Abstract
One of the biggest challenges in neuroscience is illuminating the architecture of developmental brain disorders, which include structural malformations of the brain and nerves, intellectual disability, epilepsy, as well as some psychiatric conditions like autism and potentially schizophrenia. Ongoing gene identification reveals a great diversity of genetic causes underlying abnormal brain development, illuminating new biochemical pathways often not suspected based on genetic studies in other organisms. Our greater understanding of genetic disease also shows the complexity of “allelic diversity”, in which distinct mutations in a given gene can cause a wide range of distinct diseases or other phenotypes. These diverse alleles not only provide a platform for discovery of critical protein-protein interactions in a genetic fashion, but also illuminate the likely genetic architecture of as yet poorly characterized neurological disorders.
Human genetic diseases represent “saturation mutagenesis”
The accelerating pace of human disease gene identification continues to amaze and impress. The notion that the 6-7 billion humans on our planet can be conceptualized as a “saturation mutagenesis” experiment of Nature, in which every gene in the genome has been mutated at least once and can potentially be scored for phenotypes, is not new (Brenner, 2003; Walsh, 1999). However, it has been brought into clearer focus by the accelerating pace of disease gene identification. In an experimental situation in which mutations are deliberately created in animal models (e.g, worms, flies, or zebrafish), a conventional Poisson statistic provides a rough guide that, when 3 independent alleles of an average-size gene have been observed, 95% of other genes in the genome have been mutated; when 5 independent mutant alleles are observed, 99% of other genes have been mutated. Extrapolating these animal model experiments to humans, where there are many well-characterized diseases with dozens or hundreds of independent alleles identifiable as causing a similar disease phenotype (Figure 1), instructs us that humans far exceed the criteria needed to be certain that all genes in the genome have been mutated repeatedly.
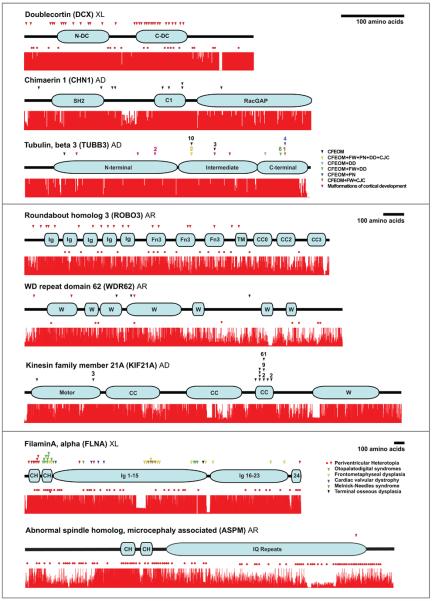
Figure 1.
Schematics of each protein are illustrated with the name of the protein and its abbreviation to the upper left followed by mode of inheritance of disease (XL = X-linked, AD = autosomal dominant, AR = autosomal recessive). Proteins are ordered according to length with the shortest on the top and the longest on the bottom, and are presented in three differently scaled groups, each based on the amino acid scale bar found to the upper right of the group. ‘Stop’ mutations (including nonsense, frameshift, intragenic deletions, or splicing alleles that cause frameshifts) are represented by red dots positioned below the protein corresponding to the location of the mutation. Missense mutations are represented by arrowheads positioned above the protein corresponding to the location of the mutation. Each dot and arrowhead represents a unique mutation. Additional arrowheads aligned directly above (or red dots aligned directly below) one another indicate distinct mutations have been reported at the same amino acid residue (i.e. in KIF21A, the mutation altering an amino acid residue in the distal motor domain has been identified in three unrelated patients). Some arrowheads or red dots are unevenly stacked; they represent unique mutations that, for reasons of scale, were not able to be aligned horizontally along the protein. When multiple unrelated patients have been reported with the identical mutation, the number of times that mutation has been independently observed appears above the arrow (or below the red dot). Within each protein, each arrowhead color corresponds to a distinct disease phenotype, and red arrowheads mean that the missense mutation results in the same phenotype as the Stop mutation(s). When more than one phenotype results from mutations in a given protein, the arrowheads/circles are color-coded with a corresponding key to the right of the protein. For example, in the intermediate domain of TUBB3, 10 unrelated patients with the same phenotype have been reported to harbor the identical missense mutation, while 2 other unrelated patients share a distinct phenotype and a different missense mutation that alters the same amino acid but to a different residue. Conservation tracts were made by aligning the most homologous amino acid sequences available for each gene for human, chimpanzee, dog, mouse, chicken, and zebrafish using the ClustalX software package with default settings. Accession numbers of the sequences used are listed below. Complete sequence data for FLNA, KIF21A and ROBO3 was not available for chimpanzee and so rhesus monkey FLNA sequence, and orangutan KIF21a and ROBO3 sequences were substituted. Chicken ROBO3 and WDR62 sequences as well as zebrafish DCX sequence were not available and no substitutions for these were made. Several sequences for CHN1 and ROBO3 were predicted using the UCSC genome browser multiple sequence alignment tool based on the comparative genomics conservation tracts (uc# noted below) and the dog sequence for KIF21a was predicted from genome sequence with N-SCAN.
Protein motifs, key abbreviations and sequence accession numbers for conservation tracts
Doublecortin (DCX) Protein motifs: N-terminal and C-terminal doublecortin domains; Accession numbers: NP_000546.2, XP_529107.2, XP_853182.1, NP_034155.2, NP_989666.1.
Chimaerin 1 (CHN1) Protein motifs: N-terminal Src homology 2 (SH2), phorbol esters/diacylglycerol binding (C1), and Rac GTPase activating (RacGAP) domains; Accession numbers: uc002uji.1_hg18, uc002uji.1_panTro2, uc002uji.1_canFam2, uc002uji.1_mm9, NP_001012970.1, NP_998165.1.
Tubulin, beta 3 (TUBB3) Key abbreviations: CFEOM (congenital fibrosis of the extraocular muscles), FW (facial weakness), PN (peripheral neuropathy), DD (intellectual and social developmental delay), and CJC (congenital joint contractures); Accession numbers: NP_006077.2, NP_001038974.1, XP_860049.1, NP_075768.1, NP_998655.1, NP_001026769.1.
Roundabout homolog 3 (ROBO3) Protein motifs: Immunoglobulin-like (Ig), fibronectin-like (Fn3), transmembrane, and conserved cytoplasmic (CC) domains; Accession numbers: ROBO3 NP_071765.2, uc001qbc.1_ponAbe2, XP_546425.2, XP_001476890.1, NP_571557.1.
WD repeat domain 62 (WDR62) Protein motifs: WD40 (W) domains; Accession numbers: NP_001077430.1, XP_512609.2, XP_853669.1, XP_896469.1, XP_699579.2.
Kinesin family member 21A (KIF21A) Protein motifs: Motor (M), coiled-coil (CC), and WD40 (W) domains; Accession numbers: NP_060111.2, XP_002823145.1, N-SCAN (chr27.18.003.a), NP_060111.2, XP_001920258.1, XP_415936.2.
