Abstract
The efficiency and fidelity of nucleotide incorporation and next-base extension by DNA polymerase (pol) κ past N2-ethyl-Gua were measured using steady-state and rapid kinetic analyses. DNA pol κ incorporated nucleotides and extended 3′ termini opposite N2-ethyl-Gua with measured efficiencies and fidelities similar to that opposite Gua indicating a role for DNA pol κ at the insertion and extension steps of N2-ethyl-Gua bypass. The DNA pol κ was maximally activated to similar levels by a twenty-fold lower concentration of Mn2+ compared to Mg2+. In addition, the steady state analysis indicated that high fidelity DNA pol κ-catalyzed N2-ethyl-Gua bypass is Mg2+-dependent. Strikingly, Mn2+ activation of DNA pol κ resulted in a dramatically lower efficiency of correct nucleotide incorporation opposite both N2-ethyl-Gua and Gua compared to that detected upon Mg2+ activation. This effect is largely governed by diminished correct nucleotide binding as indicated by the high Km values for dCTP insertion opposite N2-ethyl-Gua and Gua with Mn2+ activation. A rapid kinetic analysis showed diminished burst amplitudes in the presence of Mn2+ compared to Mg2+ indicating that DNA pol κ preferentially utilizes Mg2+ activation. These kinetic data support a DNA pol κ wobble base pairing mechanism for dCTP incorporation opposite N2-ethyl-Gua. Furthermore, the dramatically different polymerization efficiencies of the Y-family DNA pols κ and ι in the presence of Mn2+ suggest a metal ion-dependent regulation in coordinating the activities of these DNA pols during translesion synthesis.
Keywords: Y-family DNA polymerase, Translesion Synthesis, Alkylation DNA damage, Metal ion activation, Rapid kinetic analysis, DNA polymerase κ
1. Introduction
The minor groove DNA adduct N2-ethyl-Guanine (N2-ethyl-Gua)3 is generated from the reduction of acetaldehyde with 2′-deoxyguanosine 3′-monophosphate [1]. Humans are exposed to acetaldehyde from both environmental and endogenous sources. Endogenous acetaldehyde pools arise from the oxidation of ethanol to acetaldehyde during ethanol metabolism [2]. Ethanol is a classified human carcinogen and acetaldehyde has been shown to contribute to tumor formation [3]. The N2-ethyl-Gua adduct has been shown to have a high mis-coding potential during nucleotide incorporation with Klenow fragment of Escherichia coli DNA pol I resulting in G→C transversion mutations [4]. Thus, the mutagenic properties of N2-ethyl-Gua could contribute to the formation of ethanol related cancers [5].
N2-ethyl-Gua is blocking to DNA replication by the replicative DNA pols T7− DNA pol, HIV-1 reverse transcriptase, and the human B family DNA pol α [6,7]. The blocking effect of N2-ethyl-Gua on DNA pols has been observed in both in vitro and cell-based assays [6–8]. In contrast to the blocking effects on replicative DNA pols the N2-ethyl-Gua adduct is bypassed by the Y-family DNA pols η, ι, and κ [6,9–12]. The differences in lesion bypass properties result from the overall active site organization and fidelity checks mechanisms that are dramatically different between Y-family DNA pols and the higher-fidelity A- and B-family DNA pols. Replicative DNA pols have a “minor groove checking” mechanism [13,14]. A conserved K-K-K/R-Y motif identified in several B-family DNA pols and observed in the crystal structure of the bacteriophage RB69 DNA pol scans the DNA minor groove for misincorporation events and for potentially mutagenic base damage [14]. In contrast, the Y-family DNA pols lack a minor groove checking mechanism. DNA pol ι-mediated bypass of N2-ethyl-Gua occurs by a Hoogsteen base-pairing mechanism where the ethyl moiety of N2-ethyl-Gua is repositioned to the major groove by rotation to the syn conformation from the usual for B-DNA anti conformation [9]. The active sites of DNA pol η and DNA pol κ appear sufficiently open to accommodate the N2-ethyl-Gua adduct by utilizing a wobble base pairing mechanism to avoid steric interference when pairing with the correct dCTP [6,9,11].
