Abstract
In the past few years a great deal of progress has been made in studying the mechanical and structural properties of biological protein fibers. Here, we compare and review the stiffness (Young's modulus, E) and breaking strain (also called rupture strain or extensibility, εmax) of numerous biological protein fibers in light of the recently reported mechanical properties of fibrin fibers. Emphasis is also placed on the structural features and molecular mechanisms that endow biological protein fibers with their respective mechanical properties. Generally, stiff biological protein fibers have a Young's modulus on the order of a few Gigapascal and are not very extensible (εmax < 20%). They also display a very regular arrangement of their monomeric units. Soft biological protein fibers have a Young's modulus on the order of a few Megapascal and are very extensible (εmax > 100%). These soft, extensible fibers employ a variety of molecular mechanisms, such as extending amorphous regions or unfolding protein domains, to accommodate large strains. We conclude our review by proposing a novel model of how fibrin fibers might achieve their extremely large extensibility, despite the regular arrangement of the monomeric fibrin units within a fiber. We propose that fibrin fibers accommodate large strains by two major mechanisms: (1) an α-helix to β-strand conversion of the coiled coils; (2) a partial unfolding of the globular C-terminal domain of the γ-chain.
Keywords: Stiffness, Young's modulus, Breaking strain, Rupture strain, Extensibility, Fibrin fiber, Elastin, Resilin, Spider silk, Fibrillin, Fibronectin, Myofibrils, Intermediate filament, Keratin, Actin filament, Microtubules, Collagen, Mussel byssus
Introduction
Across biological systems, an array of intra- and extracellular protein fibers perform various mechanical functions. As is the case with all biological components, these fibers evolved structural and mechanical properties that are appropriate for these functions. Two important mechanical properties are the stiffness and the breaking strain (extensibility) of these fibers (detailed definitions of mechanical properties: see Section “Mechanical Measurements on Single Fibrin Fibers and Fibrinogen Molecules”). A review of the literature reveals that stiff fibers are usually not very extensible. These fibers often have a regular, paracrystalline (nearly crystalline) structure and/or are crosslinked. Examples include actin filaments, microtubules, collagen, matrix-embedded keratin fibers, and the spokes of spider webs. On the other hand, softer fibers are often very extensible. The structure of these fibers is often amorphous and/or contains less crosslinking; though these fibers utilize different molecular mechanisms to achieve high extensibility. Examples of soft, extensible fibers include elastin, resilin, fibrillin, fibronectin, intermediate filament, myofibrils, mussel byssal fibers, and the catching thread of spider webs.
It was recently discovered that fibrin fibers, which are the major structural component of a blood clot, are extraordinarily extensible and elastic [1], and that they are relatively soft [2]. This was unexpected, because fibrin fibers have a regular, paracrystalline structure [3, 4] and crosslinked monomer units.
In this review we compare the mechanical and structural properties of fibrin fibers with those of other polymerized protein fibers. This comparison leads us to suggest feasible molecular mechanisms that allow for the large extensions of fibrin fibers despite their nearly crystalline structure.
This review is divided into four sections:
-
(1)
Fibrin(ogen) structure and fibrin fiber assembly.
-
(2)
Mechanical measurements of fibrin fibers and single fibrinogen molecules.
-
(3)
Stiffness (Young's Modulus) and breaking strain (extensibility) of fibrin fibers and other polymerized protein fibers.
-
(4)
Proposed molecular mechanisms for fibrin fiber extension.
Fibrin(ogen) Structure and Fibrin Fiber Assembly
Fibrinogen is a highly abundant, soluble plasma protein. Removal of two pairs of fibrinopeptides converts it into fibrin monomers, which polymerize into a meshwork of fibrin fibers, the basic structural component of a blood clot. Fibrinogen consists of six peptide chains (2Aα, 610 residues; 2Bβ, 461 residues; 2γ, 411 residues; human numbering is used throughout this article). The recently solved crystal structures of human fragment D [5], bovine fibrinogen [6] and native chicken fibrinogen [7] (Fig. 1) added much clarity to the structure of fibrinogen and corroborated many features that had been gleaned from previous experiments. Fibrinogen has an approximately centrosymmetric, trinodular, S-shaped structure and is 46 nm in length and 4.5 nm in diameter [9–11]. Two nodules (D nodules) are at either end of the protein and one nodule (E nodule) is in the center of the protein. The nodules are connected via two, 17 nm-long coiled coils comprised of three α-helices, including residues α 51–161, β85–197, and γ33–143. The D nodule contains the globular β C-terminal domain (β197–461; called βC) and the globular γ C-terminal domain (γ143–411; called γC), both of which consist of a β-sheet core flanked by a few small α-helices. The central, globular E-nodule contains all six N-termini and also fibrinopeptides A and B. The α C-terminal threads briefly through the D nodule, rejoins the coiled coils as a fourth helix (α164–220) and ends with the segment called the αC domain (α220–610) that stretches from the distal D-nodule back towards the central E-nodule. The αC domain is mobile, and contains little well-defined secondary structure, although there is some evidence that this region consists of a flexible connector region (221–391) and a globular unit (392–610) [12–14]. This segment is shorter in chicken fibrinogen, α220–491 (chicken numbering). Although this segment is present in the crystals of chicken fibrinogen, the electron density for this region is too weak to resolve a structure, as expected for a segment with high mobility. Hence, these residues are not depicted in Fig. 1.
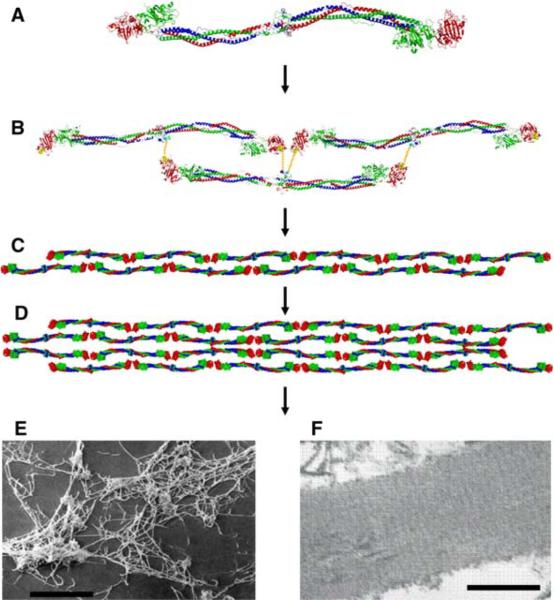
Fig. 1.
Crystal structure of chicken fibrinogen (7) and fiber assembly. (A) The fibrinogen molecule is 46 nm long and 4.5 nm in diameter and consists of six polypeptide chains; two α-chains (blue), two β-chains (green), and two γ-chains (red). (B) The monomers assemble in a half-staggered fashion to form the two-stranded protofibrils (C); the A: a interactions are shown as yellow lines. (D) The protofibrils aggregate laterally to form thicker fibers, branching occurs and eventually a full clot (E) is formed in which fiber diameters range from about 20 to 200 nm ((E) shows an SEM image of a 50% lysed thrombus containing fibrin fibers and platelets; courtesy R. R. Hantgan; reproduced with permission from [8]; scale bar: 10 μm). (F) A striated pattern with a periodicity of 22.5 nm (half the length of a fibrin monomer) is seen in the TEM image of a longitudinal cross section of a single fiber (Image: W. Liu, scale bar 200 nm). The striated pattern is due to the half-staggered arrangement of the monomers
Disulfide Bonds
Fibrinogen contains 29 disulfide bonds that stabilize the molecule. Five disulfide bonds are within the central E nodule linking together the two halves of the molecule. Prominent features are the four “disulfide rings” in each fibrinogen molecule; there is one ring at each end of the two coiled coils. Each “disulfide ring” consists of three disulfide bonds, linking the α- to the β-chain, the α- to the γ-chain, and the β- to the γ-chain, thus forming a brace that keeps the three α-helices in the coiled coils together. Last, there are twelve intra-chain disulfide bonds, three in each of the two globular βC domains (D-nodules), two in each of the two globular γC domain (D-nodules), and one in each of the two αC domains.
Fibrin fibers are assembled from fibrin monomers and the widely accepted sequence of events leading to fibrin fiber formation, starting with fibrinogen, is outlined in Fig. 1. Activated thrombin proteolytically removes fibrinopeptides A (16 residues) and B (14 residues) from the N-terminal region of the Aα- and Bβ-chains, respectively, thereby converting fibrinogen into fibrin monomers and exposing binding sites (“knobs”) A (Gly-Pro-Arg-Val) and B (Gly-His-Arg-Pro). These positively charged “knobs” fit into negatively charged binding pockets “a” and “b” in the γ- and β-chains within the distal globular D domains, respectively, thus forming the A:a and B:b interactions. Pocket “a” in γC is composed of four loops including key residues γGln329, γAsp330, γHis340, and γAsp364 [5, 15, 16]. Pocket “b” in βC is composed of key residues βGlu397, βAsp398, and βAsp432 [17]. Fibrinopeptide A is cleaved first and upon the exposure of knob A, fibrin monomers assemble in a half-staggered fashion into two-stranded protofibrils [18]. This assembly also aligns the D:D interface of abutting fibrin monomers. The key D:D interfacial residues include γArg275, γTyr280, and γSer300 [7]. The assembly is illustrated in Fig. 1B, which depicts a fibrin trimer, i.e., a nascent protofibril. Upon growing to an average length of about 15 monomers [18, 19], approximately twice the length of the protofibril shown in Fig. 1C, the protofibrils aggregate laterally to from fibers (Fig. 1D) that branch into a three-dimensional network (Fig. 1E). Mature fibers have a diameter of about 100 nm [20], i.e., about 150 monomers per cross section (assuming 30% protein content in a fiber [21]). Release of fibrinopeptide B (exposure of knob B) occurs subsequent to release of fibrinopeptide A. The B:b interaction may aid lateral aggregation; however, it is not required for lateral aggregation and is likely not a strong contributor to fiber stability. This statement is based on the observation that clotting reactions catalyzed with various snake venom enzymes, which only cleave Fibrinopeptide A (i.e., Batroxobin), produce mature and stable fibers [22].