FilaminA, alpha (FLNA) Protein motifs: Calpain-homology (CH) and immunoglobulin-like (Ig) domains; Accession numbers: NP_001104026.1, XP_001091073.1, XP_867483.1, NP_034357.2, NP_989905.1, XP_001922206.1.
Abnormal spindle homolog, microcephaly associated (ASPM) Protein motifs: Calpainhomology (CH) and isoleucine-glutamine repeat (IQ) domains; Accession numbers: NP_060606.3, NP_001008994.1, XP_537130.2, NP_033921.3, XP_001923712.1, XP_422197.2.
Many diseases represent “special” mutations, that do not merely compromise function, but might create a new, abnormal biochemical function or constitutively activate the protein, or might create a “dominant negative” allele. As we will review, these unusual mutations are often recurrent, meaning that the identical mutation has occurred multiple times in different unrelated patients around the world. Since the probability of mutation at one codon is similar to the probability of mutation at another codon (save for the greater tendency for the sequence “CG” to be mutated than other dinucleotide combinations), this implies that the other codons in the gene are mutated in different patients somewhere in the world, although potentially resulting in a different “special” mutation and a different phenotype. Moreover, for the most densely studied genes, such as globin genes, it appears that virtually every codon in the gene has had a corresponding disease-causing mutation observed, suggesting that almost every codon in the genome is present in a mutated form in someone, somewhere. Therefore, humans represent the richest sort of “saturation mutagenesis” experiment, one that we are unlikely to observe in any animal model anytime soon.
Here we will review, using examples from human developmental brain diseases, how the density and diversity of mutation -- that we are only beginning to decode -- can eventually be harnessed to go beyond merely associating a gene with a disease. The “unusual” mutations create linkages from protein to protein, by identifying special protein interaction interfaces; or can represent something like “conditional mutations” of mice, in which the expression of a gene may be removed only from a particular place or domain. This range of mutation, a delicious biological tool for dissecting mechanisms for the neuroscientist, is also an uncomfortable fact of life for the human geneticist: this mutational diversity, or “heterogeneity,” helps explain the as yet unexplored allelic diversity of human neurological disorders, and the inherent difficulty in identifying underlying causes of many neurogenetic diseases.
Loss of function mutations implicate essential gene functions
DCX illustrates perhaps a typical mutational spectrum for a gene that basically causes one disease, albeit in milder or more severe forms. Null mutations in DCX cause a profound defect in neuronal migration in males, called lissencephaly, in which the brain is smooth rather than folded, reflecting severely abnormal neuronal organization because of defects in migration of essentially all cortical neurons (des Portes et al., 1998; Gleeson et al., 1998). Since DCX is an X-linked gene, females show a milder condition in the heterozygous state; they have a relatively normal appearing cortex, with a second, “double” cortex in the subcortical white matter. Neurons appear to migrate either normally or abnormally depending upon which X-chromosome is active. Cells that transcriptionally inactivate the X-chromosome carrying the X-linked mutation seem to have normal migration because they have normal levels of DCX activity, although normal cells can occasionally be obstructed by Dcx deficient, arrested neurons (Bai et al., 2003). Cells that inactivate the normal DCX gene arrest in the subcortical white matter because they lack DCX activity. Most patients show the “full-blown” brain malformation, although mild alleles that only partially impair DCX function can cause seizures with a normal-looking brain, or even cause isolated mental retardation with a normal appearing brain (Guerrini and Marini, 2006; Guerrini et al., 2003).
Mutations in DCX that block the formation of a full-length protein include nonsense alleles, frameshift alleles, intragenic deletions, and splicing alterations that result in frameshift alleles. We will refer to these protein-truncating mutations collectively as “Stop” mutations. Alternatively, missense mutations that alter individual amino acids might preserve a full-length protein but render it dysfunctional. Of the >60 known mutations of DCX (many of which are summarized schematically in Figure 1), the missense alleles slightly predominate over the Stop alleles. Stop alleles are distributed quite evenly over the length of the protein, in no obvious pattern, and wherever the protein is truncated, the result is an equally severe phenotype. This suggests that full-length DCX is required for normal function, and that any mutation that truncates the protein blocks its normal function.
In contrast to the even distribution of Stop mutations in DCX, missense mutations are notably clustered in two repeated domains, called “doublecortin” domains (N-DC and C-DC in Figure 1). There are a few missense mutations that affect the N-terminus of the protein or the short linker separating the doublecortin domains, but strikingly there are no missense mutations over the C-terminal 100 amino acids of the protein. The importance of the doublecortin domains for binding of DCX protein to microtubules was shown on the basis that these missense mutations block the ability of DCX to bind microtubules or tubulin (Gleeson et al., 1999; Sapir et al., 2000; Taylor et al., 2000). The clustering of missense mutations in the doublecortin domains strongly suggests that the 3-dimensional structure of these domains is essential for the normal function of DCX, which is to bind and organize microtubules in migrating neurons (Kim et al., 2003; Reiner et al., 2006). On the other hand, the function of the C-terminus is less clear. The Stop mutations suggest that the C-terminus is essential, but the absence of C-terminus missense mutations suggests that the particular amino acid sequence of this region may be less important, or perhaps that missense mutations in the C-terminus might cause a different disease.
Like many X-linked mutations, autosomal mutations often act in a recessive fashion, except that for autosomal recessive genes both copies of the gene must be mutated in order to cause disease, with mutation of one allele being asymptomatic, and disease generally caused by an absence of functional protein. This pattern is nicely illustrated by homozygous or compound heterozygous mutations in the ROBO3 gene (encoding the axon guidance receptor ROBO3/Rig-1), which cause the disorder ‘horizontal gaze palsy with progressive scoliosis’ (HGPPS) (Chan et al., 2006; Jen et al., 2004). HGPPS is characterized by absent horizontal gaze from birth (an inability to move the eyes to the left or right) followed by development of scoliosis, or curvature of the spine, within the first decade of life. In addition, axons in the descending corticospinal and ascending sensory tracts fail to cross the midline in the medulla, and thus do not decussate to the opposite side as they normally do in unaffected individuals (Jen et al., 2004). This is similar to the Robo3−/− mouse, in which both axons and neurons in the developing hindbrain and spinal cord fail to decussate across the midline (Marillat et al., 2004; Sabatier et al., 2004).
Similar to DCX, human mutations in ROBO3 represent a roughly equal mixture of Stop mutations and missense mutations (Figure 1). The clinical features of patients with missense mutations cannot be distinguished from those with Stop mutations, suggesting that all mutations eliminate the function of the ROBO3 protein. Also similar to DCX, the missense changes predominate at the N-terminus of the protein, where they highlight extracellular domains of this transmembrane receptor that are essential for protein-protein interactions. Only one mutation, a Stop, has been identified in the C-terminus of ROBO3 (again implying that the full-length protein is essential for normal function). Once again, the notable absence of missense mutations at the C-terminus of the protein suggests that the primary amino acid sequence of the C-terminal intracellular portion may be less important, or that mutations in this portion of the protein cause a different disease.
