Mikkelsen et al., 2008).
Similarity in Genes and Regions Occupied by Modified Histones in ESCs and iPSCs
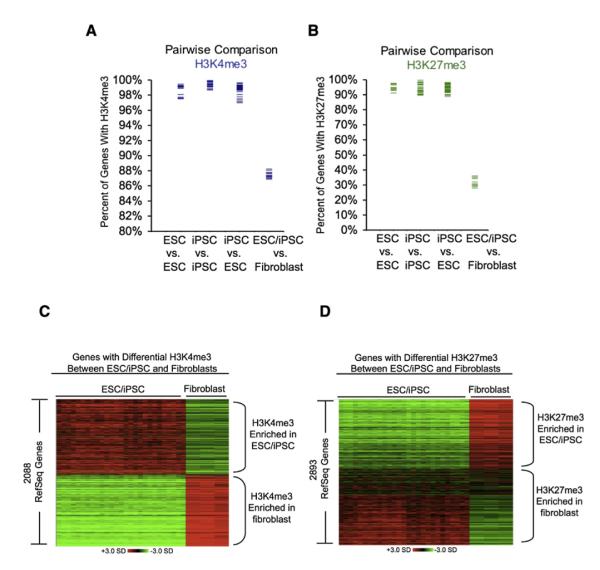
Inspection of gene tracks revealed some variation in H3K4me3 and H3K27me3 nucleosome occupancy among these cells lines, prompting us to systematically compare the sets of genes occupied by these histone modifications in each cell line with the set occupied in all other lines (Figure 2; Supplemental Experimental Procedures). We first performed all pairwise comparisons among the ESC lines and found that 1.4% ± 0.8% of genes had different H3K4me3 occupancy and 5.5% ± 2.0% of genes had different H3K27me3 occupancy. Similarly, 0.7% ± 0.3% genes varied for H3K4me3 and 6.0% ± 2.6% varied for H3K27me3 among the iPSC lines. Pairwise comparison of ESC and iPSC lines revealed that the variation for H3K4me3- (1.3% ± 0.7%) and H3K27me3- (6.0% ± 2.4%) occupied genes was not significantly different from the variation observed within ESC lines or within iPSC lines. In contrast, comparisons of the genes enriched for H3K4me3 or H3K27me3 between pluripotent cells (ESCs and iPSCs) and adult fibroblast cells revealed significantly larger differences (12.3% ± 0.4% for H3K4me3; 67.8% ± 2.8% for H3K27me3) (Figure 2; Table S2). Thus, we observed no more variation between ESCs and iPSCs than was evident within the ESC lines or within the iPSC lines. We also examined the magnitude of the ChIP-Seq peaks associated with each gene and again found that differences between ESCs and iPSCs were no greater than the differences observed within ESC lines and within iPSC lines (Table S2). These results suggest that there were few, if any, consistent differences in the set of genes occupied by H3K4me3 or H3K27me3 between these human ESCs and iPSCs.
Figure 2. Similarity in Genes and Regions Occupied by Modified Histones in ESCs and iPSCs.
(A) Pairwise comparisons of genes occupied by H3K4me3 in ESCs, iPSCs, and fibroblast cells. Each blue bar represents an individual pairwise comparison of the set of genes identified as enriched in one cell line with the set enriched in a second cell line. Comparisons between two ESC lines (ESC versus ESC), between two iPS lines (iPSC versus iPSC), between an ESC line and an iPSC line (ESC versus iPSC), and between an ESC or iPSC line and fibroblast cells (ESC/iPSC versus fibroblast) are shown in separate columns. Gene occupancy was determined as described in Supplemental Experimental Procedures.
(B) Pairwise comparisons of genes occupied by H3K27me3 in ESCs, iPSCs, and fibroblast cells. Each green bar represents an individual pairwise comparison of the set of genes identified as enriched in one cell line with the set enriched in a second cell line as in (A).