Covalent Crosslinking
Fibrin fibers are stabilized by the formation of covalent crosslinks between specific glutamine and lysine residues (ε-amino(γ-glutamyl)lysine isopeptide bonds) [23]. The formation of these bonds is catalyzed by the transglutaminase factor XIIIa (FXIIIa). Initially, and concomitantly with protofibril formation, two reciprocal crosslinks are formed between the γ-chains of the fibrin monomers within a protofibril at residues Lys406 and Gln398/399, forming γ chain dimers. At about ten times slower rate than the γ–γ crosslink formation, Lys-Gln crosslinks are also formed between several sites in the αC domains, forming α-chain oligomers [24]. The main potential acceptor and donor sites within the α-chain may include Lys 418, 448, 508, 539, 556, 580, 601 and Gln 221, 237, 328, 366 or a subset of those [25–27]. The α–α crosslinks may form between fibrin units of different protofibrils, i.e., the protofibrils are bound together within fibrin bundles by α chain crosslinking. There is also some crosslinking between α- and γ-chains; these crosslinks may occur internally within one fibrin molecule [28], such that they will not contribute much to fiber stability.
Although establishing exactly how fibrin molecules pack together within a fiber is still an active area of research, the major interaction sites have been determined. It is becoming clear that the main non-covalent stabilizing inter-fibrin interactions are the D:D interface interaction, the A:a interaction, and to a lesser extent the B:b interaction; the main covalent interactions are the γ–γ crosslinks and perhaps to a lesser extent the polymeric α-crosslinks. The putative D:D interface does not exhibit any specific hydrogen bonds or salt bridges; the apparent driving force is the burying of 7.5 nm2 of surface area [5]. Thus, this interaction may not be very strong and the stability of fibrin fibers may mainly depend on the A:a interaction that supports protofibril formation and a large number of weak interactions, including D:D, that support the assembly of protofibrils into fibers [19]. The covalent cross-links, of course, greatly stabilize the noncovalent interactions. It is interesting to note that, except for the weak, hydrophobic D:D interaction, all other interactions involve flexible and mobile regions whose structures have not been discerned in any crystal [29]. This mobility may be functionally important. The A-knobs are attached to a mobile 10 residue-long tether (α17–27); the B-knobs are attached to a mobile 48 residue-long tether (β15–63). The reciprocal γ–γ links between Lys406 and Gln398/399 combine the two 19 residue-long, mobile C-γ-terminal domains which results in the formation of a 21 residue-long flexible γ–γ crosslink. The α–α crosslinks occur between the large, highly mobile αC domains.
It is not yet entirely clear how protofibrils aggregate laterally, though some putative interfaces on the γ-chains have been inferred from protein contacts within crystals, having given rise to the (speculative) multibundle model of fibrin packing within fibers [30]. In this model, fibrin molecules arrange longitudinally in the well-established, half-staggered fashion to form protofibrils, as outlined above. The two strands of a protofibril are 8 nm apart (Fig. 1B, C). It is suggested that the protofibrils then arrange in a complex, four-tiered, half-staggered fashion in which the tiers are apart by 4.75 nm, roughly the diameter of a protofibril (Fig. 1D). Such an arrangement would result in a “pseudo-unit cell” with dimensions of 19 nm × 19 nm × 45 nm containing eight fibrin molecules. The longitudinal and lateral distances of this suggested unit cell are consistent with energy dispersive X-ray diffraction techniques [31], giving credence to this model. Nevertheless, the data show real fibrin fibers will not assemble in such a uniform, perfectly crystalline fashion, as uniform crystals would give extremely sharp peaks, rather than the much broader peaks observed in the X-ray studies. Moreover, branching would become difficult if the fibers assembled in perfectly crystalline fashion.
Electron microscopy images of negatively stained fibrin fibers show a striated pattern with a periodicity of 22.5 nm (half the length of a monomer) along the fiber axis (Fig. 1F; [3, 4]). This observation clearly supports the model of fibrin monomers assembling in a regular, half-staggered, nearly crystalline fashion.
Mechanical Measurements on Single Fibrin Fibers and Fibrinogen Molecules
Before reviewing recent measurements of the mechanical properties of fibrin fibers and their building block, fibrinogen, we will define some of the terminology of force measurements. When a force, F, is applied longitudinally to a uniform, isotropic fiber with cross-sectional area, A, and length, L, it will stretch by a distance ΔL. In an elastic deformation, the fiber will return to its original length upon the release of force. In a linearlyelastic deformation, the strain ε = ΔL/L is proportional to the stress, σ (force per unit area; F/A): σ = E·ε, where E is the Young's modulus (Fig. 2). The Young's modulus is a measure of fiber stiffness; the higher the Young's modulus, the stiffer the fiber. If a material is non-linearly elastic, the elastic modulus (slope of stress–strain curve) changes as a function of strain. The material may become stiffer (strain-hardening) or softer (strain-softening) with increasing strain and one may define the modulus either as the secant modulus ((total stress)/(total strain)) or the tangent modulus ((differential stress)/(differential strain)).
Fig. 2.
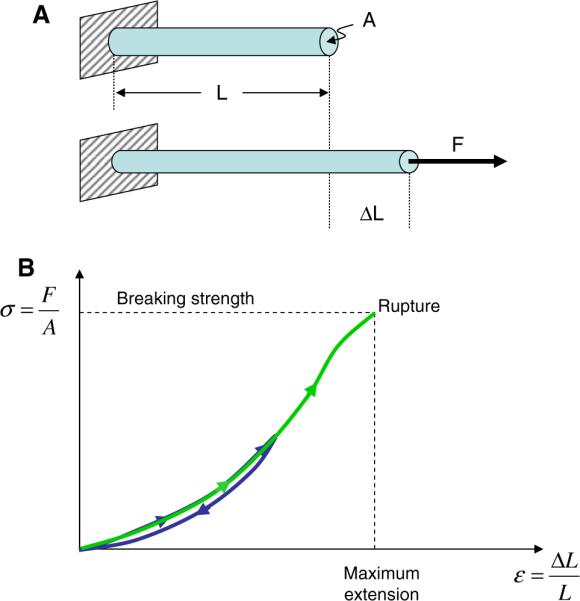
Stress–strain curves of stretched fibers. (A) A force F is applied in the longitudinal direction to a fiber with length L and cross sectional area A. The fiber extends by an amount ΔL. (B) A schematic stress–strain curve of the stretching of a fiber. The slope of the curve corresponds to the stiffness of the fiber. In a linear, elastic model, the stiffness (slope) is the called Young's modulus of the material. The maximum extension at which the fiber ruptures is called breaking strain (or extensibility) of the fiber
The extensibility (also called breaking strain or strain at rupture) of a fiber is defined as the strain, εmax, at which it will rupture. The elastic limit is the largest strain a fiber can sustain and still return to its original length upon the release of the applied force. This property is important, because fibers can recover without any permanent deformations from strains below the elastic limit, whereas strains above this limit cause permanent damage in the fiber.
Using an Atomic Force Microscope (AFM), Guthold et al. performed the first direct force measurements on single fibrin fibers as they determined the radius-dependence of the rupture force (breaking strength or ultimate strength) of fibrin fibers [32]. They found that the rupture force increases as R1.3, where R is the radius of the fibers. This was unexpected, since the rupture force would increase as R2 for a uniform cylindrical fiber with a solid cross section. This finding suggests that the density of fibrin fibers may be proportional to R−0.7 (not uniform), i.e., the fibers may be denser in the center than on the periphery. These force measurements were complicated by the fact that the fibers were adsorbed onto mica, so friction with the surface added resistance. Moreover, these measurements were done on dried fibers, whereas under physiological conditions fibrin fibers contain 70–80% aqueous solvent [21].
Subsequently, Collet et al. used laser tweezers to determine the Young's modulus of fibrin fibers in buffered saline analogous to plasma [2]. For small strains they found that uncrosslinked and crosslinked fibrin fibers have a Young's modulus of 1.7 and 14.5 MPa, respectively. These values are of the same order of magnitude as those for other soft biological fibers; however, they are about a 1,000 times less than those of stiff biological fibers (Table 1). Thus, the data at small strains indicate fibrin fibers are “soft”.
Table 1.