A pattern of mutation distinct from ROBO3 is seen in another autosomal recessive disorder of brain development, microcephaly (i.e. a very small brain). The gene most commonly mutated in human microcephaly is ASPM (abnormal spindle microcephaly) (Bond et al., 2002; Bond et al., 2003; Nicholas et al., 2009). ASPM encodes a huge protein with many tandem repeats of 20-24 amino acids that begin with isoleucine-glutamine, hence named “IQ” repeats. The Aspm protein localizes to the mitotic spindle and appears essential for normal mitotic spindle function (Fish et al., 2006; Kouprina et al., 2005). In striking contrast to ROBO3 and DCX mutations however, all but one of the >90 known mutations in ASPM associated with microcephaly represent Stop mutations of one kind or another (Figure 1) (Bond et al., 2002; Kousar et al.; Nicholas et al., 2009). Once again, these Stop mutations occur virtually anywhere along the coding region of the gene, with no apparent relationship between the location of the mutation and the severity of the disease. Only a single missense mutation has ever been reported in ASPM, and it alters one of the highly conserved “Q” codons in one of the IQ repeats (Gul et al., 2006). A similar pattern of mutation is seen in several other microcephaly genes that comprise other components of the mitotic spindle: CDK5RAP2, CENPJ (Bond et al., 2005), and STIL (Kumar et al., 2009) in which most or all known mutations are Stop mutations.
A potential explanation for the apparent absence of missense mutations in ASPM is provided by recent analysis of another microcephaly gene, WDR62. This gene has been identified as causative for several distinct but related conditions: Stop mutations cause severe phenotypes, in which severe microcephaly is associated with abnormal gyral pattern of the brain, clefts in the brain (schizencephaly), and abnormal neuronal migration, suggesting markedly abnormal histological organization of the brain (Bilguvar et al., 2010) (Nicholas et al., 2010; Yu et al., 2010). In contrast, some missense mutations in the same gene are responsible for a milder form of microcephaly, with less severe reduction of brain size and less evidence for abnormal brain histology (Bilguvar et al., 2010; Nicholas et al., 2010; Yu et al., 2010) (Figure 1). Hence, it appears that complete loss of gene function (due to Stop mutations or occasional missense changes that severely block protein function) and partial loss of function lead to different conditions that are not always easily recognized as allelic. Therefore, one possibility is that missense mutations in ASPM, STIL, CDK5RAP2, or CENPJ also exist, but cause a milder disease that has so far not been recognized as allelic to the more severe syndromes. An alternate explanation is that perhaps the length of these proteins is more critical to their function than is their primary amino acid sequence.
Comparative genomics provides some insight into the patterns of disease-causing mutations, since interspecies conservation of primary amino acid sequence reflects evolutionary selection, i.e., the extent to which the exact amino acid sequence is essential to normal function. Interspecies conservation of amino acid sequence reflects the fact that most changes of conserved amino acids are deleterious in that they reduce reproductive fitness, reflecting a decreased likelihood of their transmission to the next generation. The most common cause of reduced human fitness is of course disease, especially when we are talking about the brain. As we recover more and more disease mutations, the theoretical of concept of “negative evolutionary selection,” in which amino acid sequence tends to be conserved because alterations cause reduced fitness, begins to come alive. In the case of DCX, the relative absence of disease causing mutations in the C-terminus correlates with a relative lack of amino acid conservation in this region between species. This contrast suggests that there may be diseases associated with missense mutations of the C-terminus, but that these diseases have not yet been recovered, i.e. patients with these other diseases have not yet had their DCX gene sequenced. Alternatively, many polymorphisms at the C-terminus of DCX may not deleterious, i.e., don't cause any disease. A very similar pattern is seen with ROBO3, where the lack of missense mutations at the C-terminus is matched by a relatively lower level of amino acid conservation at the C-terminus of the protein.
Interspecies comparisons of the ASPM microcephaly gene have generated an entire field unto itself, with evidence from multiple labs suggesting that the overall amino acid sequence is under less strong negative selection than most human proteins. In fact, ASPM may have been a target of “positive” evolutionary selection, with greater changes in amino acid sequence between humans and nonhuman primates than most genes, suggesting a potential role of ASPM in the evolution of the larger brains that characterize humans (Evans et al., 2004; Kouprina et al., 2004; Mekel-Bobrov et al., 2005; Yu et al., 2009; Yu et al., 2007; Zhang, 2003). Analysis of amino acid conservation of ASPM among vertebrates shows remarkably low conservation compared to other neurological disease genes (Figure 1). Large chunks of the protein near the N terminus, and within the “IQ” repeats, show extremely low conservation because these segments of the protein are lacking in rodents and nonmammals altogether. On the other hand, the region surrounding the calponin homology (CH) domains is extremely highly conserved, yet still no missense mutations have been seen here yet. Thus, the degree of amino acid differences between species, however, does not suffice to explain the scarcity of missense mutations identified in all of these genes in patients with known phenotypes, suggesting that some missense changes may have other phenotypic consequences.
Dominant mutations can implicate specific protein-protein interactions
While some dominant mutations function through reduced gene dosage, many are missense mutations that cause disease by creating new or aberrant functions in a protein. Thus, the patterns of dominant mutations are often very different from those of autosomal or X-linked recessive mutations. This is illustrated nicely by the “special” mutations identified in CHN1, KIF21A, and TUBB3, each of which alters rather than eliminates the function of the encoded protein, causes aberrant axon growth and guidance, and results in an autosomal dominant complex eye movement disorder.
Heterozygous mutations in CHN1 cause stalling of axons of the abducens nerve, one of the three cranial nerves that control eye movement, and result in a stereotypical pattern of abnormal horizontal gaze referred to as Duane retraction syndrome (Chan et al., 2010; Miyake et al., 2008). CHN1 encodes α2-chimaerin, a RacGAP signaling molecule that turns active GTP-bound Rac off by enhancing the conversion of GTP-bound Rac to inactive GDP-bound Rac, and has been shown to serve as an effector for axon guidance (Brown et al., 2004; Iwasato et al., 2007). CHN1 mutations are all missense, the opposite of ASPM. At first glance, the mutations appear to be somewhat randomly scattered onto the 2D structure of the protein, residing both within and between its known functional domains (Figure 1). These are, however, “special” mutations as each has been shown to hyperactivate α2-chimaerin's normal function and to pathologically lower RacGTP levels in the cell (Miyake et al., 2008). Most mutations appear to do this by altering amino acid residues involved in intramolecular interactions that normally stabilize the closed, inactive conformation of the molecule. By substituting a different amino acid, the inactive conformation is destabilized and the activity of the signaling molecule is pathologically enhanced.