(C) Expression data for genes differentially occupied by H3K4me3 in pluripotent cells (ESCs and iPSCs) and fibroblast cells. Genes are ordered by the magnitude of differential H3K4me3 occupancy and relative gene expression is shown. Samples with higher than average expression are shown in red and samples with lower than average expression are shown in green (scale in standard deviations).
(D) Expression data for genes differentially occupied by H3K27me3 in pluripotent cells (ESCs and iPSCs) and fibroblast cells. Genes are ordered by the magnitude of differential H3K27me3 occupancy and relative gene expression is shown. Samples with higher than average expression are shown in red and samples with lower than average expression are shown in green (scale in standard deviations). See also Figure S1 and Tables S1–S4.
We developed a statistical method (Supplemental Experimental Procedures) to scan the entire genome and identify regions with significant differential H3K4me3 and H3K27me3 occupancy between ESCs and iPSCs. To confirm the sensitivity and specificity of this method, we compared all male ESC and iPSC lines to all female ESC and iPSC lines and found a number of regions with significant differences in histone modifications, which were located almost exclusively on the sex chromosomes, as expected (Table S3). We then compared female ESCs to female iPSCs and found that iPSCs showed increased H3K27me3 occupancy relative to ESCs at several X-linked genes including Xist. We believe these differences in occupancy are likely a consequence of exposing female iPSCs to high oxygen conditions during derivation, since high oxygen growth conditions induce X-inactivation in human pluripotent cells (Lengner et al., 2010). To further validate our ability to detect chromatin differences, we compared all pluripotent cells (ESCs and iPSCs) to donor fibroblasts and observed a large number of regions with differences in histone modifications (Table S3) that were strongly associated with differences in gene expression (Figures 2C and 2D).
We then applied this method to identify statistically significant differences in chromatin structure between ESCs and iPSCs and found 50 genomic regions (29 genes) with differential H3K4me3 occupancy and four regions (two genes) with differential H3K27me3 occupancy (Table S3). These regions of differential occupancy represent a tiny fraction of the genome (0.003%), and although there was no obvious theme associated with them, we considered several possible causes for the differential modification. First, we investigated whether these differences were due to the presence of exogenous reprogramming factors in iPSCs, but there were no significant differences in these chromatin modifications between transgene-containing and transgene-excised iPSCs (Table S2; Soldner et al., 2009). Second, we investigated whether the chromatin differences between ESCs and iPSCs were due to residual epigenetic signatures left from the parental fibroblast cell line, but found no evidence that iPSCs contain H3K4me3 or H3K27me3 signatures that reflect their cell of origin (Table S4). Lastly, we examined whether any gene expression changes were associated with differences in histone modification between ESCs and iPSCs, but found that this was not the case (Figure S1). We conclude that there are a small number of regions in these human ESCs and iPSCs that show differences in H3K4me3- and H3K27me3-modified nucleosomes. These differences involve a small fraction of the genome and have little or no influence on gene expression. However, we cannot exclude the possibility that these small chromatin differences observed in undifferentiated cells may exert subtle effects on cells upon differentiation.
