Abstract
Cell-cell interactions and cross-talk between signaling pathways specify Caenorhabditis elegans vulval precursor cells (VPCs) to adopt a spatial pattern: a central “1°” VPC, in which epidermal growth factor receptor (EGFR)–mitogen-activated protein kinase (MAPK) activity is high and LIN-12/Notch activity is low, flanked by two “2°” VPCs, in which LIN-12/Notch activity is high and EGFR-MAPK activity is low. Here, we identify a microRNA gene, mir-61, as a direct transcriptional target of LIN-12 and show that expression of mir-61 promotes the 2° fate. We also identify vav-1, the ortholog of the Vav oncogene, as a target of mir-61, and show that down-regulation of VAV-1 promotes lin-12 activity in specifying the 2° fate. Our results suggest that lin-12, mir-61, and vav-1 form a feedback loop that helps maximize lin-12 activity in the presumptive 2° VPCs.
Six multipotential VPCs, numbered P3.p to P8.p, adopt an invariant pattern of fates termed 3°-3°-2°-1°-2°-3° (Fig. 1A). Two signaling events specify this pattern: “inductive” signaling, mediated by an EGFR-Ras-MAPK pathway, and “lateral” signaling, mediated by LIN-12 (Fig. 1A) (1). The inductive signal from the gonad activates an EGFR-Ras-MAPK cascade in a graded fashion in the underlying VPCs, P5.p, P6.p, and P7.p. The centralmost VPC, P6.p, has the highest level of EGFR-Ras-MAPK activation and becomes the presumptive 1° VPC; it produces the lateral signal, which activates LIN-12 in P5.p and P7.p. When LIN-12 is activated, proteolysis releases its intracellular domain, which translocates to the nucleus and forms a transcriptional activation complex with the DNA binding protein LAG-1 (2). Transcriptional targets of LIN-12 in P5.p and P7.p can be identified by the presence of LAG-1 binding sites (LBSs) in their 5′ flanking regions and include genes that encode negative regulators of EGFR-Ras-MAPK activity in P5.p and P7.p, which inhibit the expression of 1° fate features in these cells (3).
Fig. 1.
mir-61 is a direct target of LIN-12 in presumptive 2° VPCs. (A) An inductive signal from the anchor cell (AC) of the gonad activates EGFR-MAPK signaling primarily in P6.p, and a lateral signal from P6.p activates LIN-12 in P5.p and P7.p. The descendants of the 1° and 2° VPCs form the vulva, and the progeny of 3° VPCs fuse with the hypodermal syncytium. We used the 1° fate marker arIs92[egl-17p::cfp-lacZ] (3), the 2° fate marker nIs106[lin-11p::gfp] (21), and the 3° fate marker arIs101[K09H11.1p::yfp] (22). (B) mir-61 is expressed in P5.p and P7.p. The mir-61 promoter contains two LBSs that are conserved in the C. briggsae ortholog of mir-61 (5). A reporter containing 1 kb upstream of mir-61 fused to YFP is expressed in P5.p and P7.p (22) and their daughters (shown here). Prominent expression in cells of the gonad in which LIN-12 is active is also seen. (C) LBSs are required for mir-61 expression in P5.p and P7.p. Two LBSs (YRTGRGAA) (3, 23) that are conserved in C. briggsae were mutated to YRAGRGAA; a third nonconserved sequence, RTGGGAA, was also mutated to RAGGGAA. In three individual lines analyzed, expression of YFP in P5.p and P7.p disappeared. Expression in cells in which lin-12 is not known to play a role in cell fate specification was normal, but gonadal expression was also abolished.