Heterozygous mutations in KIF21A result in a different autosomal dominant stereotypical congenital eye movement disorder, CFEOM (congenital fibrosis of the extraocular muscles) type 1, which likely results from the stalling or misguidance of axons in another ocular cranial nerve (the oculomotor nerve) that normally innervates several extraocular muscles important for horizontal and vertical gaze (Yamada et al., 2003). KIF21A encodes a kinesin motor protein that transports cargo from the neuronal cell body to the developing axon's growth cone by ‘walking’ along microtubules (Marszalek et al., 1999). KIF21A mutations are all missense, are typically recurrent, and often arise as de novo mutations in children of unaffected parents around the world. As highlighted in Figure 1, these rare mutations repeatedly alter specific highly conserved amino acid residues located within two regions of this very large protein, the 3rd coiled-coil domain of the stalk region and the distal motor domain. Remarkably, the most common missense mutation, 2860C>T (R954W), is present in 61 of the 84 patients reported to date, and 72 of the 84 have mutations altering the KIF21A R954 residue. This specificity leads to the prediction that the mutations disrupt specific protein-protein interactions, and thus that they provide a biological tool for dissecting both disease mechanism and the sub-functions of the altered kinesin domains. Notably, unaffected control individuals have been found to harbor missense polymorphisms that map to the distal stalk of KIF21A. Unlike the disease mutations, these non-pathogenic changes alter amino acids that are poorly conserved in other species, and thus are not under negative evolutionary selection and are not critical to KIF21A function (Yamada et al., 2003). In contrast, no KIF21A Stop mutations have been reported in CFEOM1 patients or controls, suggesting that heterozygous Stop mutations may be embryonic lethal or result in a different, unrecognized human disorder.
Finally, heterozygous missense mutations in TUBB3 cause a third congenital eye movement disorder, CFEOM type 3, that, in some patients, is indistinguishable from the KIF21A phenotype, and also results from aberrant guidance of oculomotor cranial nerve axons (Tischfield et al., 2010). TUBB3 encodes the neuronal specific β-tubulin isotype III, a component of the microtubule cytoskeleton on which kinesins walk. Similar to KIF21A disease mutations, TUBB3 mutations are also “special”: they are missense, often arise de novo, and the same mutation is found among multiple unrelated patients (Figure 1). Unlike mutations in CHN1 and in KIF21A that result in isolated and stereotypical ocular phenotypes, these TUBB3 mutations demonstrate ‘allelic diversity’, with specific missense mutations causing additional phenotypes, including facial weakness, progressive peripheral neuropathy, congenital joint contractures, and/or developmental disabilities (Figure 1) (Tischfield et al., 2010). Studies of both humans and a mouse model reveal aberrant axon growth and guidance without evidence of errors in cortical neuronal migration. All mutations increase microtubule stability, while a subset appears to alter microtubule-kinesin interactions. Thus, these repetitive human mutations highlight and hence identify interfaces on TUBB3 that are essential to specific sub-functions of this tubulin isoform that are critical to the development and maintenance of axons in both the central and peripheral nervous system.
Some genes are subject to multiple types of mutations associated with different diseases
If specific dominant heterozygous mutations in CHN1, KIF21A, or TUBB3, all three of which encode proteins expressed in neurons throughout the developing and mature nervous system, lead to new enhanced or aberrant protein function that cause specific eye movement defects, what would we expect to be the effects of different mutations altering other conserved residues in these same genes? For TUBB3, this question was answered, in part, by the recent report of a second set of dominant missense human mutations that result in less, rather than more stable microtubules, and cause malformations of cortical development secondary to neuronal migration defects, in the absence of CFEOM type 3 (Figure 1) (Poirier et al., 2010). Thus, this new set of missense mutations likely alter residues essential to different sub-functions of the TUBB3 protein.
Although human loss-of-function mutations have not been identified for CHN1, KIF21A, or TUBB3, we might get insights of what to expect from engineered mouse models. Indeed, Chn1−/− mice survive and have a completely different phenotype from the phenotype found to result from missense mutations in humans (Miyake et al., 2008); the mice have misguidance not of cranial but of corticospinal axons, resulting in an abnormal hopping gait (Iwasato et al., 2007). This suggests that human CHN1 Stop mutations might underlie a yetto-be-identified neuromotor disorder.
There are other developmental examples where different sorts of mutations in the same gene can cause two or more different diseases, often with almost no overlap, due to differing biochemical mechanisms. One nice example are mutations in FLNA: FLNA mutations include loss of function, gain of function, and partial loss of function, and each mutation type results in a distinct phenotype (Feng and Walsh, 2004b) (Figure 1). FLNA encodes FilaminA, and the first known disease associated with loss of FLNA function was periventricular heterotopia, a neuronal migration disorder in which neurons fail to migrate out of the ventricular zone during prenatal development (Fox et al., 1998). The disorder is X-linked, typically prenatally lethal in males, and is associated with many Stop mutations that block normal protein translation from that locus (as opposed to making a truncated protein) (Sheen et al., 2001). FLNA is essential for normal heart and vascular development as well, explaining the prenatal lethality of the condition (Feng et al., 2006; Hart et al., 2006). Several FLNA missense mutations cause an indistinguishable phenotype, presumably by also acting as heterozygous null mutations (Sheen et al., 2001; Sole et al., 2009), and these tend to cluster in the exons encoding the first calpain-homology (CH) domain, required for actin binding (Parrini et al., 2006). Occasionally male patients are found to harbor FLNA mutations who survive and have periventicular heterotopia; these mutations are often missense changes, or alleles that only truncate the extreme C-terminus, suggesting that they may create hypomorphic proteins that retain some residual function (Parrini et al., 2006; Sheen et al., 2001; Sole et al., 2009).
After the initial discovery of null mutations associated with periventricular heterotopia, an amazing array of skeletal dysplasias have been described in association with FLNA mutations, including many with unusual or recurrent missense mutations; these include otopalataldigital (OPD) syndrome I and II, frontometaphyseal dysplasia, and Melnick-Needles syndrome (Robertson, 2005; Robertson et al., 2003). Even more recently, additional specific mutant alleles of this same gene have been associated with inherited X-linked myxomatous valvular dystrophy (XMVD) affecting primarily the heart (Kyndt et al., 2007) and with terminal osseous dysplasia (TOD), which has so far been seen with a single mutant allele that has recurred in separate families at least 6 times (Sun et al., 2010). Many of these disorders do not show periventricular heterotopia, suggesting that the mutations do not remove FilaminA function but may alter it. Only one reported allele causes both OPD and periventicular heterotopia (Zenker et al., 2004). Many OPD mutations cluster in the second CH domain, which also regulates actin binding. Recent work suggests that at least some of the OPD missense mutations actually enhance the binding of FilaminA to F-actin in vitro, resulting in a gain of function mechanism (Clark et al., 2009), whereas the periventricular heterotopia missense mutations presumably disrupt actin binding.