Limited Variation in Gene Expression between Human ESCs and iPSCs
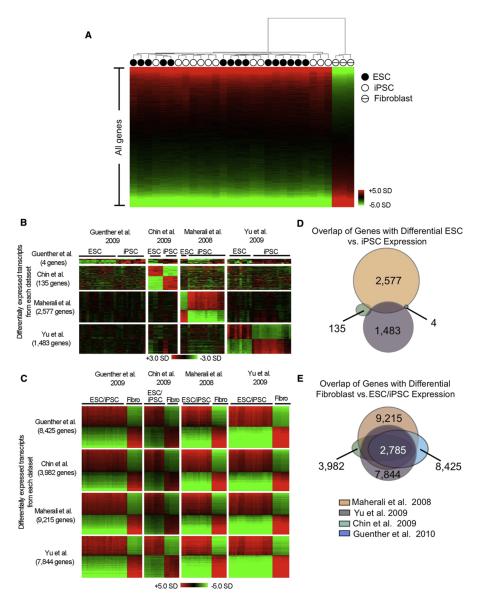
Although the H3K4me3 and H3K27me3 profiles of the human ESCs and iPSCs were nearly identical, we investigated the possibility that there were differences in the gene expression profiles between these ESCs and iPSCs. All 12 ESC and iPSC lines, in addition to donor fibroblast cells, were subjected to expression profiling and the data were analyzed with a single-factor analysis of variance for testing statistical significance with a Bayesian model of measurement error and a false discovery rate correction for multiple hypothesis testing (Sharov et al., 2005; Supplemental Experimental Procedures). We found zero genes with statistically significant differential expression between ESCs and iPSCs using this method. To gain greater statistical power to identify small differences in transcript levels between ESCs and iPSCs, we included expression data sets from several additional ESC and iPSC lines that were not used in the chromatin studies and repeated the analysis. In this panel of 39 samples, we found only four genes with statistically significant differential expression between ESCs and iPSCs (Figures 3A and 3B; Table S5). These results are consistent with a study that shows the overall mRNA and microRNA expression patterns of isogenic mouse ESCs and iPSCs are nearly indistinguishable within the exception of a few transcripts on chromosome 12qF1 (Stadtfeld et al., 2010).
Figure 3. Limited Variation in Gene Expression between Human ESCs and iPSCs.
(A) Unsupervised hierarchical clustering of whole genome expression data from ESCs (closed circle), iPSCs (open circle), and fibroblasts (hashed circle). Expression data was ordered by the magnitude of differential expression between pluripotent cells (ESCs or iPSCs) and fibroblast cells. Normalization and analysis for all expression data is described in Supplemental Experimental Procedures.
(B) Differential gene expression between ESCs and iPSCs across multiple expression data sets. For each data set (top to bottom) the transcripts with statistically significant differential expression between ESCs and iPSCs are shown. Within the set of differentially expressed transcripts from each data set, expression data was ordered by the statistical significance of differential expression between ESCs and iPSCs and then aligned to all other data sets for comparison. Samples with higher than average expression are shown in red and samples with lower than average expression are shown in green (scale in standard deviations).
Previous reports have observed that ESCs and iPSCs exhibit considerable differences in gene expression (Chin et al., 2009; Marchetto et al., 2009). To determine whether these gene expression differences were consistently observed in multiple laboratories, we re-examined a large collection of previously published expression data comparing ESCs and iPSCs (Table S5) (Maherali et al., 2008; Chin et al., 2009; Yu et al., 2009). If there were truly consistent gene expression differences between ESCs and iPSCs that were not a product of laboratory-specific biases in cell culture conditions, passage number, RNA preparation methods, or data processing methods, it would be expected that similar sets of genes would be identified as differentially expressed in more than one of these studies. However, we found that overlap between the genes identified in each of these studies was extremely low (Figures 3B and 3D) and conclude that there are very few, if any, consistent differences in the gene expression programs of ESC and iPSC lines. In contrast, the differential expression observed between pluripotent (ESC and iPSC) lines and fibroblast lines was highly reproducible across laboratories (Figures 3C and 3E; Table S5).