DNA pol κ is a member of the DinB polymerase family and is the only human Y-family DNA pol with homologues in bacteria (DinB) and archaea (Dpo4) [15–17]. DNA pol κ exhibits the highest fidelity of the Y-family DNA pols when copying unadducted DNA templates but maintains a low level of fidelity compared to replicative DNA pols [15,18,19]. The higher fidelity of DNA pol κ likely relates to its higher level of selectivity of DNA adduct bypass compared to other Y-family DNA pols. DNA pol κ proficiently extends mis-matched primer termini leading to the proposal that DNA pol κ may act mainly as an extender polymerase of adducted 3′ termini during lesion bypass [20–22]. However, DNA pol κ efficiently inserts and extends nucleotides during bypass of some DNA adducts including several minor groove adducts at the N2 position of Gua [11,23–25]. Structural studies of DNA pol κ in complex with DNA show that this polymerase encircles the DNA with a unique N-terminal domain, the “N-clasp”, which is not found in other Y-family DNA pols [26]. The “N-clasp” lies along the DNA major groove potentially disrupting polymerase binding of major groove DNA adducts but accommodating adducts that position into the DNA minor groove [25].
DNA pols require divalent metal ions for activation [27,28]. Mg2+ is the preferred metal for most DNA pols [29]. Activation of DNA pols using divalent metal ions other than Mg2+ results in diminished effects on polymerization efficiency and fidelity [30,31]. However, it was recently demonstrated that DNA pol ι exhibits increased activity and fidelity when copying opposite a template Thy in the presence of Mn2+ [32]. We subsequently demonstrated that Mn2+ activation of DNA pol ι results in increased polymerization efficiency during nucleotide incorporation opposite the N2-ethyl-Gua adduct [9].
The blocking and potentially mutagenic effects of the N2-ethyl-Gua adduct on replicative DNA pols with the potential for N2-ethyl-Gua-mediated carcinogenesis indicate cellular bypass of this adduct by a specialized Y-family DNA pol. In our continuing efforts to better understand translesion synthesis past N2-ethyl-Gua a series of kinetic studies was performed using human DNA pol κ. The data indicate that DNA pol κ is Mg2+ activated and is efficient at both the insertion and extension steps when bypassing N2-ethyl-Gua. These results indicate that DNA pol κ catalyzes efficient nucleotide insertion and extension during bypass of this minor groove N2-alkyl-Gua DNA adduct. Furthermore, the data support a wobble base pairing mechanism for DNA pol κ during dCTP incorporation opposite the N2-ethyl-Gua lesion. The preference for Mg2+ activation in this DNA pol κ bypass process suggests that metal ion availability at the replication fork could regulate the activity of Y-family DNA pols during translesion synthesis. These experiments demonstrate the contrasting abilities of DNA pol κ and DNA pol ι to utilize Mn2+ for efficient N2-ethyl-Gua bypass providing new insights into the molecular mechanisms of N2-Gua adduct translesion synthesis.
2. Material and methods
2.1 Oligonucleotides
N2-ethyl-Gua phosphoramidites and template oligonucleotides were prepared as described previously [6]. The DNA primer oligonucleotides, 5′-(6-FAM)-GCTCCGGAACCC-3′, 5′-(6-FAM)-GCTCCGGAACCCTT-3′, 5′-(6-FAM)-GCTCCGGAACCCTTC-3′, and 5′-(6-FAM)-GCTCCGGAACCCTTT-3′ were purchased from Operon Biotechnologies, Inc. (Huntsville, AL).