The large number of disease-associated alleles found in FLNA might be predicted by the very high conservation of its amino acid sequence between species. Moreover, the seven or more distinct genetic disorders resulting from different mutations in the same gene suggest that FilaminA has diverse interaction with many different other protein networks in distinct developmental contexts, and the long list of filamin-interacting proteins further supports this hypothesis (Feng and Walsh, 2004a). These allelic disorders provide direct pointers into distinct signaling pathways. They also prompt the question of the hidden allelic diversity of other genes, in which a single gene can be affected in many ways to cause widely divergent phenotypes.
How do the “missing” mutant alleles map onto uncharacterized diseases?
Comparisons of the sorts of mutations that cause developmental phenotypes reveal remarkable differences from gene to gene, given that the pattern of disease-associated mutation reflects not only patterns of DNA mutation, but also structures and functions of proteins, and the extent to which amino acid sequence is conserved. Some genes (ROBO3, DCX, FLNA) cause developmental disorders due to apparently simple loss of function mechanisms, either by Stop mutations or missense changes that are presumably also disabling. FLNA shows one phenotype due to loss of function, but other phenotypes due to gain or alteration of function, while TUBB3 shows distinct phenotypes due to different gain or alteration in function, with specific missense mutations targeting specific protein-protein interactions. Dominant mutations in KIF21A, CHN1 apparently also act by disrupting normal protein-protein interactions. Notably, simple loss-of-function mutations in TUBB3, KIF21A, and CHN1 have not yet been observed, and might have completely different phenotypes. Thus, if we assume that humans are “saturated” for mutations at most codons (as well as presumably some, but certainly not all, noncoding segments) the depth of mutational analysis in humans allows one to begin to ask questions about what alleles we are missing. For diseases that are familiar as dominant, gain-of-function conditions, are there additional unrecognized phenotypes resulting from gain-of-function mutations altering a different set of conserved amino acids? And what is the loss-of-function phenotype in that same gene, and does it look at all like the dominant disease? In some cases it might, yet in others it could be embryonically lethal, or have a totally different phenotype. Or, if we have observed the null phenotype for a gene, then what are that gene's hypomorphic phenotypes, since our previous analysis suggests that hypomorphic mutations frequently exist. For many genes with essential roles in brain development, it is likely that the possible range of alleles has not yet been identified, and presumably many of these other types of alleles disrupt brain function, but remain so far unrecognized.
So then we might ask, what diseases, or other phenotypes, do these “missing alleles” cause? Where does the burden of unexplained neurogenetic disease lie? In the case of severe developmental disorders of the brain, associated with neonatal presentations with epilepsy, brain malformations, or mental retardation, these disorders are remarkably well-characterized, with clinicians having a >60% chance of identifying a specific causative condition or responsible gene (Rimoin and Emery, 2007). For milder forms of intellectual disability, the yield of genetic investigation is lower, approximately 50% (Rimoin and Emery, 2007). For autism, which is a milder and more heterogeneous condition, intensive genetic investigation typically reveals a specific (genetic) cause (or a specific, complex interaction of rare and/or common alleles) in 15-20% of cases (Pinto et al., 2010; Shen et al., 2010) leaving the great majority as yet unexplained. And finally, for the mildest “learning disorders” and psychiatric conditions, it is safe to say that a specific genetic contribution of <5% can presently be explained, leaving these disorders almost completely uncharacterized genetically (Faraone and Mick, 2010; Owen et al., 2010).
Recent studies of the genetics of autism spectrum disorders (ASD), described in greater detail in another review in this issue (State, this issue), show how heterogeneous the milder developmental disorders might be. Perhaps 5% of children with ASD have mutations in “Mendelian” autism genes that typically cause ASD in some children and intellectual disability in other children; these genes include FMR1, MECP2, NLGN2, NLGN3, ARX, SHANK3, TSC1, TSC2, and others (Walsh et al., 2008). Heterozygous “copy number variants” (CNVs) appear to be collectively the most common cause of ASD, though estimates of the proportion of the disorder that they are responsible for range widely from more than 20% in early studies to closer to 5% in more recent and systematic studies (Sebat et al., 2007; Shen et al., 2010; Weiss et al., 2009). About a half dozen of these rare CNVs are recurrent, meaning that deletions or duplications of the same regions occur in more than one family (16p11.2, 15q, 22q11, 15q13, NRXN1, MECP2), and so are recognizable as causative when observed in isolation; others appear to be unique in each family. “Common alleles” have been described that might affect predisposition to ASD, but to date they would account for even less of the known genetic risk (Anney et al., 2010; Arking et al., 2008; Glessner et al., 2009). These sorts of heterogeneous disorders might be where we find many of the miscellaneous “missing” mutations, and the specific mutations involved could be individually rare, present in just one or small numbers of families, and very diverse in their action.
An interesting sort of mutation, found in a few ASD patients whose parents share common ancestry (and who hence share more than the usual proportion of their rare genetic mutations), has recently been described and may point to a whole new category of alleles. These mutations are homozygous deletions, removing both copies of a stretch of DNA. Homozygous deletions can remove genes, making them conventional Stop mutations, but in some patients appear to delete noncoding DNA containing conserved predicted promoter-enhancer elements near genes with prominent brain expression (Morrow et al., 2008). These homozygous noncoding mutations show an appealing resemblance to “conditional” mutant alleles in mice, where the function of a gene might not be compromised in all places and times, but only where that gene's expression would have been controlled by deleted promoter elements. Thus, the possible mutations that appear to cause ASD are not only diverse and heterogeneous in the genes involved, but in the mechanisms involved (deletion, duplication, conventional point mutation) and mode of inheritance or absence of inheritance.
With the advent of high-throughput sequencing of the entire exome, or genome, one of the biggest challenges will be interpreting which rare polymorphisms in a person's DNA are likely to be causative of disease. There is little doubt that one of the biggest immediate impacts of high throughput sequencing in the clinical setting will be in expanding the range of mutation of known genes, in addition to new gene identification. Finding new alleles of known genes is simpler, since we already have some knowledge of overall gene function, but will provide a rapid bounty of biological information about interactions and pathways as well as causes of disease.