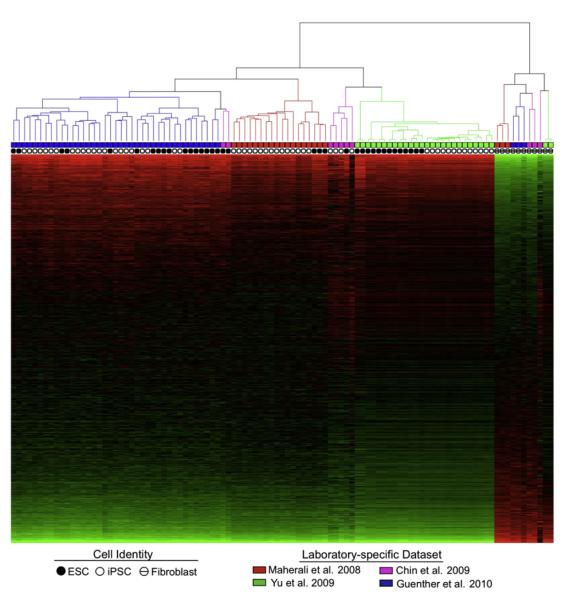
Several studies have described a few hundred to several thousand genes that show statistically significant differential expression between ESCs and iPSCs (Chin et al., 2009; Marchetto et al., 2009; Ghosh et al., 2010). There are several possible reasons that this result may have been obtained even if there is not, in reality, a unique and consistent expression signature that distinguishes iPSCs from ESCs. Cell culture conditions, derivation method, passaging technique, reagents, amount of time in culture, microarray methods, and operator-specific microarray assay biases can affect gene expression profiles. It is likely that uncontrolled variables such as these contributed to the observation of differential gene expression between the ESC and iPSC samples, given that we observe that ESC and iPSC expression data cluster by laboratory and not by ESC/iPSC identity (Figure 4). Our own data indicate that differences in gene expression do exist between various ESC and iPSC lines, but these differences do not consistently distinguish iPSCs from ESCs.
Figure 4. Human ESC and iPSC Expression Data Clusters More by Laboratory than by ESC/iPSC Identity.
Gene expression data sets for human ESCs, iPSCs, and fibroblast cells from four laboratories (Guenther et al., present study; Maherali et al., 2008; Chin et al., 2009; Yu et al., 2009) were quantile normalized as one group. Genes were Z score normalized and data were subjected to hierarchical clustering (centered correlation distance, centroid linkage) of samples. Genes were ordered from greatest to least magnitude of differential expression between pluripotent and fibroblast cells. Solid circles indicate ESC samples, empty circles indicate iPSC samples, and hashed circles represent fibroblast cell samples. Data from individual labs are coded by color as Guenther et al. (blue), Maherali et al. (red), Chin et al. (purple), and Yu et al. (green). See also Table S5.
(C) Differential gene expression between fibroblasts and pluripotent cells (ESCs and iPSCs). Expression data were ordered by the statistical significance of differential expression between fibroblasts and ESCs/iPSCs in each data set and then aligned to all other data sets for comparison. Samples with higher than average expression are shown in red and samples with lower than average expression are shown in green (scale in standard deviations).
(D) Overlap of differentially expressed genes between ESCs and iPSCs in various expression data sets. The numbers of genes differentially expressed between ESCs and iPSCs are indicated in black. The total overlap of all gene sets is zero.
(E) Overlap of differentially expressed genes between fibroblast and pluripotent cells (ESCs and iPSCs) in various expression data sets. The numbers of genes differentially expressed between fibroblast and pluripotent cells are indicated in black. The total overlap of all gene sets is shown in white. See also Figure S2 and Table S5.
DISCUSSION
ESCs and iPSCs have been shown to share key features of pluripotency, including expression of pluripotency markers, teratoma formation, cell morphology, ability to differentiate into germ layers, and tetraploid complementation (Okita et al., 2007; Wernig et al., 2007; Boland et al., 2009; Kang et al., 2009; Smith et al., 2009; Zhao et al., 2009; Stadtfeld et al., 2010). Human iPSCs offer great promise for regenerative medicine and the establishment of patient or subgroup-specific disease models, but some reports suggest that ESCs and iPSCs may not be equivalent (Chin et al., 2009; Marchetto et al., 2009; Ghosh et al., 2010). We have mapped two histone modifications that are critical for cell state and development in human ESCs and iPSCs and find a very small number of consistent differences between ESCs and iPSCs for these marks. These differences in chromatin structure are not associated with differential gene expression. Furthermore, the consistent differences between ESCs and iPSCs are considerably smaller than the overall variability among these cell lines. In our analysis of gene expression data across several studies, we find that variations in gene expression occur among different ESC and iPSC lines, but we do not observe a consistent signature that distinguishes iPSC lines from ESC lines.