lists the stiffnees (Young's modulus, E); breaking strain (extensibility εmax); and the proposed molecular mechanisms of extensibility for various protein fibers
| Material | E (MPa) | εmax (%) | Proposed extensibility model | References |
|---|---|---|---|---|
| High extensibility, soft fibers | ||||
| Crosslinked fibrin fibers | 1–10 | 332 | α ↔ β transition, domain unfolding, protofibril sliding | [1, 2, 33] |
| Uncrosslinked fibrin fibers | 1–10 | 226 | α ↔ β transition, domain unfolding, protofibril sliding | [1, 2, 33] |
| Spider silk (Araneus Flag silk) | 3 | 270 | Modular composition; large extensible amorphous or β-spiral regions connected by stiff crystallites | [34–39] |
| Elastin (Bovine ligament) | 1 | 150 | Compact, amorphous, hydrophobic domains (random or β-spiral), which are crosslinked together, entropic, rubber-like elasticity | [40–6] |
| Resilin (Dragonfly tendon)a | 1–2 | 190a | Compact, amorphous, hydrophobic domains, which are crosslinked | [40, 46, 47]a |
| Cloned resilinb | 313b | together, entropic, rubber-like elasticity | Cloned [48]b | |
| Matrix-free Intermediate filament (mammalianc, hagfishd) | 6–300 | 160c–220d | α ↔ β transition and/or fibril sliding | [49–53] |
| Fibrillin | 0.2–100 | > 185 | Unstacking of pleated domains | [54–58] |
| Myofibrils (sarcomere) Titin (Connectin) | 1 | 200 | Unfolding of PEVK and Immunoglobulin domains in Titin | [59–61] |
| Mussel byssus (distal) | 10–500 | 109 | Block copolymer: collagen-like central domain (stiff) flanked by crystalline βsheets (stiff) and amorophous or β-sprial motifs (elastic) | [62–64] |
| Fibronectin | - | 200–300 | Extension of bent and looped molecules and/or domain unfolding | [65–67] [68] |
| Low extensibility, stiff fibers | ||||
| Spider silk (Araneus MA silk) | 10, 000 | 27 | Modular composition; small, extensible amorphous or β spiral regions, connected by stiff crystallites | [34–39] |
| Uncrosslinked, self-assembled collagen-I | - | 24–68 | Sliding of collagen fibrils | [69, 70] |
| Cross-linked, self-assembled collagen | 5,000–7,500 | 12–16 | Highly regular, paracrystalline structure, small, reversible molecular deformations | [69, 70] |
| Tendon collagen (mammalian tendon) | 160–7,500 | 12 | Highly regular, paracrystalline structure, small, reversible molecular deformations | [71–74] |
| Actin | 1,800–2,500 | ≤ 15 | Highly regular, paracrystalline structure | [75–80] |
| Microtubules | 1,000–1,500 | ≤ 20 | Highly regular, paracrystalline structure | [76, 81–84] |
| Wet, hard α-keratin in high-sufur matrix (hair, wool) | 2,000 | 45 | α ↔ β transition of keratin (embedded in crosslinked, high-sulfur matrix) | [50, 85–90] |
There are two reasons for the range of values in the Young's modulus of a given fiber: (1) Authors report a range of values. (2) Strain hardening/softening; the fibers become stiffer/softer as the strain increases. The table includes microscopic fibers (diameter: order of micrometer or less) and macroscopic fibers (diameter: order of several micrometers or larger). Microscopic fibers are fibrin fibers, elastin fibers, resilin fibers, intermediate filaments, fibrillin, myofibrils, fibronectin, actin filaments, microtubules; macroscopic fibers are spider silk, mussel byssus, collagen fibers, wool fibers
Dragonfly tendon
Cloned resilin
Mammalian, matrix free intermediate filament
Hagfish threads, a model fiber for matrix-free intermediate filament
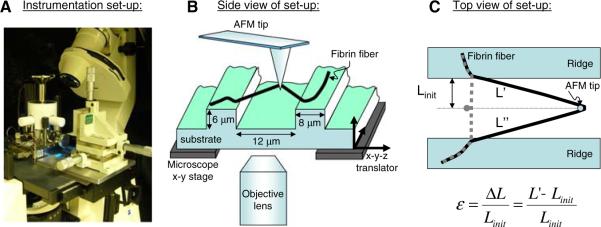
Recently Liu et al. used a combined atomic force/fluorescence microscopy instrument to measure the breaking strain (extensibility) and elastic limit of crosslinked and uncrosslinked, single fibrin fibers formed at low fibrinogen concentrations in buffered saline [1] (γ–γ crosslinking was 50–5% complete and α-crosslinking was 25–5% complete). In these experiments, fluorescently labeled fibrin fibers were suspended over 12-μm wide groves in a striated substrate. The AFM tip was used to stretch out these fibers, while the stretching process was observed from underneath with the fluorescence microscope (Fig. 3). These studies showed fibrin fibers have extremely large extensibilities and are able to recover elastically from very large strains.
Fig. 3.
Experimental set-up to determine fibrin fiber breaking strain (extensibility) (Figure adopted from [1] with permission). The experimental set-up to stretch single fibrin fibers is depicted photographically in Fig. 2A and schematically in Fig. 2B. The atomic force microscope (AFM) was used to stretch fibers that were suspended across ~12 μm-wide channels; the fluorescence microscope was used to record movies of this stretching process. This set-up has several advantageous features for stretching out fibers. Somewhat fortuitously, we found that the fibers were very well anchored on the ridges of the striated substrate as the anchoring points rarely changed; even at the most extreme fiber extensions. This implies that the observed deformations are not due to fiber slipping on the ridges. We selected fibers that bridged the grooves at a right angle with respect to the ridge edge. Thus, our set-up yielded a well-defined, easy-to-analyze geometry. By suspending the fibers over grooves, the substrate did not interfere with the measurement. Being able to record movies of the stretching process allowed us to accurately determine the lengths of the fibers and ascertain that the fibers did not slip at the anchoring points. The technique may also be applied to other fibers
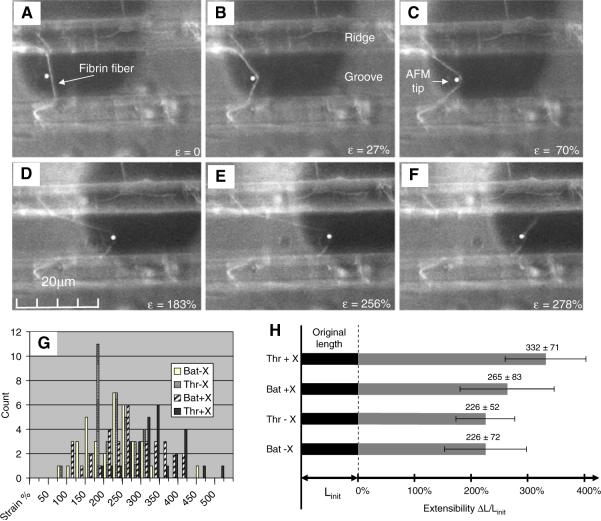
Figure 4A–F shows movie frames of an experiment that measured breaking strain (extensibility). The AFM tip was brought in contact with the fiber and moved to the right, parallel to the ridges, until the fiber ruptured. The fiber did not break at the point where the AFM tip contacted the fiber, but instead the two segments stretching from the tip to the two anchoring points broke independently. This observation indicates that the fiber did not break due to bending stress at the tip, but due to stretching between the tip and the anchoring point. After rupture, the fiber arms contract to nearly their original length (Fig. 4F), indicating that the deformations were mainly elastic. Thus, significant elasticity was retained even after the large extension and ultimate rupture. The distribution of extensibilities and the average extensibilities of the different fiber types are shown in Fig. 4G, H. Uncrosslinked fibers formed with thrombin and batroxobin showed a breaking strain of 226 ± 52% and 226 ± 72%, respectively (average ± standard deviation). Crosslinked fibers formed with thrombin or batroxobin showed a breaking strain of 332 ± 71%, and 265 ± 83%, respectively. A breaking strain (extensibility) of 332% corresponds to a fiber that is stretched to 4.32 times its original length as is schematically depicted in Fig. 4H.
Fig. 4.
Extensibility of fibrin fibers (adopted from [1] with permission and modified). (A–F) Snapshots from a fluorescence microscopy movie of a fiber extensibility experiment (crosslinked batroxobin fiber). A fluorescently labeled fiber is suspended between two ridges that appear as two horizontal bands. The AFM cantilever is seen from underneath as a dark, ~35 μm-wide rectangle; the location of the AFM tip is indicated by a white dot. The AFM tip is moved to the right, thus stretching the fiber; the lower segment of the fiber breaks at 183% strain and the upper segment of the fiber breaks at 278%. Histogram (G) and bar graph (H) representations of the extensibilities of the four different types of fibrin fibers; Thr = thrombin, Bat = Batroxobin, X = crosslinking
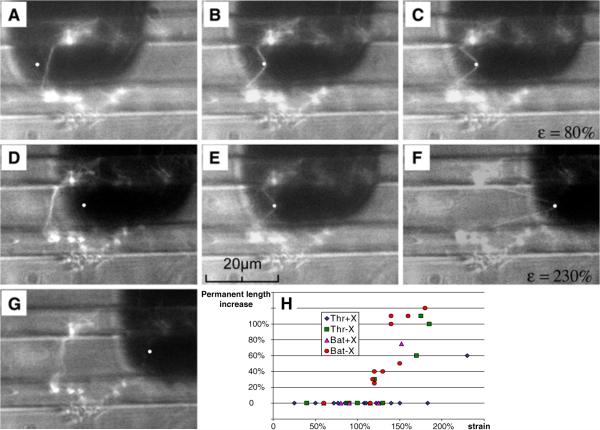
The elastic limit of fibrin fibers was tested by stretching them to a certain strain and releasing the applied force. In the experiment shown in Fig. 5, both elastic fiber deformation and deformation that induced permanent damage to the fiber were seen. First, the fiber was strained 80% after which it returned to its original length with no discernable permanent lengthening (Fig. 5A–D). The fiber was then strained 230% (Fig. 5E–G). When it snapped back under the tip, the fiber showed permanent damage with an increased length (Fig. 5G). A quantitative analysis (Fig. 5H) showed that the elastic limit of uncrosslinked fibers (batroxobin and thrombin) and crosslinked batroxobin fibers was at strains of about 120%; that is, these fibers were stretched to 2.2 times their original length and recovered completely elastically. Remarkably, crosslinked thrombin fibers were stretched to over 2.8 times their length (180% strain) and still recovered without any discernable permanent damage.
Fig. 5.
Elastic limit of fibrin fibers (adopted from [1] with permission and modified). (A–D) A fibrin fiber (thrombin, crosslinked) was strained 80% (C), from which it snapped back to its original length (D) without any permanent lengthening. (D–G) The same fiber was strained to 230% (F); at this strain it did suffer some permanent damage as the fiber is significantly deformed in image (G). (H) Plot of the amount of permanent deformations (% length increase) upon the release of force versus strain. Data points along the horizontal axis of 0%-permanent length increase indicate elastic deformations. Crosslinked thrombin fibers could be strained over 180% and the other fiber types about 120% without suffering permanent lengthening. None of the fibers analyzed here were ruptured
Force-Induced Fibrinogen Unfolding
In an very recent study, Brown et al. investigated the force-induced unfolding of single fibrinogen molecules [33]. When pulling on short oligomers of γ–γ crosslinked fibrinogen molecules with an AFM tip, they observed the typical sawtooth pattern of unfolding proteins. The observed peak-to-peak distance of 23 nm between unfolding events suggests that application of about 100 pN of force caused the unfolding of the 17-nm long α-helical coiled coils into 40-nm long extended β-strands. Since each fibrinogen molecule has two α-helical coiled coils (Fig. 1A) such a α-helix to β-strand conversion would extend the molecule by 2 × 23 nm = 46 nm. This lengthening corresponds to a strain of about 100%, since the initial length of a fibrinogen molecule is 46 nm.
Stiffness (Young's Modulus) and Breaking Strain (Extensibility) of Fibrin Fibers and Other Polymerized Protein Fibers
Table 1 lists the stiffness (Young's modulus, E), maximum breaking strain (extensibility, εmax) and putative extensibility mechanism of many important, polymerized protein fibers. Following the table we summarize the major, general features of the fibers listed in Table 1. An overview of the table reveals that high extensibility fibers (εmax > 100%) have a lower Young's modulus, usually on the order of 1–10 MPa. Conversely, low extensibility fibers are usually much stiffer with a Young's modulus on the order of 1,000–10,000 MPa. By way of contrast to well-known man-made materials, elastic rubber bands have a Young's modulus of about 1–10 MPa and nylon fishing line of about 1,000–10,000 MPa.