Acknowledgements
The authors thank Brenda J. Barry, Wai-Man Chan, R. Sean Hill, Jennifer N. Partlow, and Allison M. Pelger for preparing the figure and Michelle Cirioni for technical support. C. A. W. is supported by grants from the National Institute of Neurological Disease and Stroke, the National Institute of Mental Health, and the Simons Foundation. E.C.E. is supported by grants from the National Eye Institute. C.A.W. and E.C.E. are supported by the Manton Center for Orphan Disease Research and are Investigators of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anney R, Klei L, Pinto D, Regan R, Conroy J, Magalhaes TR, Correia C, Abrahams BS, Sykes N, Pagnamenta AT, et al. A genome-wide scan for common alleles affecting risk for autism. Hum. Mol. Genet. 2010 doi: 10.1093/hmg/ddq307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arking DE, Cutler DJ, Brune CW, Teslovich TM, West K, Ikeda M, Rea A, Guy M, Lin S, Cook EH, Chakravarti A. A common genetic variant in the neurexin superfamily member CNTNAP2 increases familial risk of autism. Am. J. Hum. Genet. 2008;82:160–164. doi: 10.1016/j.ajhg.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai J, Ramos RL, Ackman JB, Thomas AM, Lee RV, LoTurco JJ. RNAi reveals doublecortin is required for radial migration in rat neocortex. Nat. Neurosci. 2003;6:1277–1283. doi: 10.1038/nn1153. [DOI] [PubMed] [Google Scholar]
- Bilguvar K, Ozturk AK, Louvi A, Kwan KY, Choi M, Tatli B, Yalnizoglu D, Tuysuz B, Caglayan AO, Gokben S, et al. Whole-exome sequencing identifies recessive WDR62 mutations in severe brain malformations. Nature. 2010 doi: 10.1038/nature09327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond J, Roberts E, Mochida GH, Hampshire DJ, Scott S, Askham JM, Springell K, Mahadevan M, Crow YJ, Markham AF, et al. ASPM is a major determinant of cerebral cortical size. Nat. Genet. 2002;32:316–320. doi: 10.1038/ng995. [DOI] [PubMed] [Google Scholar]
- Bond J, Roberts E, Springell K, Lizarraga SB, Scott S, Higgins J, Hampshire DJ, Morrison EE, Leal GF, Silva EO, et al. A centrosomal mechanism involving CDK5RAP2 and CENPJ controls brain size. Nat. Genet. 2005;37:353–355. doi: 10.1038/ng1539. [DOI] [PubMed] [Google Scholar]
- Bond J, Scott S, Hampshire DJ, Springell K, Corry P, Abramowicz MJ, Mochida GH, Hennekam RC, Maher ER, Fryns JP, et al. Protein-truncating mutations in ASPM cause variable reduction in brain size. Am. J. Hum. Genet. 2003;73:1170–1177. doi: 10.1086/379085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. Nature's gift to science (Nobel lecture) Chembiochem. 2003;4:683–687. doi: 10.1002/cbic.200300625. [DOI] [PubMed] [Google Scholar]
- Brown M, Jacobs T, Eickholt B, Ferrari G, Teo M, Monfries C, Qi RZ, Leung T, Lim L, Hall C. Alpha2-chimaerin, cyclin-dependent Kinase 5/p35, and its target collapsin response mediator protein-2 are essential components in semaphorin 3A-induced growth-cone collapse. J. Neurosci. 2004;24:8994–9004. doi: 10.1523/JNEUROSCI.3184-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan W-M, Traboulsi E, Arthur B, Friedman N, Andrews C, Engle E. Horizontal gaze palsy with progressive scoliosis can result from compound heterozygous mutations in ROBO3. J. Med. Genet. 2006;43:e11. doi: 10.1136/jmg.2005.035436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan WM, Miyake N, Zhu-Tam L, Andrews C, Engle EC. Two novel CHN1 mutations in two families with Duane's retraction syndrome. Arch. Ophthalmol. 2010 doi: 10.1001/archophthalmol.2011.84. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AR, Sawyer GM, Robertson SP, Sutherland-Smith AJ. Skeletal dysplasias due to filamin A mutations result from a gain-of-function mechanism distinct from allelic neurological disorders. Hum. Mol. Genet. 2009;18:4791–4800. doi: 10.1093/hmg/ddp442. [DOI] [PubMed] [Google Scholar]
- des Portes V, Pinard JM, Billuart P, Vinet MC, Koulakoff A, Carrie A, Gelot A, Dupuis E, Motte J, Berwald-Netter Y, et al. A novel CNS gene required for neuronal migration and involved in X-linked subcortical laminar heterotopia and lissencephaly syndrome. Cell. 1998;92:51–61. doi: 10.1016/s0092-8674(00)80898-3. [DOI] [PubMed] [Google Scholar]
- Evans PD, Anderson JR, Vallender EJ, Choi SS, Lahn BT. Reconstructing the evolutionary history of microcephalin, a gene controlling human brain size. Hum. Mol. Genet. 2004;13:1139–1145. doi: 10.1093/hmg/ddh126. [DOI] [PubMed] [Google Scholar]
- Faraone SV, Mick E. Molecular genetics of attention deficit hyperactivity disorder. Psychiatr. Clin. North Am. 2010;33:159–180. doi: 10.1016/j.psc.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Chen MH, Moskowitz IP, Mendonza AM, Vidali L, Nakamura F, Kwiatkowski DJ, Walsh CA. Filamin A (FLNA) is required for cell-cell contact in vascular development and cardiac morphogenesis. Proc. Natl. Acad. Sci. U. S. A. 2006;103:19836–19841. doi: 10.1073/pnas.0609628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Walsh CA. Mitotic spindle regulation by Nde1 controls cerebral cortical size. Neuron. 2004a;44:279–293. doi: 10.1016/j.neuron.2004.09.023. [DOI] [PubMed] [Google Scholar]
- Feng Y, Walsh CA. The many faces of filamin: a versatile molecular scaffold for cell motility and signalling. Nat Cell Biol. 2004b;6:1034–1038. doi: 10.1038/ncb1104-1034. [DOI] [PubMed] [Google Scholar]
- Fish JL, Kosodo Y, Enard W, Paabo S, Huttner WB. Aspm specifically maintains symmetric proliferative divisions of neuroepithelial cells. Proc. Natl. Acad. Sci. U. S. A. 2006;103:10438–10443. doi: 10.1073/pnas.0604066103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox JW, Lamperti ED, Eksioglu YZ, Hong SE, Feng Y, Graham DA, Scheffer IE, Dobyns WB, Hirsch BA, Radtke RA, et al. Mutations in filamin 1 prevent migration of cerebral cortical neurons in human periventricular heterotopia. Neuron. 1998;21:1315–1325. doi: 10.1016/s0896-6273(00)80651-0. [DOI] [PubMed] [Google Scholar]
- Gleeson JG, Allen KM, Fox JW, Lamperti ED, Berkovic S, Scheffer I, Cooper EC, Dobyns WB, Minnerath SR, Ross ME, Walsh CA. Doublecortin, a brain-specific gene mutated in human X-linked lissencephaly and double cortex syndrome, encodes a putative signaling protein. Cell. 1998;92:63–72. doi: 10.1016/s0092-8674(00)80899-5. [DOI] [PubMed] [Google Scholar]