2.2 Expression and Purification of Human DNA Pol κ
The recombinant catalytic fragment of human DNA pol κ (amino acids 19-526) was made as an MBP-DNA pol κ fusion protein with a PreScission Protease (GE Healthcare, Piscataway, NJ) cleavage site seven residues from the DNA pol κ N-terminal methionine. The PreScission Protease recognition sequence and DNA pol κ coding sequence were verified by DNA sequencing. The plasmid constructs were transformed into E. coli BL21(DE3) Rosetta 2 cells (Stratagene, Santa Clara, CA) for overexpression. Cells were grown to an O.D.600 = 0.5 at 37 °C and quickly cooled on ice to 17 °C. After induction with 1 mM isopropyl-β-D-thiogalactopyranoside, the cells were allowed to grow for 15 h at 17 °C. Cell extracts were prepared and the MBP-DNA pol κ fusion protein was bound to an amylose resin in 20 mM Tris-HCL buffer (pH 7.5) containing 1 mM EDTA and 200 mM NaCl. The fusion protein was cleaved overnight by on-column incubation with PreScission Protease at 4 °C. The recovered DNA pol κ was purified to homogeneity using phosphocellulose chromatography.
2.3 Assays
For primer extension assays the DNA primer (12-mer) was hybridized to the 32-mer DNA template and added to reactions containing 20 mM Tris-HCl buffer (pH 7.5) and 2 mM DTT, 0.1 mg/ml BSA, 100 μM dNTP’s, 10 nM DNA pol κ, and the amount of MgCl2 or MnCl2 indicated in the figure legends. Incubations were 15 min at 37 °C and reactions were quenched with EtOH. Samples were dried and resuspended in 5 μl of a 95% formamide/dye solution. Extension products were separated on 8 M urea/23% polyacrylamide gels, imaged using a PhosphoImager (GE Healthcare, Piscataway, NJ), and quantified using ImageQuant software (GE Healthcare, Piscataway, NJ).
For steady-state kinetic assays, the site specific insertion procedure of Boosalis et al. [33] was used. The DNA primers (14-mer for insertion and 15-mer for extension) were hybridized to the 32-mer DNA template and added to reactions containing 20 mM Tris-HCl buffer (pH 7.5) and 2 mM DTT, 0.1 mg/ml BSA, 2 mM MgCl2 or 0.075 mM MnCl2, 50 nM template-primer, and 0.625–3.75 nM DNA pol κ. The amounts of DNA pol κ in reactions yielded less than 20% extended product maximally. Incubations were 5–10 min at 37 °C and reactions were processed as described above. All extended product bands were used to determine kinetic parameters (Km and kcat values) by non-linear regression using SigmaPlot 8.02 software (Systat Software, Inc., San Jose, CA) and GraphPad Prism 5.0 software (GraphPad, San Diego, CA). Relative insertion frequencies were calculated as 1/[(kcat/Km)correct/(kcat/Km)incorrect].
For rapid kinetic assays a Bio-Logic SFM-400 (Bio-Logic USA, LLC, Knoxville, TN) configured for quench-flow or a KinTek RQF-3 quench-flow apparatus (KinTek Corp., Austin, TX) was used. Reactions were as described for the steady-state experiments and were initiated by rapid mixing of a pre-incubated template-primer and DNA pol κ mixture from one syringe with dNTP’s and Mg2+ or Mn2+ chloride salts from a second syringe. Reactions were at 37 °C for time intervals ranging from 5 to 5000 msec and quenched in 0.3 M EDTA. A portion of the quenched reaction mixture (10 μl) was mixed with 30 μl of 95% formamide/dye solution and processed as described above. The amount of product formed was plotted versus time and fit by non-linear regression to the burst equation y = A(1 − e−kobs·t) + kss·t, where A = burst amplitude and indicates the amount of product formed in the first binding event, kobs = pre-steady-state rate of nucleotide incorporation, kss = steady-state rate of nucleotide incorporation and t = time.