Despite the fact that detailed structural information is missing for most high extensibility fibers (many contain unstructured or high mobility motifs which hamper crystallization), there is reasonable experimental evidence to support a specific extension mechanism. The following mechanisms, correlated with each protein's structural features, have been proposed for high extensibility fibers: (i) Extension of amorphous, unstructured or β-spiral regions (spider silk, elastin, resilin); (ii) α-helix to β-strand transition (intermediate filament (keratin), hair (keratin), fibrin fibers) (iii) unstacking of intramolecular pleats (fibrillin fibers); (iv) unfolding of immunoglobulin or other protein domains (myofibrils (titin in sacromers), fibronectin); (v) straightening of bent or looped molecules (fibronectin); (vi) sliding of fibrils past each other (uncrosslinked collagen). Among these fibers, several contain coiled coil motifs [91]. For example, fibrinogen and the intermediate filament proteins, keratin and vimentin, and hair fibers (matrix-embedded keratin) have a long α-helical coiled coil, which may convert to an extended β-strand conformation upon the application of force.
The common theme for most low extensibility fibers (actin, microtubules, collagen) is a highly regular, nearly crystalline arrangement of monomer units without regions that can extend, change secondary structure or unfold.
At this point we should also mention that in the text below `crosslinks' usually though refers to covalent bonds; the exception is spider silk in which `crosslinks' refer to bonds formed by crystallites.
High Extensibility Fibers
Fibrin Fibers
Fibrin fibers are very stretchable with an average breaking strain (extensibility) of 332% and 226% for crosslinked and uncrosslinked fibers, respectively (1). To our knowledge, crosslinked fibrin fibers display the largest extensibility of all protein fibers. They are also rather soft, with a Young's modulus of 1–10 MPa (2). This is unusual, because the fibrin monomers within fibrin fibers are arranged in a relatively regular, half-staggered, almost crystalline pattern along the fiber axes [18, 19, 30], Fig. 1). As will be outlined in Section “Proposed Molecular Mechanisms for Fibrin Fibers Extension,” where we propose molecular mechanisms, these seemingly conflicting properties may be partly reconciled by assuming that the fibrin monomers within a fiber are very extensible while at the same time maintaining the most important fibrin–fibrin interactions.
Spider Silk
Of the many different types of spider silk fibers, the most relevant ones for examining extensibility and stiffness are those excreted by the major ampullate (MA) gland and the flagelliform (Flag) gland of araneoidea (web-spinning spiders) [34]. MA fibers, which form the frame and spokes of the web, are much stiffer and less extensible than Flag fibers, which form the catching threads spiraling around the frame. MA fibers have a maximum breaking strain of 27% and a Young's modulus of 10,000 MPa. Flag fibers have a maximum breaking strain of 270% and an initial (low strain) Young's modulus of 3 MPa [35, 36].
It is thought that the mechanical properties of these fibers originate from the modular composition of the silk fibroin proteins comprising each fiber. The fibroin proteins are composed of two main domains: (i) poly-alanine repeats, which form crystallites of β-pleated sheets, and (ii) glycine-rich domains which form amorphous domains. The crystallites may act as “cross-links” between the amorphous regions, which can unravel and extend upon the application of stress [37–39]. The proportion of the fibroin protein devoted to each structural module affects the fibers' mechanical behavior. Crystalline regions confer tensile strength and stiffness; and the amorphous regions provide high extensibility. The stiff and less extensible MA fibers have a high proportion of crystallite domains, whereas the Flag fibers contain few crystallites and consist mainly of large amorphous domains [39].
Elastin Fibers
Elastin fibers are found in the extracellular matrix of many mammalian tissues, such as skin, lung, and large blood vessels (reviews [44–46]). Tropoelastin, the precursor of elastin is deposited into the extracellular matrix, where it is cross-linked at its lysine residues, thus rapidly forming elastic fibers with the help of several scaffolding microfibril proteins, such as fibrillin. Elastin fibers have a Young's modulus of 1 MPa and a large breaking strain (extensibility) of 150% [41].
Due to the extensive cross-linking, elastin is nearly insoluble, and thus, structural information for elastin fibers remains limited. Elastin probably consists of two alternating domains: (i) a hydrophilic domain, rich in lysine and alanine, which forms cross-links; (ii) a hydrophobic domain, rich in glycine, proline, and valine, which forms repeats of poly (VPGVG) and poly (VGGVG) and endows the fiber with its elasticity and large extensibility. The structure of the hydrophobic domain may be a compact amorphous structure of random chains (similar to rubber) [42] or β-spirals [43] (review [44]). Both the models (random chain or β-spiral) could explain the observation that elastin fibers display entropic elasticity, similar to a typical rubber.
Resilin Fibers
Resilin is a protein found in the cuticles of many insects. It forms fibers that display elastic properties similar to elastin fibers [40, 46, 47]. Recently, Elvin et al. expressed resilin from Drosophila melanogaster in E. coli and formed resilin fibers with the purified recombinant protein [48]. These fibers have a breaking strain of up to 313% and a Young's modulus of 1–2 MPa.
Though there is still ongoing debate on the structural basis for the elastic properties of these fibers, there seems to be consensus that spider silk, elastin, and resilin fibers consist of an amorphous, hydrophobic region of random chains or β-spirals, which can unravel, and an ordered region that links these amorphous regions together.
Intermediate Filament
Intermediate filaments are one of the three cytoskeletal fibers (in addition to actin filaments and microtubules) and they are prominent in cells that are exposed to mechanical stresses. A number of proteins, including keratin, vimentin and desmin, polymerize into intermediate filaments. These proteins exhibit a characteristic “tripartite” structure consisting of a central elongated, α-helical domain, flanked by a globular head (N-terminal), and a tail (C-terminal) domain [92]. Monomers twist around each other to form an about 45 nm long and 2–3 nm wide dimer such that the two α-helices form a coiled coil. The dimers assemble, in a not yet fully understood geometry, into the 8- to 12-nm wide intermediate filament. These filaments then form bundles. Kreplak and Fudge recently wrote an excellent review about the mechanical properties of intermediate filaments [53].
Fudge and Gosline have measured the breaking strain of hagfish threads to be 220%. These fibers can be considered bundles of matrix-free intermediate filaments [49] (as opposed to matrix-embedded keratin-fibers, see below). These authors also showed that the Young's modulus of these filaments is 6 MPa for small strains; when stretched further, the fibers become stiffer (strain hardening) displaying an instantaneous stiffness (or tangent modulus = slope of the stress–strain curve) of 300 MPa for strains over 100% [49]. The lower initial value (6 MPa) is consistent with the persistence length of 1 lm that was determined for single intermediate filaments from AFM images [52]. The persistence length, P, and Young's modulus, E, are related for isotropic materials; E · I = kB · T · P, where I is the areal moment of inertia, kB is the Boltzmann constant and T is the temperature [93]. Assuming the filament is cylindrical such thatI = 0.25·π·R4 E ~ 8 MPa for a filament with persistence length of 1 μm and radius R = 5 nm.
The extensibilities of three different types of intermediate filaments (desmin, keratin, and neurofilaments) were determined by Kreplak et al. [51], by rupturing these filaments laterally with an AFM tip. They found an average breaking strain of 160%, and a maximum value of 260%. Guzman et al. used an AFM tip to bend single vimentin filaments, which were suspended over holes in a substrate. They determined a bending modulus of 300 MPa for uncrosslinked vimentin fibers [94].
Fudge and Gosline proposed that the large extension in intermediate filaments is made possible by a transition of the central, α-helical coiled coil rod to an elongated β-strand structure [50].
Fibrillin fibers
Fibrillin fibers are widely distributed in elastin-containing tissues, such as blood vessels, lung, and skin, where they act as a lattice for elastin deposition during elastin fiber formation. Fibrillin-rich fibrils also occur in tissue that does not contain elastin, such as the ciliary zonules, where they hold the lens of the eye in dynamic suspension [95]. Fibrillin fibrils resemble beads-on-a-string with a periodicity of 56 nm and a diameter of 10–14 nm. They can be extended to a periodicity of at least 160 nm, which amounts to a strain of at least 186% [54]. Using molecular combing techniques, Sherratt et al. found a Young's modulus of 78–98 MPa for fibrillin-rich microfibrils (zonular filaments) [56]. Megill et al. report a Young's modulus of 0.9 MPa for fibrillin-containing fibers in the mesoglea of the hydromedusa Polyorchis penicillatus. Megill et al. argue that the stress–strain curve for fibrillin, similar to hagfish threads, is J-shaped with an initial low value, in the 1 MPa range, at low strains and a much higher value, up to 100 MPa, at higher strains (strain hardening) [57]. Values of 0.19–1.88 MPa were also obtained for the zonular filaments from cows [58].
Though fiber assembly is still poorly understood, the model that best accounts for the observed data is the intramolecular pleating model [54, 55]. Each fibrillin molecule consists of rod-like regions that fold upon themselves, forming pleats. Upon stretching the pleats become undone.
Myofibrils
Myofibrils, which consist of concatenated structures called sarcomeres, are the stretchable fibers of striated muscle. Myofibrils are a highly ordered assembly of three myofilaments: the thick filament, which is mainly myosin, the thin filament, which is mainly actin, and titin, also called connectin. Sarcomeres have a large, 200% breaking strain [53]. A Young's modulus of about 1 MPa has been determined for myofibrils from the Drosophila indirect flight muscle; this report argues that other muscles should have similar stiffness [61]. Titin is the myofilament that is mainly responsible for the large extensibility of myofibrils in relaxed muscle fibers. Titin is a giant, single-peptide-chain protein (~0.6–3.7 MDa, depending on isoform) that stretches half the length of the sarcomere (for excellent review see [96]). Titin consists of four different regions (Z-disc region, I-band, A-band, M-line region), but only the I-band, consisting of immunoglobulin repeats and the PEVK region (rich in proline, glutamate, valine, and lysine) is functionally extensible. Extension of the sarcomere first causes straightening of the compact, relaxed I-band section of titin. Further extension induces unfolding of the PEVK region and the immunoglobulin domains of titin. Titin refolds upon the release of force [60].