Short regulatory microRNAs (miRNAs), first identified in C. elegans (4), mediate posttranscriptional down-regulation of target genes. The profound and pervasive roles that miRNAs play as critical regulators of developmental gene expression are only now becoming fully appreciated. We obtained an indication that a miRNA may be involved in lateral signaling after observing a lateral signaling defect when a miRNA-processing gene was depleted (5). We then computationally identified mir-61 as a potential miRNA that is transcribed when the lateral signal from P6.p activates LIN-12 in P5.p and P7.p (5). A mir-61 transcriptional reporter is specifically expressed in P5.p and P7.p and their daughters (Fig. 1B) (6), consistent with a function for mir-61 as a direct target of LIN-12 during lateral signaling. This inference was confirmed by mutating the LBSs in the mir-61 promoter and finding that expression in P5.p and P7.p was lost (Fig. 1C) (7).
Ectopic expression has been a successful approach to elucidating the role of miRNAs for which null alleles are not available and obviates potential problems that may be posed by functional redundancy (8-10). Expression of mir-61 ectopically in P6.p, the presumptive 1° VPC, causes expression of the canonical 2° fate marker lin-11::gfp, whereas expression of an unrelated miRNA does not (Fig. 2). These observations suggest that mir-61 activity promotes the 2° fate.
Fig. 2.
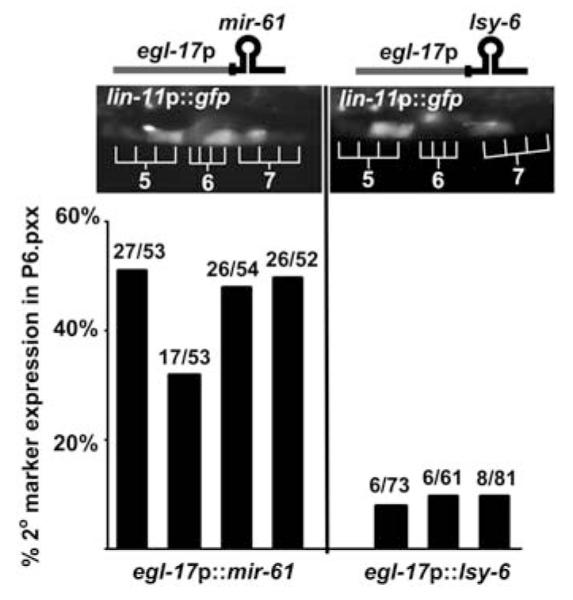
Ectopic expression of mir-61 in P6.p confers 2° fate characteristics. Expression of the 2° fate marker nIs106[lin-11p::gfp] (see Fig. 1A) was assessed when egl-17p (3, 24) was used for ectopic expression of mir-61 or the unrelated miRNA lsy-6 (25) in P6.p and its descendants. Ectopic lin11p::gfp expression in P6.p descendants was observed only when mir-61 was expressed. Each bar represents an independent transgenic line. The number of individuals expressing the 2° marker out of the total is given above each bar.
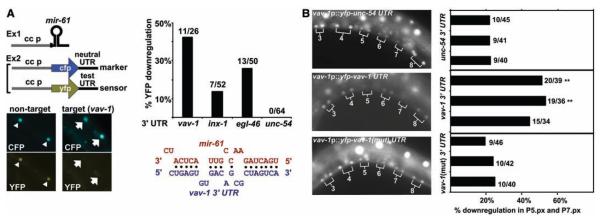
miRNAs bind to sites in 3′ untranslated regions (UTRs) of target mRNAs and inhibit their translation (8). Thus, we hypothesized that mir-61 is expressed in presumptive 2° VPCs in order to down-regulate potential target gene products that would interfere with specification of the 2° fate. We identified potential target genes of mir-61 computationally (5), requiring that 3′ UTRs have at least seven bases of perfect complementarity to the 5′ end of mir-61 and that the binding sites be conserved in C. briggsae orthologs. This analysis yielded three candidates, vav-1, inx-1, and egl-46 (Fig. 3A) (5).