Previous studies comparing the gene expression profiles of human ESCs and iPSCs argued that a recurrent gene expression signature appears in iPSCs regardless of their origin or the method by which they were generated and suggested that iPSCs should be considered a unique subtype of pluripotent cell (Chin et al., 2009). This interpretation is inconsistent with our own. This discrepancy is probably due to four features of the analytic methods used by Chin et al. (2009) that, in our view, are key to accurate data interpretation. First, a correction for multiple hypothesis testing was not used, so the number of statistically significant differentially expressed genes was greatly overestimated. Second, there was not a requirement that gene expression change in the same direction. Third, the same ESC expression data was compared to both “early” and “late” passage iPSCs, negating the assumption that these sets of differentially expressed genes would be independent. Fourth, biases were introduced into the identification of differentially expressed genes that violate the assumption of independent assortment, which is relied upon to calculate the statistical significance of gene list overlaps. The use of a fold change threshold creates a bias toward identifying genes with larger measurement error, and collapsing measurements for several probe sets into a single measurement for each gene could cause some genes to be more likely identified as differentially expressed across several data sets. In studies that came to the conclusion that ESCs and iPSCs have different gene expression signatures (Chin et al., 2009; Marchetto et al., 2009; Ghosh et al., 2010), we believe uncontrolled laboratory-specific variables probably contributed to the observation of differential gene expression. When we repeat the analysis of published data with standard methods, we donot observe significant overlap between the genes that are differentially expressed between ESCs and iPSCs across several laboratories (Figures 3B and 3D). Instead, we find that ESC and iPSC expression data cluster more by laboratory than by ESC/iPSC identity (Figure 4). Similarly, in mouse, most expression differences between ESCs and iPSCs are not consistently observed across laboratories and are probably caused by variations in genetic background or method of iPSC production (Chin et al., 2009; Stadtfeld et al., 2010).
In summary, our experiments and analysis do not demonstrate a significant difference between the H3K4me3 or H3K27me3 modifications or a consistent difference in the gene expression programs of ESCs and iPSCs when examined after extended culture. It is possible that there are small differences between ESCs and iPSCs that we lacked the statistical power to observe or that differences may occur in noncoding or regulatory RNAs. Additionally, it is possible that there exist important epigenetic differences between ESCs and iPSCs that are not reflected in the chromatin marks that we examined, such as DNA methylation events (Meissner et al., 2008; Doi et al., 2009; Kim et al., 2010). These possible changes in histone modification, DNA methylation, or other epigenetic marks may result in subtle functional differences that could affect differentiation or other cell processes (Feng et al., 2010; Hu et al., 2010). Nevertheless, our results and the phenotypic similarities shared by ESCs and iPSCs (Smith et al., 2009; Zhao et al., 2009) support the view that ESC and iPSCs are nearly identical cell types.
EXPERIMENTAL PROCEDURES
Human ESC and iPSC Culture
All primary fibroblast cell lines described in this paper were purchased from the Coriell Cell Repository (Camden, NJ). Fibroblasts were cultured in fibroblast medium (Dulbecco’s modified Eagle’s medium [DMEM] supplemented with 15% fetal bovine serum [FBS; Hyclone], 1 mM glutamine [Invitrogen], 1% non-essential amino acids [Invitrogen], and penicillin/streptomycin [Invitrogen]).