3. Results
3.1 Primer Extension Reactions
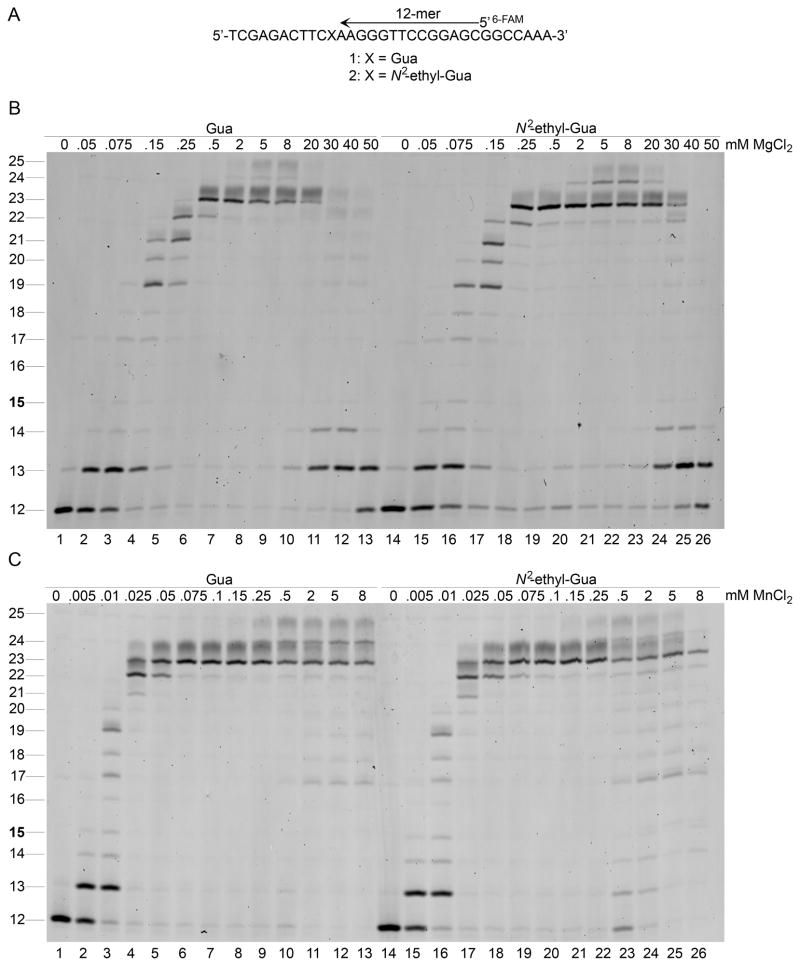
DNA pol κ bypasses N2-ethyl-Gua the same as unadducted Gua using Mg2+ or Mn2+ as the activating divalent metal ion. Different DNA pols utilize Mg2+ and Mn2+ for activation with varying effects on catalytic efficiency and fidelity [31,34]. For example, the human Y-family DNA pol ι exhibits increased catalytic efficiencies with increased fidelities during nucleotide incorporation opposite unadducted DNA and increased bypass efficiency of some DNA adducts [9,32]. Our results have demonstrated that DNA pol ι exhibits increased efficiency but reduced fidelity opposite N2-ethyl-Gua in the presence of Mn2+ compared to Mg2+ [9,32]. The replicative DNA pols exhibit decreased efficiency and fidelity using Mn2+ as the activating metal [30,31]. The metal-dependant effects on DNA pol κ activity during N2-ethyl-Gua bypass were determined in primer extension reactions. A 12-mer primer annealed to a 32-mer DNA template with the 3′-end positioned three nucleotides before the target N2-ethyl-Gua or Gua was used for a substrate (Fig. 1A). Incubation of the template-primer with DNA pol κ in the presence of increased concentrations of MgCl2 (Fig. 1B) or MnCl2 (Fig. 1C) generated extension products ranging from 13 to 25 nucleotides in length. DNA pol κ extended the 12-mer primer past the target N2-ethyl-Gua- and Gua-containing DNA templates without detectable polymerase pausing before, at, or after the target site indicating that both nucleotide incorporation and extension past N2-ethyl-Gua occur similarly to that observed opposite Gua.
Figure 1.