Fig. 3.
vav-1 is a target of mir-61. (A) Rapid assay to validate predicted miRNA targets. Components are expressed in coelomocytes to determine whether a miRNA causes down-regulation of YFP in a sensor construct containing a test UTR without affecting CFP in a marker construct with the neutral unc-54 3′ UTR (26). Here, an array carrying unc-122p::mir-61 was combined with an array carrying the unc-122p::cfp::unc-54 3 ′UTR marker and hlh-8p::yfp::vav-1 3 ′UTR sensor (right) or an array carrying hlh-8p::yfp::unc-54 3 ′UTR in lieu of the sensor (left) (6). The presence of the mir-61–expressing array does not affect expression of YFP produced from a sensor construct that contains a nontarget UTR (in this case, the unc-54 UTR) (triangles). In contrast, YFP expressed from a sensor construct that contains a target UTR (shown here, the vav-1 UTR) is not seen, whereas the CFP marker shows that the array is present and expressed (arrows). The graphs indicate the percentage of worms that show the down-regulation of YFP signal. The alignment shows predicted configuration of mir-61 (in red) binding to its target site in the 3′ UTR of vav-1 (in blue). (B) VAV-1 is posttranscriptionally down-regulated in P5.p and P7.p. The 8.4-kb upstream region of vav-1 drives expression of YFP in all VPCs and their daughters. When the unc-54 3′ UTR is replaced by vav-1 3′ UTR, down-regulation of YFP expression in P5.px and P7.px is evident. Mutation of the mir-61–complementary sequence in the vav-1 3′UTR from TAGTCA to GTCGAC causes persistent YFP expression. In the graph, each bar represents an individual line. **P < 0.01 by Fisher’s exact test. We minimized the potential lack of expression in P5.px or P7.px due to genetic mosaicism by including data only for animals in which expression could be seen in P3.px, P4.px, and P8.px.
To assess candidate genes in vivo, we developed a simple heterologous assay to circumvent potential detection problems due to weak or transient mir-61 expression in the VPCs. This assay may be used to test whether any miRNA can target the 3′ UTR of any candidate target gene. We expressed mir-61 in coelomocytes, distinctive cells for which strong promoters are available (11, 12). On a second transgene, we expressed two reporters in coelomocytes: one a yellow fluorescent protein (YFP) reporter with the 3′ UTR of the putative target gene and the other a cyan fluorescent protein (CFP) reporter with the unc-54 3′ UTR, which does not contain any mir-61–binding sites (13). If a candidate is a bona fide target, then we would expect to see coelomocytes displaying CFP expression and down-regulation of YFP expression. Using this assay, we obtained evidence that mir-61 can regulate the expression of the three candidate genes identified by using the criteria described above (Fig. 3A) (5).
mir-61 is also expressed in cells other than the VPCs, and lin-12 activity specifies many other cell fate decisions (14), so even bona fide targets of mir-61 may not be relevant to lateral signaling. The desired target genes should be transcribed in the VPCs but posttranscriptionally down-regulated by way of their 3′ UTRs in P5.p and P7.p and their daughters. We fused the 5′ upstream sequence of vav-1, inx-1, or egl-46 to yfp::unc-54 3′UTR and found that only vav-1 is expressed in the VPCs and their daughters (Fig. 3B). When we replaced the unc-54 3′UTR with the vav-1 3′UTR, creating a sensor construct, vav-1 expression was lost in P5.p and P7.p in a significant proportion of hermaphrodites; this loss depends on an intact mir-61 target site (Fig. 3B). These observations indicate that vav-1 is posttranscriptionally regulated in P5.p and P7.p, consistent with regulation by endogenous mir-61, and suggest that VAV-1 may be down-regulated in presumptive 2° VPCs to promote lin-12 activity.
VAV-1 is an ortholog of the Vav oncoprotein, which has guanine nucleotide exchange factor (GEF) activity and additional domains that mediate interactions with other proteins (15); thus, there are many different potential mechanisms by which Vav proteins may modulate the activity of signaling pathways in presumptive 2° VPCs. In mammalian cells, in some contexts, Vav appears to be a positive regulator of MAPK signaling, but in others, it has no effect (15). Loss of vav-1 activity does not prevent vulval induction, which indicates that vav-1 is not required for EGFR-MAPK signaling in the cellular context of VPCs (16) and that down-regulation of VAV-1 by mir-61 may not specifically attenuate EGFR-MAPK signaling in presumptive 2° VPCs in response to the low level of inductive signal (3).