hiPSCs iPS A1, iPS C1, iPS4, and iPS A6 (Hockemeyer et al., 2008); hiPSCs iPS PDB2lox-17 and iPS PDB2lox-21 (Soldner et al., 2009); hESC lines BG01 and BG03 (National Institutes of Health code: BG01 and BG03; BresaGen, Inc., Athens, GA); and hESC cell lines WIBR1, WIBR2, WIBR3, and WIBR7 (White-head Institute Center for Human Stem Cell Research) (Lengner et al., 2010) were maintained on mitomycin C (MMC)-inactivated mouse embryonic fibroblast feeder layers in hESC medium (DMEM/F12 [Invitrogen] supplemented with 15% FBS [Hyclone], 5% KnockOut Serum Replacement [Invitrogen], 1 mM glutamine [Invitrogen], 1% nonessential amino acids [Invitrogen], 0.1 mM β-mercaptoethanol [Sigma], and 4 ng/ml FGF2 [R&D Systems]). Cultures were passaged every 5 to 7 days either manually or enzymatically with collagenase type IV (Invitrogen; 1.5 mg/ml).
ChIP-Seq
Detailed descriptions of antibodies, antibody specificity, ChIP, and ChIP-Seq analysis methods used in this study have been published previously and are provided in the Supplemental Experimental Procedures. The antibodies for ChIP were specific for H3K4me3 (ab 8580; Abcam) and H3K27me3 (ab 6002; Abcam). Purified immunoprecipitated DNA was prepared for sequencing according to a modified version of the Solexa Genomic DNA protocol, applied to a flow-cell with the Solexa Cluster Station fluidics device, and sequenced in accordance with Illumina’s standard protocols. Images acquired from the Solexa sequencer were processed through the bundled Solexa image extraction pipeline and aligned to the March 2006 build (NCBI36.1/hg18) of the human genome with Bowtie software (Langmead et al., 2009).
Expression Analysis
For RNA analysis, hESC and hiPSC colonies were mechanically isolated and pooled for RNA extraction. Total RNA was isolated from ESCs, iPSCs, and fibroblast cells with RNeasy MiniKit (QIAGEN). Five micrograms of total RNA was labeled in accordance with standard Affymetrix protocols and hybridized to Affymetrix HG-U133 2.0 plus arrays. The data were analyzed with Affymetrix Gene Chip Operating Software using default settings. Expression data were quantile normalized and analyzed for differential expression with the NIA Array Analysis Tool (http://lgsun.grc.nia.nih.gov/ANOVA/). A more detailed description of the gene expression analysis methods is provided in the Supplemental Experimental Procedures.
Supplementary Material
ACKNOWLEDGMENTS
We thank Tom Volkert, Sumeet Gupta, Jen Love, and Jeong-Ah Kwon of the WICMT for help with direct sequencing and data analysis; Ping Xu and Raaji Alagappan for help with human ESC culture; and Bingbing Yuan, George Bell, and Whitehead Institute BaRC for computational analysis. We acknowledge the generous support of Lilianne and Hillel Bachrack, Landon Clay, and Susan Whitehead. D.H. is a Merck Fellow of the Life Science Research Foundation. RJ is an advisor to Stemgen and a cofounder of Fate Therapeutics.
Footnotes
ACCESSION NUMBERS Complete ChIP-Seq and gene expression data are available from the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE22499.
SUPPLEMENTAL INFORMATION Supplemental Information includes two figures, five tables, and Supplemental Experimental Procedures and can be found with this article online at doi:10.1016/j.stem.2010.06.015.