DNA pol κ catalyzed primer extension opposite N2-ethyl-Gua- or Gua-containing DNA templates. Primer extension assays were carried out as described in Materials and Methods. The primer (12-mer) was designed with the 3′-end three nucleotides from the target Gua or N2-ethyl-Gua (A). DNA pol κ (10 nM) catalyzed primer extension opposite N2-ethyl-Gua or Gua with increasing concentrations of MgCl2 (B). The maximum level of primer extension was observed at MgCl2 concentrations ranging from 2 mM to 20 mM for both templates. DNA pol κ (10 nM) catalyzed primer extension opposite N2-ethyl-Gua or Gua with increasing concentrations of MnCl2 (C). Full-length primer extension products were observed in the presence of MnCl2 at concentrations of 0.075 mM and higher.
The DNA pol κ activities in these primer extension analyses demonstrate different sensitivities to Mg2+ and Mn2+ concentrations. The DNA pol κ extends the DNA primer to completion upon addition of increased levels of each activating metal ion with maximal Mn2+-activated DNA pol κ activity detected at levels ~26-fold lower than those for MgCl2 (Fig. 1B and C). The metal ion concentration dependent activities of DNA pol κ are similar to those previously described with other Y-family DNA pols [9]. The maximal Mg2+-activated DNA pol κ activity is detected in the presence of 2–20 mM MgCl2 (Fig. 1B, lanes 6–10 and 19–23). At MgCl2 concentrations above 20 mM, DNA pol κ generates reduced levels of full-length extension products and less-than-full-length primer extension products are detected on the gel as bands 13–14 nucleotides in length (Fig. 1B, lanes 10–13 and 23–26). DNA pol κ catalyzes full-length primer extension in the presence of 0.075–0.25 mM MnCl2 as products are observed on the gel as bands 25 nucleotides in length (Fig. 1C, lanes 6–9 and 19–22). At concentrations of MnCl2 above 0.25 mM, less-than-full-length primer extension products are detected on the gel ranging from 17–22 nucleotides in length (Fig. 1C, lanes 10–13 and 23–26). The accumulation of shorter primer extension products at these higher metal ion concentrations indicates decreased levels of DNA pol κ activity.
3.2 Efficiency and Fidelity of N2-Ethyl-Gua Bypass by DNA Pol κ
A steady-state kinetic assay was performed to precisely quantify the efficiency and fidelity of nucleotide incorporation by DNA pol κ opposite N2-ethyl-Gua and Gua in the presence of MgCl2 or MnCl2. Single nucleotide incorporation experiments were performed with DNA pol κ using 2 mM MgCl2 or 0.075 mM MnCl2 and steady-state kinetic parameters (Km and kcat) were determined. The metal ion concentrations in these steady state experiments are the lowest levels that result in the maximal amount of primer extension observed by DNA pol κ (Fig. 1). For single nucleotide incorporation reactions, a 14-mer primer was annealed to a 32-mer DNA template to position the 3′ terminus one nucleotide before the target N2-ethyl-Gua or Gua. Separate reactions were initiated by addition of increased concentrations of each of the four dNTP’s. The data were quantified and summarized in Table 1 (Supplemental Fig. S1).
Table 1.