- Gleeson JG, Lin PT, Flanagan LA, Walsh CA. Doublecortin is a microtubule-associated protein and is expressed widely by migrating neurons. Neuron. 1999;23:257–271. doi: 10.1016/s0896-6273(00)80778-3. [DOI] [PubMed] [Google Scholar]
- Glessner JT, Wang K, Cai G, Korvatska O, Kim CE, Wood S, Zhang H, Estes A, Brune CW, Bradfield JP, et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459:569–573. doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrini R, Marini C. Genetic malformations of cortical development. Exp. Brain Res. 2006;173:322–333. doi: 10.1007/s00221-006-0501-z. [DOI] [PubMed] [Google Scholar]
- Guerrini R, Moro F, Andermann E, Hughes E, D'Agostino D, Carrozzo R, Bernasconi A, Flinter F, Parmeggiani L, Volzone A, et al. Nonsyndromic mental retardation and cryptogenic epilepsy in women with doublecortin gene mutations. Ann. Neurol. 2003;54:30–37. doi: 10.1002/ana.10588. [DOI] [PubMed] [Google Scholar]
- Gul A, Hassan MJ, Mahmood S, Chen W, Rahmani S, Naseer MI, Dellefave L, Muhammad N, Rafiq MA, Ansar M, et al. Genetic studies of autosomal recessive primary microcephaly in 33 Pakistani families: Novel sequence variants in ASPM gene. Neurogenetics. 2006;7:105–110. doi: 10.1007/s10048-006-0042-4. [DOI] [PubMed] [Google Scholar]
- Hart AW, Morgan JE, Schneider J, West K, McKie L, Bhattacharya S, Jackson IJ, Cross SH. Cardiac malformations and midline skeletal defects in mice lacking filamin A. Hum. Mol. Genet. 2006;15:2457–2467. doi: 10.1093/hmg/ddl168. [DOI] [PubMed] [Google Scholar]
- Iwasato T, Katoh H, Nishimaru H, Ishikawa Y, Inoue H, Saito YM, Ando R, Iwama M, Takahashi R, Negishi M, Itohara S. Rac-GAP alpha-Chimerin Regulates Motor-Circuit Formation as a Key Mediator of EphrinB3/EphA4 Forward Signaling. Cell. 2007;130:742–753. doi: 10.1016/j.cell.2007.07.022. [DOI] [PubMed] [Google Scholar]
- Jen JC, Chan WM, Bosley TM, Wan J, Carr JR, Rub U, Shattuck D, Salamon G, Kudo LC, Ou J, et al. Mutations in a human ROBO gene disrupt hindbrain axon pathway crossing and morphogenesis. Science. 2004;304:1509–1513. doi: 10.1126/science.1096437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MH, Cierpicki T, Derewenda U, Krowarsch D, Feng Y, Devedjiev Y, Dauter Z, Walsh CA, Otlewski J, Bushweller JH, Derewenda ZS. The DCX-domain tandems of doublecortin and doublecortin-like kinase. Nat. Struct. Biol. 2003;10:324–333. doi: 10.1038/nsb918. [DOI] [PubMed] [Google Scholar]
- Kouprina N, Pavlicek A, Collins NK, Nakano M, Noskov VN, Ohzeki J, Mochida GH, Risinger JI, Goldsmith P, Gunsior M, et al. The microcephaly ASPM gene is expressed in proliferating tissues and encodes for a mitotic spindle protein. Hum. Mol. Genet. 2005;14:2155–2165. doi: 10.1093/hmg/ddi220. [DOI] [PubMed] [Google Scholar]
- Kouprina N, Pavlicek A, Mochida GH, Solomon G, Gersch W, Yoon YH, Collura R, Ruvolo M, Barrett JC, Woods CG, et al. Accelerated evolution of the ASPM gene controlling brain size begins prior to human brain expansion. PLoS Biol. 2004;2:E126. doi: 10.1371/journal.pbio.0020126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kousar R, Nawaz H, Khurshid M, Ali G, Khan SU, Mir H, Ayub M, Wali A, Ali N, Jelani M, et al. Mutation analysis of the ASPM gene in 18 Pakistani families with autosomal recessive primary microcephaly. J. Child Neurol. 25:715–720. doi: 10.1177/0883073809346850. [DOI] [PubMed] [Google Scholar]
- Kumar A, Girimaji SC, Duvvari MR, Blanton SH. Mutations in STIL, encoding a pericentriolar and centrosomal protein, cause primary microcephaly. Am. J. Hum. Genet. 2009;84:286–290. doi: 10.1016/j.ajhg.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyndt F, Gueffet JP, Probst V, Jaafar P, Legendre A, Le Bouffant F, Toquet C, Roy E, McGregor L, Lynch SA, et al. Mutations in the gene encoding filamin A as a cause for familial cardiac valvular dystrophy. Circulation. 2007;115:40–49. doi: 10.1161/CIRCULATIONAHA.106.622621. [DOI] [PubMed] [Google Scholar]
- Marillat V, Sabatier C, Failli V, Matsunaga E, Sotelo C, Tessier-Lavigne M, Chedotal A. The slit receptor Rig-1/Robo3 controls midline crossing by hindbrain precerebellar neurons and axons. Neuron. 2004;43:69–79. doi: 10.1016/j.neuron.2004.06.018. [DOI] [PubMed] [Google Scholar]
- Marszalek JR, Weiner JA, Farlow SJ, Chun J, Goldstein LS. Novel dendritic kinesin sorting identified by different process targeting of two related kinesins: KIF21A and KIF21B. J. Cell. Biol. 1999;145:469–479. doi: 10.1083/jcb.145.3.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekel-Bobrov N, Gilbert SL, Evans PD, Vallender EJ, Anderson JR, Hudson RR, Tishkoff SA, Lahn BT. Ongoing adaptive evolution of ASPM, a brain size determinant in Homo sapiens. Science. 2005;309:1720–1722. doi: 10.1126/science.1116815. [DOI] [PubMed] [Google Scholar]
- Miyake N, Chilton J, Psatha M, Cheng L, Andrews C, Chan WM, Law K, Crosier M, Lindsay S, Cheung M, et al. Human CHN1 mutations hyperactivate alpha2-chimaerin and cause Duane's retraction syndrome. Science. 2008;321:839–843. doi: 10.1126/science.1156121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow EM, Yoo SY, Flavell SW, Kim TK, Lin Y, Hill RS, Mukaddes NM, Balkhy S, Gascon G, Hashmi A, et al. Identifying autism loci and genes by tracing recent shared ancestry. Science. 2008;321:218–223. doi: 10.1126/science.1157657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas AK, Khurshid M, Désir J, Carvalho OP, Cox JJ, Thornton G, Kausar R, Ansar M, Ahmad W, Verloes A, et al. WDR62 is a spindle pole protein essential for neurogenesis. Nat. Genet. 2010 doi: 10.1038/ng.682. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas AK, Swanson EA, Cox JJ, Karbani G, Malik S, Springell K, Hampshire D, Ahmed M, Bond J, Di Benedetto D, et al. The molecular landscape of ASPM mutations in primary microcephaly. J. Med. Genet. 2009;46:249–253. doi: 10.1136/jmg.2008.062380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen MJ, Craddock N, O'Donovan MC. Suggestion of roles for both common and rare risk variants in genome-wide studies of schizophrenia. Arch. Gen. Psychiatry. 2010;67:667–673. doi: 10.1001/archgenpsychiatry.2010.69. [DOI] [PubMed] [Google Scholar]