Mussel Byssal Fibers
Mussels are tethered to solid supports by a byssus composed of collagen-like threads. The byssal threads have three serially arranged parts: a corrugated proximal segment, a smooth distal segment and an adhesive plaque. The breaking strain of the distal and proximal segment are 109% and 200%, respectively [62]. The Young's modulus for byssal threads ranges from 10 to 500 MPa [64]. The exact secondary structure of byssal fibers is not well known, but it appears to be a block copolymer consisting of a central collagen-like domain flanked by more elastic regions [63]. Some of the flanking domains have sequence motifs similar to the crystalline β-sheets and amorphous domains in spider silk and the β-spiral motifs of elastin.
Fibronectin
Fibronectin fibers are found in embryonic tissue, in healing wounds and in the extracellular matrix. Fibronectin fibers can be stretched to a strain of 200–300% as they are being assembled on the cell surface [66, 97]. The Young's modulus of fibronectin fibers has not been determined and it is also unknown how fibronectin molecules are arranged to comprise a fibril. Agreement on the stretching mechanism has not yet been achieved. There are models proposing that fibronectin domains may unfold upon stretching [68, 98]; these models are based on FRET experiments [68] and NMR and steered molecular dynamics simulations [98]. However, there are also models proposing that fibronectin may not unfold upon stretching [65, 67]. In those models fibronectin molecules are bent and looped into a compact conformation. Stretching then pulls the molecules into an extended conformation, but protein domains would remain folded.
Low Extensibility Fibers
Collagen
There are over 20 different types of collagen, some of which form fibrils (e.g., types I, II, III, V, and X) that provide the structural framework of many tissues. These fibrils are the major stress-bearing component in connective tissues such as bone, teeth, cartilage, tendon, ligament and the fibrous matrices of skin and blood vessels. Given the many different types and functions of collagen fibers, their properties are varied, though none are very extensible. All the fibril-forming collagens self-assemble into cross-striated fibrils with a characteristic 67 nm repeat [70]. The major structural feature in these collagens is an about 300 nm long, right-handed “collagen triple helix' consisting of three left-handed proline helices (not α-helices) wrapped around each other. The collagen triple helix should not be confused with the coiled coils consisting of α-helices. The collagen triple helix is designed to not be very extensible as the rise per residue is 0.29 nm, vs. 0.34 nm for a β-strand. The rise per residue in an α-helix is 0.15 nm.
Type-I collagen molecules self-assemble into quarter-staggered arrays of fibrils 20 to several hundred nanometers in diameter. Bundles of fibrils pack into collagen fibers and higher order tissue structures.
The breaking strain (extensibility) of self-assembled, cross-linked and uncrosslinked type-I collagen (tendon is predominantly composed of crosslinked type I collagen) was found to be 10% and 50%, respectively [69, 70]. The authors attribute the larger extensibility of uncrosslinked collagen to sliding of collagen fibrils past each other. The total stretch modulus (including viscous components) of type-I collagen fibers is on the order of 300–500 MPa, as determined for both mineralized turkey tendons and for 6-month aged, cross-linked, self-assembled type-I collagen fibers. The Young's modulus (elastic part of the total stretch modulus) is on the order of 250–400 MPa for these fibers [71]. This study employed incremental stress–strain curves to separate the elastic and viscous components of the total stretch modulus [71]. These numbers were then corrected for the facts that fibrils slide within a fiber and that fibers are porous and a Young's modulus of 5,000–7,500 MPa was obtained for “solid” collagen fibers that do not slide. Values of 160 and 1,660 MPa, and 2,900 MPa were reported for newborn and mature pig digital tendons [72] and for bovine Achilles tendon [73], respectively.
Actin
Soluble G-actin polymerizes into helical actin filaments (F-actin). Actin- filaments have a variety of roles in cells that include forming a scaffold to maintain cell shape, being part of the mechanism for cell motility, and serving as “rails” for myosin in muscle contraction (the thin filaments of muscle (see myofibrils above) are composed of filamentous actin). Actin often interacts with a number of other structural proteins resulting in the formation of fibers with somewhat different properties. The breaking strain of actin filaments is small, 15% or less (Figs. 5 and 6 in [77], [78, 79]). The Young's modulus of actin filaments is on the order of 1,800–2,500 MPa [75–77, 80].
Fig. 6.
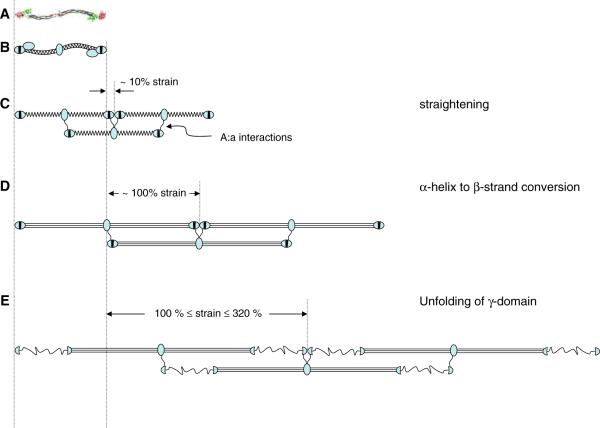
Schematic molecular mechanism models for fibrin fiber extensions. (A) Crystal structure and (B) schematic model of fibrinogen. (C) Half-staggered assembly of three fibrin monomers; molecules are rotated 90° from view in (B); βC domain is no longer depicted; straightening and slightly stretching the molecule may accommodate 10% strain. (D) An α-helix to β-strand conversion of the coiled coil and a slight straightening and alignment of the molecules could accommodate ~100% strain. (E) Higher strains, up to 320% could be accommodated by a partial unfolding of the γC domains; 230% strain is depicted
Microtubules
Microtubules are hollow fibers whose cylindrical shells have a diameter of 25 nm. They provide the framework that organizes the cell and guides the movement of organelles within cells. They are formed of alternating, noncovalently joined helices of globular α- and β-tubulin with 13 staggered α-β tubulin heterodimers per turn. Microtubules have a very regular, paracrystalline structure, and a breaking strain of less than 20% [81] and a Young's modulus of 1,000–1,500 MPa [76, 82, 83] as determined from the flexural rigidity of microtubules and in the 100 MPa range or higher as determined by bending suspended microtubules with an AFM tip [84].
Hard α-keratin
Hard α-keratin is a tough composite material that forms structures such as hair, hooves, and claws in mammals. The composite consists of keratin microfibrils, (very similar in structure to the intermediate filament), embedded in a sulfur matrix. The breaking strain of hard, wet α-keratin fibers, such as hair and wool, is about 45% and their Young's modulus is about 2,000 MPa [86]. It has long ago been suggested [87, 88] and experimentally verified [89, 90], that these fibers undergo an α-helix to β-strand transition, when stretched. The increased stiffness and decreased extensibility of hard α-keratin fibers as compared to intermediate (keratin) filaments is mainly due to the matrix.
Proposed Molecular Mechanisms for Fibrin Fibers Extension
The molecular mechanisms that allow fibrin fibers to accommodate large and reversible strains are unknown. Nevertheless, utilizing the crystal structure of native chicken fibrinogen [7], studies of the other proteins listed in Table 1, and the single molecule stretching experiments by Brown et al. [33], it is possible to suggest several likely mechanisms (Fig. 6). These mechanisms are not mutually exclusive, such that they may all contribute to the overall mechanical properties.
1. Small strains of about 10% may be accommodated by small, reversible structural changes in the fibrin monomer, protofibril and protofibril arrangement within a fiber. Fibrin monomers may straighten their slight S-shape (Fig. 6C). Protofibrils are shaped like a wavy ribbon [30] and the major interactions which hold the two strands of a protofibrils together utilize flexible linkers [29]. Upon stretching, the wavy shape of the protofibrils may straighten and the flexible linkers may align along the stretch direction, increasing the distance between monomer units. Though it is still not known how protofibrils aggregate laterally to from fibrin fibers, the open structure (fibrin fibers contain 70–80% aqueous solvent [21]) suggests that the interactions between protofibrils are mediated through long linkers. Upon stretching, those linkers could also align along the stretch direction.
2. At larger strains, the two 17 nm α-helical coiled coils may be converted to extended β-strands, resulting in an increased rise per residue from 0.15 to 0.34 nm (Fig. 6D). Completely converting the coiled coils would result in a 43-nm length increase of the 46-nm long fibrin molecule (~93% strain). This mechanism is supported by early wide-angle X-ray scattering experiments that indicated an α to β conversion in stretched fibrin clots [99, 100]. Additional support for this mechanism comes from recent experiments showing that force-induced unfolding of fibrinogen monomers results in a fibrinogen length increase that is consistent with the α to β conversion model [33].
A similar α to β-conversion has been observed in stretched keratin fibers. This conversion is reversible in hydrated, hard keratin (e.g., wool) [86, 89], though it appears to be irreversible in hagfish threads, a model for uncrosslinked, matrix-free intermediate (keratin-like) filaments [49]. Interestingly, coils of protein helices are a structural feature in several fiber-forming proteins that are exposed to stress, such as fibrinogen, intermediate filament, keratin (α-helical coiled coils), collagen (not alpha-helical) and other load-bearing proteins, such as tropomyosin and laminin. The detailed structural features of the coiled coils in those proteins are different. In fibrinogen the coiled coil is left-handed (about one turn) and consists of three right-handed α-helices [7]. In keratin, the coiled coil is also left-handed but consists of only two α-helices. In collagen, the “collagen triple helix” coil is right-handed and consists of three left-handed helices (not α-helices); this coil is not very extensible. It is intriguing to think of coiled coils as mechanical springs in proteins [101].
Scenarios 1 and 2 might account for the 120% elastic limit of uncrosslinked fibrin fibers [1].
3. At extreme strains (larger than the combined strains of scenarios 1 and 2) one or more of the globular domains might partially unfold (Fig. 6E). The γC domain in the D-nodule would be the most likely candidate, because the stress forces are transmitted through the D:D contacts, A:a interactions and the γ–γ crosslinks within the protofibril. The unfolded domain might involve a stretch of about 140 residues (from γGlu 183 to γAsn 325) in the globular γ-domain, which does not contain key residues of the A:a interaction (γGln329, γAsp330, γHis340, γAsp364) or γ–γ crosslinks (γLys406 γGln398/399). Since there are no disulfide bonds in this segment, it can be fully extended, resulting in a 50-nm length increase for each of the two γC domain; this unfolding could account for 220% additional strain. Including the 100% from scenarios 1 and 2, a total of 320% strain could be accommodated. Similar, reversible unfolding has been observed for the immunoglobulin-like (and PEVK) domains in titin [60], and reversible unfolding has been proposed as a possible mechanism in fibronectin extensions [65].