Alternatively, VAV-1 may be a negative regulator of LIN-12. If so, then down-regulation of VAV-1 by mir-61 would increase lin-12 activity in P5.p and P7.p, independent of any input from the inductive signaling pathway. We therefore looked at whether loss of vav-1 activity enhances lin-12 activity under conditions where inductive signaling does not occur. The alleles lin-12(n379) and lin-12(n676) result in mild constitutive activity: Hermaphrodites lack an anchor cell, and all VPCs generally adopt the 3° fate, as there is no inductive signal to specify a 1° fate and insufficient constitutive lin-12 activity to promote anchor cell–independent 2° fates (14). In such backgrounds, loss of some negative regulators of LIN-12 increases LIN-12 stability or activity, which causes all six VPCs to adopt the 2° fate and to generate multiple pseudovulvae (17). Negative regulators that behave in this manner include SEL-10/Fbw7, which promotes ubiquitin-mediated turnover of LIN-12/Notch (18), and SEL-9, which functions in secretory protein quality control (19). We found that vav-1(RNAi) significantly enhances the constitutive activity of lin-12(n379) and lin-12(n676), which increases the number of hermaphrodites with multiple pseudovulvae (Fig. 4A). These results suggest that vav-1 is a negative regulator of lin-12 activity.
Fig. 4.
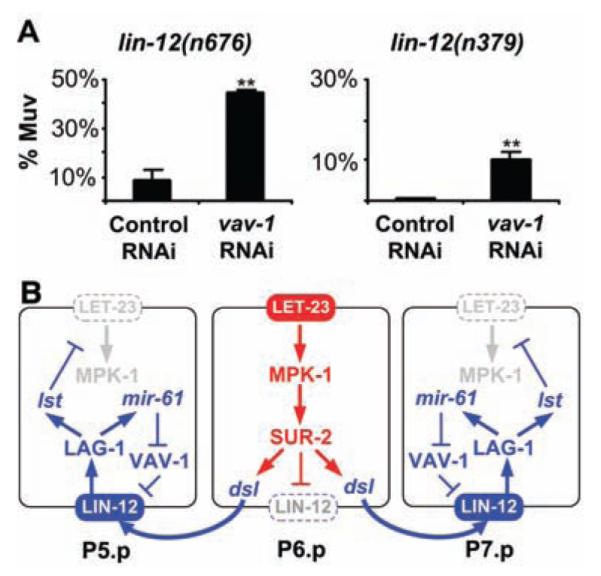
vav-1 is a negative regulator of lin-12 activity in the VPCs. (A) vav-1(RNAi) enhances lin-12 activity in VPCs. The average percentage of worms with a Multivulva (Muv) phenotype (three or more pseudovulvae) on three independent plates is shown. For lin-12(n676); vav-1(RNAi), the number of Muv hermaphrodites out of the total per plate was 50 out of 109, 49 out of 112, and 54 out of 125, with the control lin-12(n676); gfp(RNAi) values of 8 out of 157, 17 out of 168, and 18 out of 152. For lin-12(n379); vav-1(RNAi), the numbers were 20 out of 187, 25 out of 208, and 13 out of 167, with lin-12(n379); gfp(RNAi) values of 1 out of 204, 1 out of 228, and 2 out of 225. Error bar indicates SD. **P < 0.01 by Student’s t test. (B) mir-61, vav-1, and the circuitry underlying specification of the 2° VPC fate. Activation of the EGFR-MAPK pathway in P6.p has two consequences for lateral signaling: transcription of the three Delta/Serrate/LAG-2 (DSL) ligands that constitute the lateral signal, which activates LIN-12/Notch in P5.p and P7.p (27), and internalization of LIN-12, which is necessary for lateral signal activity (28). Activation of LIN-12 in P5.p and P7.p activates a set of lst genes that counteract the EGFR-MAPK pathway (3), and mir-61, which posttranscriptionally down-regulates VAV-1 to promote lin-12 activity.