Nucleotide insertion opposite Gua and N2-ethyl-Gua by DNA pol κa
| Metal Ion | dNTP | Km (μM) | kcat (min−1) | kcat/Km (min−1 μM−1) | relative insertion frequencyb |
|---|---|---|---|---|---|
| At Template N2-ethyl-Gua | |||||
| 0.075mM Mn2+ | |||||
| Cyt | 1000 ± 300 | 270 ± 50 | 2.7 × 10−1 | 1 | |
| Gua | 70 ± 40 | 1.6 ± 0.4 | 2.2 × 10−2 | 1/12 | |
| Ade | 3.0 ± 2.3 | 0.29 ± .05 | 9.8 × 10−2 | 1/2.7 | |
| Thy | 130 ± 60 | 12.1 ± 2.9 | 9.3 × 10−2 | 1/3 | |
| 2mM Mg2+ | |||||
| Cyt | 1.5 ± 0.1 | 150 ± 3 | 1.0 × 102 | 1 | |
| Gua | 120 ± 15 | 1.9 ± 0.06 | 1.5 × 10−2 | 1/6 700 | |
| Ade | 1300 ± 800 | 2.4 ± 0.82 | 2.0 × 10−3 | 1/50 000 | |
| Thy | 630 ± 140 | 6.4 ± 0.82 | 1.0 × 10−2 | 1/10 000 | |
| At Template Gua | |||||
| 0.075mM Mn2+ | |||||
| Cyt | 470 ± 130 | 240 ± 30 | 5.1 × 10−1 | 1 | |
| Gua | 35 ± 18 | 1.8 ± 0.3 | 5.1 × 10−2 | 1/10 | |
| Ade | 5.5 ± 1.6 | 0.58 ± 0.05 | 1.0 × 10−1 | 1/5 | |
| Thy | 40 ± 15 | 12.2 ± 2.0 | 3.0 × 10−1 | 1/2 | |
| 2mM Mg2+ | |||||
| Cyt | 1.6 ± 0.17 | 150 ± 5 | 0.9 × 102 | 1 | |
| Gua | 160 ± 30 | 1.3 ± 0.06 | 8.1 × 10−3 | 1/12 000 | |
| Ade | 660 ± 150 | 1.6 ± 0.16 | 2.4 × 10−3 | 1/47 000 | |
| Thy | 490 ± 260 | 3.8 ± 0.92 | 7.7 × 10−3 | 1/12 000 | |
Km and kcat values were determined by quantifying gel band intensities using ImageQuant software and non-linear regression analysis of product vs [dNTP] curves using SigmaPlot 8.0.2.
Relative insertion frequency was calculated as 1/([kcat/Km]correct/[kcat/Km]incorrect).
The steady-state kinetic analysis indicates that DNA pol κ bypasses N2-ethyl-Gua with the same efficiency as Gua. The DNA pol κ incorporates the correct dCTP opposite N2-ethyl-Gua with an efficiency approximately 400-fold higher when Mg2+ is the activating divalent metal ion compared to when Mn2+ is the activating metal. In the presence of MgCl2, the kcat/Km value for dCTP incorporation opposite N2-ethyl-Gua is 1.0 × 102 min−1 μM −1 compared to 2.7 × 10−1 min−1 μM−1 in the presence of MnCl2. Similarly, the metal ion effects on the efficiencies of DNA pol κ polymerization opposite Gua parallel those opposite N2-ethyl-Gua. In the presence of MgCl2, the efficiency of DNA pol κ-catalyzed dCTP incorporation opposite Gua, at 0.9 × 102 min−1 μM−1, is ~200-fold higher than that observed in the presence of MnCl2 at 5.1 × 10−1 min−1 μM−1. The higher efficiencies for correct nucleotide incorporation by DNA pol κ in the presence of MgCl2 can be attributed to increased nucleotide binding as is reflected in the lower Km values for dCTP insertion (Table 1). Interestingly, the lower efficiency of DNA pol κ-catalyzed nucleotide incorporation in the presence of MnCl2 contrasts the increased efficiency observed with DNA pol ι using Mn2+ as the activating divalent metal ion [9,32].
The steady-state analysis further indicates that high fidelity DNA pol κ-catalyzed N2-ethyl-Gua bypass is Mg2+-dependent. In the presence of MgCl2, the relative insertion frequencies by DNA pol κ for correct dCTP compared to incorrect dGTP (1/6,700), dATP (1/50,000), and dTTP (1/10,000) opposite N2-ethyl-Gua indicate a relatively high level of fidelity for this Y-family DNA pol during adduct bypass. This level of fidelity is very similar to that measured during bypass of the unadducted template Gua (Table 1). The fidelity of DNA pol κ in the presence of MgCl2 can be attributed to the high Km values and low kcat values for incorporation of the incorrect nucleotides compared to those for dCTP insertion. In contrast, Mn2+ activation of DNA pol κ showed low insertion frequencies for correct dCTP compared to incorrect dGTP (1/12), dATP (1/2.7), and dTTP (1/3) opposite N2-ethyl-Gua indicating a low level of fidelity during adduct bypass and similarly low levels of fidelity during Gua bypass (Table 1). The dramatically lower levels of correct versus incorrect nucleotide incorporation by DNA pol κ in the presence of MnCl2 compared to MgCl2 were the result of a ~600-fold change in the Km value for correct dCTP when Mn2+ is the activating metal compared to when Mg2+ is used (Table 1). Surprisingly, the Km values for incorrect nucleotides measured in the presence of MnCl2 are ~10- to 300-fold lower than those for the correct nucleotide opposite N2-ethyl-Gua or Gua. These data indicate a lack of nucleotide discrimination by DNA pol κ during polymerization opposite N2-ethyl-Gua or Gua upon Mn2+ activation. These Mn2+-related effects on polymerization fidelity by DNA pol κ are similar to those observed for replicative DNA pols [31,34–36]. In particular, this Mn2+-dependent decrease in the Km values for mismatched deoxynucleotides in the polymerase-template complex has been described in fidelity measurements of the T4 DNA pol [37].