4. If there is little lateral crosslinking between them, protofibrils might slide past each other [102, 103] and fibrin monomers within a protofibril might slide past each other. Without a restoring mechanism which pulls those units back, sliding will not be reversible, though. From studies on whole clots, it has been proposed that the protofibrils within a fibrin fiber [102, 103] and the fibrin monomers within a protofibril may slide past each other [104].
5. Several protein fibers with very large, mainly reversible extensibilities (spider silk, elastin fibers, resilin fibers) have amorphous, unstructured regions, which unravel under stress. Fibrinogen has a large unstructured domain, the 170 residue flexible connector region of the αC-domain. As the αC-domain may have a role in fibrin fiber assembly [105], stress forces may unravel this unstructured domain upon stretching. Since the length of this domain varies across species, one would anticipate that extensibility would vary among species if unraveling of the αC domains is important.
Scenarios 1 and 2 are likely reversible and could account for elastic deformations of up to ~120%; however, it cannot entirely account for the 180% elastic limit of crosslinked fibrin fibers. In this case, scenario 3 might have to be invoked. We also noted that the broken arms of the ruptured fibrin fibers contract back to nearly their original length (see Fig. 4). This implies that even in the ruptured fibers most of the deformations are reversible, which argues against significant sliding and for scenarios 1, 2, and perhaps 3.
The rupture force of one A:a interaction was recently determined with laser tweezers and found to be 130 pN [106]. The force to rupture both A:a interactions in each half-staggered fibrin dimer is thus on the order of 260 pN. This is one of the largest ruptures forces for protein–protein interactions. The unfolding force (as determined by AFM force spectroscopy) of T4 lysozyme (mainly a-helical) and the titin Ig-domain (mainly β-structure) are 64 pN [107] and 150–300 pN [60], respectively. Thus, judging by these forces, it is certainly possible that the α to β transition, which occurs at 100 pN, and perhaps the (partial) unfolding of globular domains occur before the A:a interactions break, giving credence to scenarios 2 and 3 above. Here, it should be mentioned that the rupture force depends on the pulling speed; thus some of the force values may not be directly comparable if they were obtained at different pulling speeds. Nevertheless, the dependence of the rupture force, Frupt, on the pulling speed, vpull, is weak (Frupt.α ln vpull) [108] and these experiments are often carried out a similar pulling speeds. Thus, the comparison of the rupture force values is most likely warranted.
Two years ago it was unknown how far fibrin fibers can extend and from how large a strain they can recover elastically. Due to this lack of knowledge, the prevalent assumption was that the main mechanism by which a whole clot responds to stress is by an alignment of its fibers in the stress direction. Now we know that even uncrosslinked fibrin fibers can be strained 120% and still recover elastically. Most likely, this is important in a physiological context, where fibrin fibers are subjected to the stresses of flowing blood and retracting clots [109]. Large extensibilities and elastic recovery endow fibrin fibers with toughness, so they can absorb a great deal of energy before breaking.
Acknowledgments
We thank M. C. Stahle, J. L. Moen, and O. V. Gorkun for advice and technical assistance. We acknowledge support from NIH: P41 EB002025 (RS), R01 HL31048 (STL), R41 CA103120 (MG); NSF: CMMI-0646627 (MG); Research Corporation: RI0826 (MG); American Cancer Society: IRG-93-035-6 (MG) and the American Heart Association, Mid-Atlantic Affiliate Grant-in-Aid 055527U (RRH).
References
- 1.Liu W, Jawerth LM, Sparks EA, Falvo MR, Hantgan RR, Superfine R, Lord ST, Guthold M. Fibrin fibers have extraordinary extensibility and elasticity. Science. 2006;313:634. doi: 10.1126/science.1127317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Collet JP, Shuman H, Ledger RE, Lee ST, Weisel JW. The elasticity of an individual fibrin fiber in a clot. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:9133. doi: 10.1073/pnas.0504120102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weisel JW. The electron-microscope band pattern of hman fibrin—various stains, lateral order, and carbohydrate localization. Journal of Ultrastructure and Molecular Structure Research. 1986;96:176. doi: 10.1016/0889-1605(86)90019-4. [DOI] [PubMed] [Google Scholar]
- 4.Hantgan RR, Fowler SB, Erickson HP, Hermans J. Fibrin assembly: A comparison of electron microscopic and light scattering results. Thrombosis and Haemostasis. 1980;44:119. [PubMed] [Google Scholar]
- 5.Spraggon G, Everse SJ, Doolittle RF. Crystal structures of fragment D from human fibrinogen and its cross-linked counterpart from fibrin. Nature. 1997;389:455. doi: 10.1038/38947. [DOI] [PubMed] [Google Scholar]
- 6.Brown JH, Volkmann N, Jun G, Henschen-Edman AH, Cohen C. The crystal structure of modified bovine fibrinogen. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:85. doi: 10.1073/pnas.97.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang Z, Kollman JM, Pandi L, Doolittle RF. Crystal structure of native chicken fibrinogen at 2.7 angstrom resolution. Biochemistry. 2001;40:12515. doi: 10.1021/bi011394p. [DOI] [PubMed] [Google Scholar]
- 8.Braaten JV, Jerome WG, Hantgan RR. Uncoupling fibrin from integrin receptors Hastens fibrinolysis at the platelet-fibrin interface. Blood. 1994;83:982. [PubMed] [Google Scholar]
- 9.Doolittle RF. X-ray crystallographic studies on fibrinogen and fibrin. Journal of Thrombosis and Haemostasis. 2003;1:1559. doi: 10.1046/j.1538-7836.2003.00278.x. [DOI] [PubMed] [Google Scholar]
- 10.Williams RC. Morphology of bovine fibrinogen monomers and fibrin oligomers. Journal of Molecular Biology. 1981;150:399. doi: 10.1016/0022-2836(81)90555-6. [DOI] [PubMed] [Google Scholar]
- 11.Hall CE, Slayter HS. The fibrinogen molecule: its size, shape, and mode of polymerization. The Journal of Biophysical and Biochemical Cytology. 1959;5:11. doi: 10.1083/jcb.5.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsurupa G, Tsonev L, Medved L. Structural organization of the fibrin(ogen) alpha C-domain. Biochemistry. 2002;41:6449. doi: 10.1021/bi025584r. [DOI] [PubMed] [Google Scholar]
- 13.Doolittle RF, Kollman JM. Natively unfolded regions of the vertebrate fibrinogen molecule. Proteins-Structure Function and Bioinformatics. 2006;63:391. doi: 10.1002/prot.20758. [DOI] [PubMed] [Google Scholar]
- 14.Burton RA, Tsurupa G, Medved L, Tjandra N. Identification of an ordered compact structure within the recombinant bovine fibrinogen alpha C-domain fragment by NMRT. Biochemistry. 2006;45:2257. doi: 10.1021/bi052380c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yee VC, Pratt KP, Cote HCF, LeTrong I, Chung DW, Davie EW, Stenkamp RE, Teller DC. Crystal structure of a 30 kDa C-terminal fragment from the gamma chain of human fibrinogen. Structure. 1997;5:125. doi: 10.1016/s0969-2126(97)00171-8. [DOI] [PubMed] [Google Scholar]
- 16.Everse SJ, Spraggon G, Veerapandian L, Doolittle RF. Conformational changes in fragments D and double-D from human fibrin(ogen) upon binding the peptide ligand Gly-His-Arg-Pro- amide. Biochemistry. 1999;38:2941. doi: 10.1021/bi982626w. [DOI] [PubMed] [Google Scholar]
- 17.Doolittle RF, Chen A, Pandi L. Differences in binding specificity for the homologous gamma- and beta-chain “Holes” on fibrinogen: Exclusive binding of Ala-His-Arg-Pro- amide by the beta-chain hole. Biochemistry. 2006;45:13962. doi: 10.1021/bi061219e. [DOI] [PubMed] [Google Scholar]
- 18.Ferry JD. The mechanism of polymerization of fibrin. Proceedings of the National Academy of Sciences of the Untied States of America. 1952;38:566. doi: 10.1073/pnas.38.7.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hantgan RR, Hermans J. Assembly of Fibrin—Light-scattering study. Journal of Biological Chemistry. 1979;254:11272. [PubMed] [Google Scholar]
- 20.Ryan EA, Mockros LF, Weisel JW, Lorand L. Structural origins of fibrin clot rheology. Biophysical Journal. 1999;77:2813. doi: 10.1016/S0006-3495(99)77113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carr ME, Hermans J. Size and density of fibrin fibers from turbidity. Macromolecules. 1978;11:46. doi: 10.1021/ma60061a009. [DOI] [PubMed] [Google Scholar]
- 22.Blomback B, Blomback M, Nillson IM. Coagulation studies on reptilase, an extract of the venom from Bothrops jararaca. Thrombosis et Diathesis Haemorrhagica. 1957;1:76. [PubMed] [Google Scholar]
- 23.Chen R, Doolittle RF. Gamma-gamma crosslinking sites in human and bovine fibrin. Biochemistry. 1971;10:15610. doi: 10.1021/bi00800a021. [DOI] [PubMed] [Google Scholar]
- 24.McKee PA, Mattock P, Hill RL. Subunit structure of human fibrinogen, soluble fibrin, and cross-linked insoluble fibrin. Proceedings of the National Academy of Sciences of the United States of America. 1970;66:738. doi: 10.1073/pnas.66.3.738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lorand L. Factor XIII: Structure, activation, and interactions with fibrinogen and fibrin. Annals of the New York Academy of Sciences. 2001;936:291. doi: 10.1111/j.1749-6632.2001.tb03516.x. [DOI] [PubMed] [Google Scholar]