- Parrini E, Ramazzotti A, Dobyns WB, Mei D, Moro F, Veggiotti P, Marini C, Brilstra EH, Dalla Bernardina B, Goodwin L, et al. Periventricular heterotopia: phenotypic heterogeneity and correlation with Filamin A mutations. Brain. 2006;129:1892–1906. doi: 10.1093/brain/awl125. [DOI] [PubMed] [Google Scholar]
- Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, Conroy J, Magalhaes TR, Correia C, Abrahams BS, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010 doi: 10.1038/nature09146. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier K, Saillour YY, Bahi-Buisson N, Jaglin XH, Fallet-Bianco C, Nabbout R, Castelnau-Ptakhine L, Roubertie A, Attie-Bitach T, Desguerre I, et al. Mutations in the neuronal beta tubulin subunit TUBB3 result in malformation of cortical development and neuronal migration defects. Hum. Mol. Genet. 2010 doi: 10.1093/hmg/ddq377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner O, Coquelle FM, Peter B, Levy T, Kaplan A, Sapir T, Orr I, Barkai N, Eichele G, Bergmann S. The evolving doublecortin (DCX) superfamily. BMC Genomics. 2006;7:188. doi: 10.1186/1471-2164-7-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimoin DL, Emery AEH. Emery and Rimoin's principles and practice of medical genetics. 5th edn Churchill Livingstone Elsevier; Philadelphia, Pa.: 2007. [Google Scholar]
- Robertson SP. Filamin A: phenotypic diversity. Curr. Opin. Genet. Dev. 2005;15:301–307. doi: 10.1016/j.gde.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Robertson SP, Twigg SR, Sutherland-Smith AJ, Biancalana V, Gorlin RJ, Horn D, Kenwrick SJ, Kim CA, Morava E, Newbury-Ecob R, et al. Localized mutations in the gene encoding the cytoskeletal protein filamin A cause diverse malformations in humans. Nat. Genet. 2003;33:487–491. doi: 10.1038/ng1119. [DOI] [PubMed] [Google Scholar]
- Sabatier C, Plump AS, Le M, Brose K, Tamada A, Murakami F, Lee EY, TessierLavigne M. The divergent Robo family protein rig-1/Robo3 is a negative regulator of slit responsiveness required for midline crossing by commissural axons. Cell. 2004;117:157–169. doi: 10.1016/s0092-8674(04)00303-4. [DOI] [PubMed] [Google Scholar]
- Sapir T, Horesh D, Caspi M, Atlas R, Burgess HA, Wolf SG, Francis F, Chelly J, Elbaum M, Pietrokovski S, Reiner O. Doublecortin mutations cluster in evolutionarily conserved functional domains. Hum. Mol. Genet. 2000;9:703–712. doi: 10.1093/hmg/9.5.703. [DOI] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, Yamrom B, Yoon S, Krasnitz A, Kendall J, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen VL, Dixon PH, Fox JW, Hong SE, Kinton L, Sisodiya SM, Duncan JS, Dubeau F, Scheffer IE, Schachter SC, et al. Mutations in the X-linked filamin 1 gene cause periventricular nodular heterotopia in males as well as in females. Hum. Mol. Genet. 2001;10:1775–1783. doi: 10.1093/hmg/10.17.1775. [DOI] [PubMed] [Google Scholar]
- Shen Y, Dies KA, Holm IA, Bridgemohan C, Sobeih MM, Caronna EB, Miller KJ, Frazier JA, Silverstein I, Picker J, et al. Clinical genetic testing for patients with autism spectrum disorders. Pediatrics. 2010;125:e727–735. doi: 10.1542/peds.2009-1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sole G, Coupry I, Rooryck C, Guerineau E, Martins F, Deves S, Hubert C, Souakri N, Boute O, Marchal C, et al. Bilateral periventricular nodular heterotopia in France: frequency of mutations in FLNA, phenotypic heterogeneity and spectrum of mutations. J. Neurol. Neurosurg. Psychiatry. 2009;80:1394–1398. doi: 10.1136/jnnp.2008.162263. [DOI] [PubMed] [Google Scholar]
- Sun Y, Almomani R, Aten E, Celli J, van der Heijden J, Venselaar H, Robertson SP, Baroncini A, Franco B, Basel-Vanagaite L, et al. Terminal osseous dysplasia is caused by a single recurrent mutation in the FLNA gene. Am. J. Hum. Genet. 2010;87:146–153. doi: 10.1016/j.ajhg.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor KR, Holzer AK, Bazan JF, Walsh CA, Gleeson JG. Patient mutations in doublecortin define a repeated tubulin-binding domain. J. Biol. Chem. 2000;275:34442–34450. doi: 10.1074/jbc.M007078200. [DOI] [PubMed] [Google Scholar]
- Tischfield MA, Baris HN, Wu C, Rudolph G, Van Maldergem L, He W, Chan WM, Andrews C, Demer JL, Robertson RL, et al. Human TUBB3 mutations perturb microtubule dynamics, kinesin interactions, and axon guidance. Cell. 2010;140:74–87. doi: 10.1016/j.cell.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh CA. Genetic malformations of the human cerebral cortex. Neuron. 1999;23:19–29. doi: 10.1016/s0896-6273(00)80749-7. [DOI] [PubMed] [Google Scholar]
- Walsh CA, Morrow EM, Rubenstein JL. Autism and brain development. Cell. 2008;135:396–400. doi: 10.1016/j.cell.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss LA, Arking DE, Daly MJ, Chakravarti A. A genome-wide linkage and association scan reveals novel loci for autism. Nature. 2009;461:802–808. doi: 10.1038/nature08490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Andrews C, Chan WM, McKeown CA, Magli A, De Berardinis T, Loewenstein A, Lazar M, O'Keefe M, Letson R, et al. Heterozygous mutations of the kinesin KIF21A in congenital fibrosis of the extraocular muscles type 1 (CFEOM1) Nat. Genet. 2003;35:318–321. doi: 10.1038/ng1261. [DOI] [PubMed] [Google Scholar]
- Yu F, Keinan A, Chen H, Ferland RJ, Hill RS, Mignault AA, Walsh CA, Reich D. Detecting natural selection by empirical comparison to random regions of the genome. Hum. Mol. Genet. 2009;18:4853–4867. doi: 10.1093/hmg/ddp457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Yu TW, Mochida GH, Tischfield DJ, Sgaier SK, Flores-Sarnat L, Sergi CM, Topçu M, McDonald MT, Barry BJ, Felie J, et al. Mutations in WDR62, encoding a centrosome-associated protein, cause microcephaly with simplified gyri and abnormal cortical architecture. Nat. Genet. 2010 doi: 10.1038/ng.683. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenker M, Rauch A, Winterpacht A, Tagariello A, Kraus C, Rupprecht T, Sticht H, Reis A. A dual phenotype of periventricular nodular heterotopia and frontometaphyseal dysplasia in one patient caused by a single FLNA mutation leading to two functionally different aberrant transcripts. Am. J. Hum. Genet. 2004;74:731–737. doi: 10.1086/383094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. Evolution of the human ASPM gene, a major determinant of brain size. Genetics. 2003;165:2063–2070. doi: 10.1093/genetics/165.4.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]