3.3 Efficiency of N2-Ethyl-Gua:Cyt extension by DNA Pol κ
The ability of DNA pol κ to extend from the N2-ethyl-Gua-containing template-primer after incorporation of dCTP or dTTP opposite the adduct was measured using a steady-state kinetic assay. The Km and kcat values were determined from extension reactions using two different 15-mer DNA primers containing either Cyt or Thy positioned directly opposite the target N2-ethyl-Gua or Gua. Reactions contained increasing concentrations of dGTP in the presence of MgCl2 or MnCl2 to measure incorporation opposite the next template Cyt positioned 5′ to the target Gua and N2-ethyl-Gua. The data were quantified and the results summarized in Table 2 (Supplemental Fig. S2).
Table 2.
Extension from Gua:Cyt, Gua:Thy and N2-ethyl-Gua:Cyt, N2-ethyl-Gua:Thy base pairs by DNA pol κa
| Metal ion | Base pair | Km (μM) | kcat (min−1) | kcat/Km (min−1 μM−1) | relative extension frequencyb |
|---|---|---|---|---|---|
| At Template N2 -ethyl-Gua | |||||
| 0.075mM Mn2+ | |||||
| N2etGua:Cyt | 1500 ± 1200 | 430 ± 250 | 2.8 × 10−1 | 1 | |
| N2etGua:Thy | 100 ± 20 | 20 ± 1 | 2.0 × 10−1 | 1/1.4 | |
| 2mM Mg2+ | |||||
| N2etGua:Cyt | 90 ± 60 | 390 ± 110 | 4.3 | 1 | |
| N2etGua:Thy | 12 ± 2 | 40 ± 0.7 | 3.3 | 1/1.3 | |
| At Template Gua | |||||
| 0.075mM Mn2+ | |||||
| Gua:Cyt | 630 ± 300 | 200 ± 60 | 3.2 × 10−1 | 1 | |
| Gua:Thy | 70 ± 20 | 30 ± 2 | 4.3 × 10−1 | 1/0.7 | |
| 2mM Mg2+ | |||||
| Gua:Cyt | 50 ± 30 | 530 ± 130 | 11 | 1 | |
| Gua:Thy | 6.9 ± 1.3 | 60 ± 2 | 8.7 | 1/1.2 | |
Km and kcat values were determined by quantifying gel band intensities using ImageQuant software and non-linear regression analysis of product vs [dNTP] curves using SigmaPlot 8.0.2.
Relative extension frequency was calculated as 1/([kcat/Km]correct/[kcat/Km]incorrect).
The steady-state analysis indicates that DNA pol κ extends N2-ethyl-Gua:Cyt 3′-termini with approximately the same efficiency as Gua:Cyt 3′-termini. The kcat/Km value of 4.3 min−1 μM−1 for extension from the N2-ethyl-Gua:Cyt base pair (measured as incorporation of the next correct dGTP nucleotide) is similar to the 11 min−1 μM−1 measured for extension from the Gua:Cyt base pair (Table 2). However, the measured efficiency of extension from the N2-ethyl-Gua:Cyt 3′-termini of 4.3 min−1 μM−1 is approximately 15-fold higher when Mg2+ is the activating divalent metal ion compared to 2.8 × 10−1 min−1 μM−1 when Mn2+ is the activating metal. Furthermore, the extension efficiency from the mismatched N2-ethyl-Gua:Thy 3′ terminus of 3.3 min−1 μM−1 is similar to the efficiency from correct 3′ Gua:Cyt termini (Table 2). These data indicate that DNA pol κ extends the N2-ethyl-Gua:Cyt template-primer substrate to effect highly efficient bypass of the N2-ethyl-Gua lesion.