We have shown that mir-61 is a direct transcriptional target of the LIN-12/Notch pathway and that vav-1 is a target of mir-61 in the VPCs. We have also shown that ectopic mir-61 promotes the 2° fate and that VAV-1 is a negative regulator of lin-12 activity. We propose that activation of mir-61 transcription by LIN-12 and the consequent down-regulation of VAV-1 constitute a positive-feedback loop that promotes LIN-12 activity in presumptive 2° VPCs (Fig. 4B).
Although there are many possible molecular mechanisms that may underlie this positive-feedback loop, it is notable that Vav has many domains that could couple it to receptors or to mediators of endocytosis (15). Indeed, Vav proteins have recently been shown to affect endocytosis of the activated Ephrin receptor (20). Perhaps down-regulation of VAV-1 in presumptive 2° VPCs decreases the rate of internalization, promotes endocytic recycling of LIN-12 or required proteases, or alters another aspect of trafficking that favors ectodomain shedding or transmembrane cleavage.
Supplementary Material
Fig. S1. Lateral signaling defect in alg-1(RNAi) hermaphrodites.
Fig. S2. 5′ flanking regions of C. elegans and C. briggsae mir-61. The 5′ flanking regions of mir-61 in both species are predicted to be about 1 kb; the 5′ flanking region is defined as the expanse from the next predicted gene to the start of the mir-61precursor. The positions of conserved LBSs are shown with respect to the start of the coding region for the mir-61 precursor, symbolized by a hairpin.
Fig. S3. Alignment of C. elegans and C. briggsae mir-61.
Fig. S4. mir-61 targets. All were verified using the assay described in Fig. 3; however, only vav-1 appears to be expressed in VPCs.
Fig. S5. 3′ UTRs that do not permit downregulation by mir-61 in the coelomocyte assay. The unc-54 3′UTR also does not permit downregulation (see text and Fig. 3A).
Acknowledgments
We are grateful to X. Zhou for performing microinjections; and to O. Hobert, R. Mann, and D. Shaye for discussion and insightful comments on the manuscript. This work was supported by NIH grant CA095389 (to I.G.). I.G. is an investigator with the Howard Hughes Medical Institute.
Footnotes
Supporting Online Material www.sciencemag.org/cgi/content/full/1119481/DC1 Materials and Methods SOM Text Figs. S1 to S5 References and Notes
References and Notes
- 1.Sternberg PW. WormBook. 2005 doi: 10.1895/wormbook.1.6.1. www.wormbook.org. [DOI] [PMC free article] [PubMed]
- 2.Greenwald I. WormBook. 2005 www.wormbook.org.
- 3.Yoo AS, Bais C, Greenwald I. Science. 2004;303:663. doi: 10.1126/science.1091639. [DOI] [PubMed] [Google Scholar]
- 4.Lee RC, Feinbaum RL, Ambros V. Cell. 1993;75:843. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 5.Supporting Online Material is available on Science Online.
- 6.All transcriptional reporters were made by polymerase chain reaction (PCR) fusion, from the predicted start to the next predicted upstream gene, and extrachromosomal arrays were marked with pha-1(+). Expression of mir-61 transcription was studied by using the arIs107 integrant, which displayed the same pattern as extrachromosomal arrays.
- 7.mir-61 is also expressed in cells of the somatic gonad in which LIN-12 is active, and this expression is also lost when the LBSs are mutated (Fig. 1, B and C). In contrast, expression in other tissues where we have no evidence that lin-12 activity is functionally relevant, such as intestinal cells, is unaffected. These observations suggest that the loss of expression in P5.p and P7.p reflects lack of response to lin-12 and not loss of a general enhancer.