- 26.Sobel JH, Gawinowicz MA. Identification of the alpha chain lysine donor sites involved in factor XIIIa fibrin cross-linking. The Journal of Biological Sciences. 1996;271:19288. doi: 10.1074/jbc.271.32.19288. [DOI] [PubMed] [Google Scholar]
- 27.Matsuka YV, Medved LV, Migliorini MM, Ingham KC. Factor XIIIa-catalyzed cross-linking of recombinant alpha C fragments of human fibrinogen. Biochemistry. 1996;35:5810. doi: 10.1021/bi952294k. [DOI] [PubMed] [Google Scholar]
- 28.Murthy SNP, Wilson JH, Lukas TJ, Veklich Y, Weisel JW, Lorand L. Transglutaminase-catalyzed crosslinking of the alpha and gamma constituent chains in fibrinogen. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:44. doi: 10.1073/pnas.97.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Doolittle RF. Structural basis of the fibrinogen-fibrin transformation: Contributions from X-ray crystallography. Blood Reviews. 2003;17:33. doi: 10.1016/s0268-960x(02)00060-7. [DOI] [PubMed] [Google Scholar]
- 30.Yang Z, Mochalkin I, Doolittle RF. A model of fibrin formation based on crystal structures of fibrinogen and fibrin fragments complexed with synthetic peptides. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:14156. doi: 10.1073/pnas.97.26.14156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caracciolo G, De Spirito M, Castellano AC, Pozzi D, Amiconi G, De Pascalis A, Caminiti R, Arcovito G. Protofibrils within fibrin fibres are packed together in a regular array. Thrombosis and Haemostasis. 2003;89:632. [PubMed] [Google Scholar]
- 32.Guthold M, Liu W, Stephens B, Lord ST, Hantgan RR, Erie DA, Taylor RM, Superfine R. Visualization and mechanical manipulations of individual fibrin fibers suggest that fiber cross section has fractal dimension 1.3. Biophysical Journal. 2004;87:4226. doi: 10.1529/biophysj.104.042333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brown AEX, Litvinov RI, Discher DE, Weisel JW. Forced Unfolding of Coiled-Coils in Fibrinogen by Single-Molecule AFM. Biophysical Journal. 2007;92:L30. doi: 10.1529/biophysj.106.101261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Craig CL. Spiderwebs and silk. Oxford University Press; New York: 2003. [Google Scholar]
- 35.Gosline JM, Guerette PA, Ortlepp CS, Savage KN. The mechanical design of spider silks: From fibroin sequence to mechanical function. Journal of Experimental Biology. 1999;202:3295. doi: 10.1242/jeb.202.23.3295. [DOI] [PubMed] [Google Scholar]
- 36.Denny M. Physical-properties of spiders silk and their role in design of orb-webs. Journal of Experimental Biology. 1976;65:483. [Google Scholar]
- 37.Termonia Y. Molecular modeling of spider silk elasticity. Macromolecules. 1994;27:7378. [Google Scholar]
- 38.Hinman MB, Jones JA, Lewis RV. Synthetic spider silk: A modular fiber. Trends in Biotechnology. 2000;18:374. doi: 10.1016/s0167-7799(00)01481-5. [DOI] [PubMed] [Google Scholar]
- 39.Hayashi CY, Lewis RV. Evidence from flagelliform silk cDNA for the structural basis of elasticity and modular nature of spider silks. Journal of Molecular Biology. 1998;275:773. doi: 10.1006/jmbi.1997.1478. [DOI] [PubMed] [Google Scholar]
- 40.Gosline JM. The elastic properties of rubber-like proteins and highly extensible tissues. Cambridge University Press; Cambridge: 1980. [PubMed] [Google Scholar]
- 41.Aaron BB, Gosline JM. Elastin as a random-network elastomer—a mechanical and optical analysis of single elastin fibers. Biopolymers. 1981;20:1247. [Google Scholar]
- 42.Hoeve CAJ, Flory PJ. Elastic properties of elastin. Biopolymers. 1974;13:677. doi: 10.1002/bip.1974.360130404. [DOI] [PubMed] [Google Scholar]
- 43.Urry DW, Hugel T, Seitz M, Gaub HE, Sheiba L, Dea J, Xu J, Parker T. Elastin: A representative ideal protein elastomer. Philosophical Transactions of the Royal Society of London Series B-Biological Sciences. 2002;357:169. doi: 10.1098/rstb.2001.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li B, Daggett V. Molecular basis for the extensibility of elastin. Journal of Muscle Research and Cell Motility. 2002;23:561. doi: 10.1023/a:1023474909980. [DOI] [PubMed] [Google Scholar]
- 45.Rosenbloom J, Abrams WR, Mecham R. Extracellular-matrix. 4. The Elastic Fiber. Faseb Journal. 1993;7:1208. [PubMed] [Google Scholar]
- 46.Gosline J, Lillie M, Carrington E, Guerette P, Ortlepp C, Savage K. Elastic proteins: Biological roles and mechanical properties. Philosophical Transactions of the Royal Society of London Series B-Biological Sciences. 2002;357:121. doi: 10.1098/rstb.2001.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weis-Fogh T. Molecular interpretation of the elasticity of resilin, a rubber-like proteinn. Journal of Molecular Biology. 1961;3:648. [Google Scholar]
- 48.Elvin CM, Carr AG, Huson MG, Maxwell JM, Pearson RD, Vuocolo T, Liyou NE, Wong DCC, Merritt DJ, Dixon NE. Synthesis and properties of crosslinked recombinant pro-resilin. Nature. 2005;437:999. doi: 10.1038/nature04085. [DOI] [PubMed] [Google Scholar]
- 49.Fudge DS, Gardner KH, Forsyth VT, Riekel C, Gosline JM. The mechanical properties of hydrated intermediate filaments: Insights from hagfish slime threads. Biophysical Journal. 2003;85:2015. doi: 10.1016/S0006-3495(03)74629-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fudge DS, Gosline JM. Molecular design of the alpha-keratin composite: Insights from a matrix-free model, hagfish slime threads. Proceedings of the Royal Society of London Series B-Biological Sciences. 2004;271:291. doi: 10.1098/rspb.2003.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kreplak L, Bar H, Leterrier JF, Herrmann H, Aebi U. Exploring the mechanical behavior of single intermediate filaments. Journal of Molecular Biology. 2005;354:569. doi: 10.1016/j.jmb.2005.09.092. [DOI] [PubMed] [Google Scholar]
- 52.Mucke N, Kreplak L, Kirmse R, Wedig T, Herrmann H, Aebi U, Langowski J. Assessing the flexibility of intermediate filaments by atomic force microscopy. Journal of Molecular Biology. 2004;335:1241. doi: 10.1016/j.jmb.2003.11.038. [DOI] [PubMed] [Google Scholar]
- 53.Kreplak L, Fudge D. Biomechanical properties of intermediate filaments: From tissues to single filaments and back. Bioessays. 2007;29:26. doi: 10.1002/bies.20514. [DOI] [PubMed] [Google Scholar]
- 54.Baldock C, Koster AJ, Ziese U, Rock MJ, Sherratt MJ, Kadler KE, Shuttleworth CA, Kielty CM. The supramolecular organization of fibrillin-rich microfibrils. The Journal of Cell Biology. 2001;152:1045. doi: 10.1083/jcb.152.5.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lu Y, Holmes DF, Baldock C. Evidence for the intramolecular pleating model of fibrillin microfibril organisation from single particle image analysis. Journal of Molecular Biology. 2005;349:73. doi: 10.1016/j.jmb.2005.03.066. [DOI] [PubMed] [Google Scholar]
- 56.Sherratt MJ, Baldock C, Haston JL, Holmes DF, Jones CJP, Shuttleworth CA, Wess TJ, Kielty CM. Fibrillin microfibrils are stiff reinforcing fibres in compliant tissues. Journal of Molecular Biology. 2003;332:183. doi: 10.1016/s0022-2836(03)00829-5. [DOI] [PubMed] [Google Scholar]
- 57.Megill WM, Gosline JM, Blake RW. The modulus of elasticity of fibrillin-containing elastic fibres in the mesoglea of the hydromedusa Pollyorchis penicillatus. Journal of Experimental Biology. 2005;208:3819. doi: 10.1242/jeb.01765. [DOI] [PubMed] [Google Scholar]
- 58.Wright DM, Duance VC, Wess TJ, Kielty CM, Purslow PP. The supramolecular organisation of fibrillin-rich microfibrils determines the mechanical properties of bovine zonular filaments. Journal of Experimental Biology. 1999;202:3011. doi: 10.1242/jeb.202.21.3011. [DOI] [PubMed] [Google Scholar]
- 59.Wang K, McCarter R, Wright J, Beverly J, Ramirezmitchell R. Viscoelasticity of the sarcomere matrix of skeletal-muscles—the titin myosin composite filament is a dual-stage molecular spring. Biophysical Journal. 1993;64:1161. doi: 10.1016/S0006-3495(93)81482-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rief M, Gautel M, Oesterhelt F, Fernandez JM, Gaub HE. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science. 1997;276:1109. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- 61.Hao YD, Bernstein SI, Pollack GH. Passive stiffness of drosophila IFM myofibrils: A novel, high accuracy measurement method. Journal of Muscle Research and Cell Motility. 2004;25:359. doi: 10.1007/s10974-004-0684-5. [DOI] [PubMed] [Google Scholar]
- 62.Bell E, Gosline J. Mechanical design of mussel byssus: Material yield enhances attachment strength. The Journal of Experimental Biology. 1996;199:1005. doi: 10.1242/jeb.199.4.1005. [DOI] [PubMed] [Google Scholar]
- 63.Waite JH, Qin X-X, Coyne KJ. The peculiar collagens of mussel byssus. Matrix Biology. 1998;17:93. doi: 10.1016/s0945-053x(98)90023-3. [DOI] [PubMed] [Google Scholar]
- 64.Waite JH, Vaccaro E, Sun CJ, Lucas JM. Elastomeric gradients: A hedge against stress concentration in marine holdfasts? Philosophical Transactions of the Royal Society of London Series B-Biological Sciences. 2002;357:143. doi: 10.1098/rstb.2001.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Erickson HP. Stretching fibronectin. Journal of Muscle Research and Cell Motility. 2002;23:575. doi: 10.1023/a:1023427026818. [DOI] [PubMed] [Google Scholar]
- 66.Ohashi T, Kiehart DP, Erickson HP. Dual labeling of the fibronectin matrix and actin cytoskeleton with green fluorescent protein variants. Journal of Cell Science. 2002;115:1221. doi: 10.1242/jcs.115.6.1221. [DOI] [PubMed] [Google Scholar]