REFERENCES
- Aoi T, Yae K, Nakagawa M, Ichisaka T, Okita K, Takahashi K, Chiba T, Yamanaka S. Generation of pluripotent stem cells from adult mouse liver and stomach cells. Science. 2008;321:699–702. doi: 10.1126/science.1154884. [DOI] [PubMed] [Google Scholar]
- Bammler T, Beyer RP, Bhattacharya S, Boorman GA, Boyles A, Bradford BU, Bumgarner RE, Bushel PR, Chaturvedi K, Choi D, et al. Members of the Toxicogenomics Research Consortium Standardizing global gene expression analysis between laboratories and across platforms. Nat. Methods. 2005;2:351–356. doi: 10.1038/nmeth754. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Humphrey EL, Erlich RL, Schneider R, Bouman P, Liu JS, Kouzarides T, Schreiber SL. Methylation of histone H3 Lys 4 in coding regions of active genes. Proc. Natl. Acad. Sci. USA. 2002;99:8695–8700. doi: 10.1073/pnas.082249499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Boland MJ, Hazen JL, Nazor KL, Rodriguez AR, Gifford W, Martin G, Kupriyanov S, Baldwin KK. Adult mice generated from induced pluripotent stem cells. Nature. 2009;461:91–94. doi: 10.1038/nature08310. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin MH, Mason MJ, Xie W, Volinia S, Singer M, Peterson C, Ambartsumyan G, Aimiuwu O, Richter L, Zhang J, et al. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell. 2009;5:111–123. doi: 10.1016/j.stem.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi A, Park IH, Wen B, Murakami P, Aryee MJ, Irizarry R, Herb B, Ladd-Acosta C, Rho J, Loewer S, et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat. Genet. 2009;41:1350–1353. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert BL, Golub TR. Genomic approaches to hematologic malignancies. Blood. 2004;104:923–932. doi: 10.1182/blood-2004-01-0274. [DOI] [PubMed] [Google Scholar]
- Feng Q, Lu SJ, Klimanskaya I, Gomes I, Kim D, Chung Y, Honig GR, Kim KS, Lanza R. Hemangioblastic derivatives from human induced pluripotent stem cells exhibit limited expansion and early senescence. Stem Cells. 2010;28:704–712. doi: 10.1002/stem.321. [DOI] [PubMed] [Google Scholar]
- Ghosh Z, Wilson KD, Wu Y, Hu S, Quertermous T, Wu JC. Persistent donor cell gene expression among human induced pluripotent stem cells contributes to differences with human embryonic stem cells. PLoS ONE. 2010;5:e8975. doi: 10.1371/journal.pone.0008975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Soldner F, Cook EG, Gao Q, Mitalipova M, Jaenisch R. A drug-inducible system for direct reprogramming of human somatic cells to pluripotency. Cell Stem Cell. 2008;3:346–353. doi: 10.1016/j.stem.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova N, Dobrin R, Lu R, Kotenko I, Levorse J, DeCoste C, Schafer X, Lun Y, Lemischka IR. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006;442:533–538. doi: 10.1038/nature04915. [DOI] [PubMed] [Google Scholar]
- Kang L, Wang J, Zhang Y, Kou Z, Gao S. iPS cells can support full-term development of tetraploid blastocyst-complemented embryos. Cell Stem Cell. 2009;5:135–138. doi: 10.1016/j.stem.2009.07.001. [DOI] [PubMed] [Google Scholar]
- Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, Ehrlich LI, et al. Epigenetic memory in induced pluripotent stem cells. Nature. 2010 doi: 10.1038/nature09342. in press. Published online July 19, 2010. 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengner CJ, Gimelbrant AA, Erwin JA, Cheng AW, Guenther MG, Welstead GG, Alagappan R, Frampton GM, Xu P, Muffat J, et al. Derivation of pre-X inactivation human embryonic stem cells under physiological oxygen concentrations. Cell. 2010;141:872–883. doi: 10.1016/j.cell.2010.04.010. [DOI] [PubMed] [Google Scholar]
- Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, Stadtfeld M, Yachechko R, Tchieu J, Jaenisch R, et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Maherali N, Ahfeldt T, Rigamonti A, Utikal J, Cowan C, Hochedlinger K. A high-efficiency system for the generation and study of human induced pluripotent stem cells. Cell Stem Cell. 2008;3:340–345. doi: 10.1016/j.stem.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetto MC, Yeo GW, Kainohana O, Marsala M, Gage FH, Muotri AR. Transcriptional signature and memory retention of human-induced pluripotent stem cells. PLoS ONE. 2009;4:e7076. doi: 10.1371/journal.pone.0007076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. Genomescale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, Okita K, Mochiduki Y, Takizawa N, Yamanaka S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat. Biotechnol. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell. 2003;11:709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- Ozsolak F, Poling LL, Wang Z, Liu H, Liu XS, Roeder RG, Zhang X, Song JS, Fisher DE. Chromatin structure analyses identify miRNA promoters. Genes Dev. 2008;22:3172–3183. doi: 10.1101/gad.1706508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan G, Tian S, Nie J, Yang C, Ruotti V, Wei H, Jonsdottir GA, Stewart R, Thomson JA. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell. 2007;1:299–312. doi: 10.1016/j.stem.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Pietersen AM, van Lohuizen M. Stem cell regulation by polycomb repressors: Postponing commitment. Curr. Opin. Cell Biol. 2008;20:201–207. doi: 10.1016/j.ceb.2008.01.004. [DOI] [PubMed] [Google Scholar]
- Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu. Rev. Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- Saha K, Jaenisch R. Technical challenges in using human induced pluripotent stem cells to model disease. Cell Stem Cell. 2009;5:584–595. doi: 10.1016/j.stem.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- Schneider R, Bannister AJ, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat. Cell Biol. 2004;6:73–77. doi: 10.1038/ncb1076. [DOI] [PubMed] [Google Scholar]
- Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Schwartz YB, Pirrotta V. Polycomb complexes and epigenetic states. Curr. Opin. Cell Biol. 2008;20:266–273. doi: 10.1016/j.ceb.2008.03.002. [DOI] [PubMed] [Google Scholar]
- Sharov AA, Dudekula DB, Ko MS. A web-based tool for principal component and significance analysis of microarray data. Bioinformatics. 2005;21:2548–2549. doi: 10.1093/bioinformatics/bti343. [DOI] [PubMed] [Google Scholar]
- Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: Knowns and unknowns. Nat. Rev. Mol. Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- Smith KP, Luong MX, Stein GS. Pluripotency: Toward a gold standard for human ES and iPS cells. J. Cell. Physiol. 2009;220:21–29. doi: 10.1002/jcp.21681. [DOI] [PubMed] [Google Scholar]
- Soldner F, Hockemeyer D, Beard C, Gao Q, Bell GW, Cook EG, Hargus G, Blak A, Cooper O, Mitalipova M, et al. Parkinson’s disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell. 2009;136:964–977. doi: 10.1016/j.cell.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Apostolou E, Akutsu H, Fukuda A, Follett P, Natesan S, Kono T, Shioda T, Hochedlinger K. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature. 2010;465:175–181. doi: 10.1038/nature09017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Okita K, Nakagawa M, Yamanaka S. Induction of pluripotent stem cells from fibroblast cultures. Nat. Protoc. 2007;2:3081–3089. doi: 10.1038/nprot.2007.418. [DOI] [PubMed] [Google Scholar]
- Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- Yamanaka S. Strategies and new developments in the generation of patient-specific pluripotent stem cells. Cell Stem Cell. 2007;1:39–49. doi: 10.1016/j.stem.2007.05.012. [DOI] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Yu J, Hu K, Smuga-Otto K, Tian S, Stewart R, Slukvin II, Thomson JA. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao XD, Han X, Chew JL, Liu J, Chiu KP, Choo A, Orlov YL, Sung WK, Shahab A, Kuznetsov VA, et al. Whole-genome mapping of histone H3 Lys4 and 27 trimethylations reveals distinct genomic compartments in human embryonic stem cells. Cell Stem Cell. 2007;1:286–298. doi: 10.1016/j.stem.2007.08.004. [DOI] [PubMed] [Google Scholar]
- Zhao XY, Li W, Lv Z, Liu L, Tong M, Hai T, Hao J, Guo CL, Ma QW, Wang L, et al. iPS cells produce viable mice through tetraploid complementation. Nature. 2009;461:86–90. doi: 10.1038/nature08267. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.