- 8.Ambros V. Nature. 2004;431:350. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 9.Chen CZ, Li L, Lodish HF, Bartel DP. Science. 2004;303:83. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 10.Chang S, Johnston RJ, Jr., Frokjaer-Jensen C, Lockery S, Hobert O. Nature. 2004;430:785. doi: 10.1038/nature02752. [DOI] [PubMed] [Google Scholar]
- 11.Harfe BD, et al. Genes Dev. 1998;12:2623. doi: 10.1101/gad.12.16.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loria PM, Hodgkin J, Hobert O. J. Neurosci. 2004;24:2191. doi: 10.1523/JNEUROSCI.5462-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.We used two separate arrays because of possible promoter interference that we have experienced when using the same promoter to drive two different gene products in the same array. For the assessment of many targets, the assay could be improved by integrating the transgene expressing the miRNA.
- 14.Greenwald IS, Sternberg PW, Horvitz HR. Cell. 1983;34:435. doi: 10.1016/0092-8674(83)90377-x. [DOI] [PubMed] [Google Scholar]
- 15.Tybulewicz VL. Curr. Opin. Immunol. 2005;17:267. doi: 10.1016/j.coi.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 16.Norman KR, et al. Cell. 2005;123:119. doi: 10.1016/j.cell.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 17.Sundaram M, Greenwald I. Genetics. 1993;135:765. doi: 10.1093/genetics/135.3.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hubbard EJ, Wu G, Kitajewski J, Greenwald I. Genes Dev. 1997;11:3182. doi: 10.1101/gad.11.23.3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wen C, Greenwald I. J. Cell Biol. 1999;145:1165. doi: 10.1083/jcb.145.6.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cowan CW, et al. Neuron. 2005;46:205. doi: 10.1016/j.neuron.2005.03.019. [DOI] [PubMed] [Google Scholar]
- 21.Reddien PW, Cameron S, Horvitz HR. Nature. 2001;412:198. doi: 10.1038/35084096. [DOI] [PubMed] [Google Scholar]
- 22.Yoo AS. unpublished observations.
- 23.Rebeiz M, Reeves NL, Posakony JW. Proc. Natl. Acad. Sci. U.S.A. 2002;99:9888. doi: 10.1073/pnas.152320899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burdine RD, Branda CS, Stern MJ. Development. 1998;125:1083. doi: 10.1242/dev.125.6.1083. [DOI] [PubMed] [Google Scholar]
- 25.Johnston RJ, Hobert O. Nature. 2003;426:845. doi: 10.1038/nature02255. [DOI] [PubMed] [Google Scholar]
- 26.From pPD95.67, gift of A. Fire (Stanford University).
- 27.Chen N, Greenwald I. Dev. Cell. 2004;6:183. doi: 10.1016/s1534-5807(04)00021-8. [DOI] [PubMed] [Google Scholar]
- 28.Shaye DD, Greenwald I. Development. 2005;132:5081. doi: 10.1242/dev.02076. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Lateral signaling defect in alg-1(RNAi) hermaphrodites.
Fig. S2. 5′ flanking regions of C. elegans and C. briggsae mir-61. The 5′ flanking regions of mir-61 in both species are predicted to be about 1 kb; the 5′ flanking region is defined as the expanse from the next predicted gene to the start of the mir-61precursor. The positions of conserved LBSs are shown with respect to the start of the coding region for the mir-61 precursor, symbolized by a hairpin.
Fig. S3. Alignment of C. elegans and C. briggsae mir-61.
Fig. S4. mir-61 targets. All were verified using the assay described in Fig. 3; however, only vav-1 appears to be expressed in VPCs.
Fig. S5. 3′ UTRs that do not permit downregulation by mir-61 in the coelomocyte assay. The unc-54 3′UTR also does not permit downregulation (see text and Fig. 3A).