- 67.Abu-Lail NI, Ohashi T, Clark RL, Erickson HP, Zauscher S. Understanding the elasticity of fibronectin fibrils: Unfolding strengths of FN-III and GFP domains measured by single molecule force spectroscopy. Matrix Biology. 2006;25:175. doi: 10.1016/j.matbio.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 68.Baneyx G, Baugh L, Vogel V. Supramolecular chemistry and self-assembly special feature: Fibronectin extension and unfolding within cell matrix fibrils controlled by cytoskeletal tension. PNAS %R 10.1073/pnas.072650799 99. 2002;5139 doi: 10.1073/pnas.072650799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pins GD, Christiansen DL, Patel R, Silver FH. Self-assembly of collagen fibers. Influence of fibrillar alignment and decorin on mechanical properties. Biophysical Journal. 1997;73:2164. doi: 10.1016/S0006-3495(97)78247-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Silver FH, Freeman JW, Seehra GP. Collagen self-assembly and the development of tendon mechanical properties. Journal of Biomechanics. 2003;36:1529. doi: 10.1016/s0021-9290(03)00135-0. [DOI] [PubMed] [Google Scholar]
- 71.Silver FH, Christiansen D, Snowhill PB, Chen Y, Landis WJ. The role of mineral in the storage of elastic energy in turkey tendons. Biomacromolecules. 2000;1:180. doi: 10.1021/bm9900139. [DOI] [PubMed] [Google Scholar]
- 72.Shadwick RE. Elastic energy-storage in tendons—mechanical differences related to function and age. Journal of Applied Physiology. 1990;68:1033. doi: 10.1152/jappl.1990.68.3.1033. [DOI] [PubMed] [Google Scholar]
- 73.Sasaki N, Odajima S. Stress-strain curve and Young's modulus of a collagen molecule as determined by the X-ray diffraction technique. Journal of Biomechanics. 1996;29:655. doi: 10.1016/0021-9290(95)00110-7. [DOI] [PubMed] [Google Scholar]
- 74.Pollock M, Shadwick RE. Relationship between body-mass and biomechanical properties of limb tendons in adult mammals. American Journal of Physiology. 1994;266:R1016. doi: 10.1152/ajpregu.1994.266.3.R1016. [DOI] [PubMed] [Google Scholar]
- 75.Kojima H, Ishijima A, Yanagida T. Direct measurement of stiffness of single actin-filaments with and without tropomyosin by in-vitro nanomanipulation. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:12962. doi: 10.1073/pnas.91.26.12962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gittes F, Mickey B, Nettleton J, Howard J. Flexural rigidity of microtubules and actin filaments measured from thermal fluctuations in shape. The Journal of Cell Biology. 1993;120:923. doi: 10.1083/jcb.120.4.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu X, Pollack GH. Mechanics of F-actin characterized with microfabricated cantilevers. Biophysical Journal. 2002;83:2705. doi: 10.1016/S0006-3495(02)75280-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kishino A, Yanagida T. Force measurements by micromanipulation of a single actin filament by glass needles. Nature. 1988;334:74. doi: 10.1038/334074a0. [DOI] [PubMed] [Google Scholar]
- 79.Kakar SK, Bettelheim FA. Birefringence of actin. Biopolymers. 1991;31:1283. doi: 10.1002/bip.360311106. [DOI] [PubMed] [Google Scholar]
- 80.Kas J, Strey H, Tang JX, Finger D, Ezzell R, Sackmann E, Janmey PA. F-actin, a model polymer for semiflexible chains in dilute, semidilute, and liquid crystalline solutions. Biophysical Journal. 1996;70:609. doi: 10.1016/S0006-3495(96)79630-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu CX, Guo HL, Xu CH, Yuan M, Li ZL, Cheng BY, Zhang DZ. Measurement of breaking force of fluorescence labelled microtubules with optical tweezers. Chinese Physics Letters. 2005;22:1278. [Google Scholar]
- 82.Felgner H, Frank R, Schliwa M. Flexural rigidity of microtubules measured with the use of optical tweezers. Journal of Cell Science. 1996;109:509. doi: 10.1242/jcs.109.2.509. [DOI] [PubMed] [Google Scholar]
- 83.Kurachi M, Hoshi M, Tashiro H. Buckling of a single microtubule by optical trapping forces—direct measurement of microtubule rigidity. Cell Motility and the Cytoskeleton. 1995;30:221. doi: 10.1002/cm.970300306. [DOI] [PubMed] [Google Scholar]
- 84.Kis A, Kasas S, Babic B, Kulik AJ, Benoit W, Briggs GAD, Schonenberger C, Catsicas S, Forro L. Nanomechanics of microtubules. Physical Review Letters. 2002;89:S. 248101. doi: 10.1103/PhysRevLett.89.248101. [DOI] [PubMed] [Google Scholar]
- 85.Wainwright SA, Biggs WD, Currey JD, Gosline JM. Mechanical design in organisms. Princeton University Press; Princeton: 1976. [Google Scholar]
- 86.Hearle JWS. A critical review of the structural mechanics of wool and hair fibres. International Journal of Biological Macromolecules. 2000;27:123. doi: 10.1016/s0141-8130(00)00116-1. [DOI] [PubMed] [Google Scholar]
- 87.Astbury WT, Street A. X-ray studies of the structure of hair, wool and related fibers. I. General Philosophical Transactions of the Royal Society of London. 1931;230A:75. [Google Scholar]
- 88.Astbury WT, Woods HJ. X ray studies of the structure of hair, wool, and related fibres. II. The molecular structure and elastic properties of hair keratin. (Series A: Mathematical and physical Sciences).Philosophical Transactions of the Royal Society of London. 1933;232:333. [Google Scholar]
- 89.Bendit EG. A quantitative X-ray diffraction study of the alpha-beta transformation in wool keratin. Textile Research Journal. 1960;30:547. [Google Scholar]
- 90.Fraser RD, MacRae TD, Rogers GE. Keratins: Their composition, structure, and biosynthesis. Charles C. Thomas; Springfield: 1972. [Google Scholar]
- 91.Cohen C. Why fibrous proteins are romantic. Journal of Structural Biology. 1998;122:3. doi: 10.1006/jsbi.1998.3975. [DOI] [PubMed] [Google Scholar]
- 92.Strelkov SV, Herrmann H, Aebi U. Molecular architecture of intermediate filaments. Bioessays. 2003;25:243. doi: 10.1002/bies.10246. [DOI] [PubMed] [Google Scholar]
- 93.Landau D, Lifschitz EM. Statistical physics part I. Pergamon Press; Oxford: 1980. Fluctuations in the curvature of long molecules. [Google Scholar]
- 94.Guzman C, Jeney S, Kreplak L, Kasas S, Kulik AJ, Aebi U, Forro L. Exploring the mechanical properties of single vimentin intermediate filaments by atomic force microscopy. Journal of Molecular Biology. 2006;360:623. doi: 10.1016/j.jmb.2006.05.030. [DOI] [PubMed] [Google Scholar]
- 95.Kielty M, Sherratt MJ, Marson A, Baldock C. Parry David A., Squire John M., editors. Fibrillin microfibrils. Fibrous proteins: Coiled-coils, collagen and elastomers. 2005;70:405. doi: 10.1016/S0065-3233(05)70012-7. [DOI] [PubMed] [Google Scholar]
- 96.Tskhovrebova L, Trinick J. Titin: Properties and family relationships. Nature Reviews Molecular Cell Biology. 2003;4:679. doi: 10.1038/nrm1198. [DOI] [PubMed] [Google Scholar]
- 97.Sivakumar P, Czirok A, Rongish BJ, Divakara VP, Wang YP, Dallas SL. New insights into extracellular matrix assembly and reorganization from dynamic imaging of extracellular matrix proteins in living osteoblasts. Journal of Cell Science. 2006;119:1350. doi: 10.1242/jcs.02830. [DOI] [PubMed] [Google Scholar]
- 98.Gao M, Craig D, Lequin O, Campbell ID, Vogel V, Schulten K. Structure and functional significance of mechanically unfolded fibronectin type III1 intermediates. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:14784. doi: 10.1073/pnas.2334390100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bailey K, Astbury WT, Rudall KW. Fibrinogen and fibrin as members of the keratin-myosin group. Nature. 1943;151:716. [Google Scholar]
- 100.Lorand L. Letters to nature. Nature. 1950;166:694. doi: 10.1038/166694a0. [DOI] [PubMed] [Google Scholar]
- 101.Howard J. Mechanics of motor proteins and the cytoskeleton. Sinauer Associates; Sunderland: 2001. [Google Scholar]
- 102.Roska FJ, Ferry JD. Studies of fibrin film. 1. Stress-relaxation and birefringence. Biopolymers. 1982;21:1811. doi: 10.1002/bip.360210910. [DOI] [PubMed] [Google Scholar]
- 103.Roska FJ, Ferry JD, Lin JS, Anderegg JW. Studies of fibrin film. 2. Small-angle X-ray-scattering. Biopolymers. 1982;21:1833. doi: 10.1002/bip.360210911. [DOI] [PubMed] [Google Scholar]
- 104.Nelb GW, Kamykowski GW, Ferry JD. Rheology of fibrin clots .5. Shear modulus, creep, and creep recovery of fine unligated clots. Biophysical Chemistry. 1981;13:15. doi: 10.1016/0301-4622(81)80020-8. [DOI] [PubMed] [Google Scholar]
- 105.Gorkun OV, Henschen-Edman AH, Ping LF, Lord ST. Analysis of A alpha 251 fibrinogen: The alpha C domain has a role in polymerization, albeit more subtle than anticipated from the analogous proteolytic fragment x. Biochemistry. 1998;37:15434. doi: 10.1021/bi981551t. [DOI] [PubMed] [Google Scholar]
- 106.Litvinov RI, Gorkun OV, Owen SF, Shuman H, Weisel JW. Polymerization of fibrin: Specificity, strength, and stability of knob-hole interactions studied at the single-molecule level. Blood. 2005;106:2944. doi: 10.1182/blood-2005-05-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yang G, Cecconi C, Baase WA, Vetter IR, Breyer WA, Haack JA, Matthews BW, Dahlquist FW, Bustamante C. Solid-state synthesis and mechanical unfolding of polymers of T4 lysozyme. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:139. doi: 10.1073/pnas.97.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Evans E, Ritchie K. Dynamic strength of molecular adhesion bonds. Biophysical Journal. 1997;72:1541. doi: 10.1016/S0006-3495(97)78802-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cohen I, Gerrard JM, White JG. Ultrastructure of clots during isometric contraction. Journal of Cell Biology. 1982;93:775. doi: 10.1083/jcb.93.3.775. [DOI] [PMC free article] [PubMed] [Google Scholar]