3.4 Rapid Kinetic Analysis of DNA pol κ-catalyzed nucleotide insertion opposite N2-ethyl-Gua
A rapid kinetic analysis was performed to further investigate the metal-dependent effects on nucleotide binding and polymerization rates of DNA pol κ using Mg2+ and Mn2+ as the activating metals. The results show that DNA pol κ-catalyzed DNA polymerization occurs in two phases of nucleotide incorporation. First, a rapid rate or “burst” (kobs) of product formation is observed. This is followed by a second, slower phase of steady-state (kss) product formation [38]. The burst rate is an observed rate of the chemical reaction at the active site, while the burst amplitude is a stoichiometric measurement of the amount of active enzyme in the reaction [38,39].
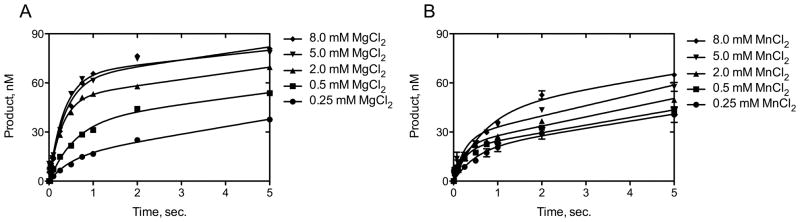
Formation of the optimal DNA pol κ–DNA template:primer-dNTP ternary complex requires Mg2+ as the activating metal as demonstrated in a rapid kinetic “burst” analysis (Fig. 2). The level of DNA pol κ activity measured in the first turnover of substrate to product is dependent upon the metal ion concentration for Mg2+ (Fig. 2A) and for Mn2+ (Fig. 2B). The measured burst of DNA pol κ–single nucleotide incorporation activity increased as the amount of metal ion was increased from 0.25 mM to 8 mM in the reaction (Fig. 2A and B). A maximum burst amplitude of ~70 ± 5 nM is observed at 5 and 8 mM MgCl2 (Fig. 2A). In contrast, a lower burst amplitude of ~40 ± 6 nM is observed using MnCl2 as the activating metal (Fig. 2B). The greater burst amplitude observed using MgCl2 compared to MnCl2 indicates that Mg2+ has greater capacity to generate the active DNA pol κ–DNA template:primer-dNTP ternary complex. The burst rates were similar for DNA pol κ (~2.4 ± 0.5 s−1 in the presence of MgCl2 and ~3.1 ± 0.7 s−1 in the presence of MnCl2) at all metal ion concentrations tested. The similar burst rates observed for DNA pol κ in the presence of MgCl2 and MnCl2 indicate that the Mg2+ or Mn2+ ions may equally facilitate chemistry at the active site.
Figure 2.
A rapid kinetic experiment was used to measure the effects of increasing Mg2+ or Mn2+ concentrations on single nucleotide incorporation by DNA pol κ. The experiment was performed under enzyme limiting conditions (50 nM DNA pol κ, 100 nM template-primer DNA, and 0.5 mM dCTP) with MgCl2 (A) or MnCl2 (B) concentrations of 0.25 mM, 0.5 mM, 2 mM, 5 mM, and 8 mM. Burst amplitudes using MgCl2 were: A = 15 ± 4, 36 ± 4, 51 ± 4, 65 ± 5, and 62 ± 4 nM at each concentration respectively; and using MnCl2 were: A= 19 ± 4, 20 ± 1, 22 ± 2, 27 ± 2, and 43 ± 6 nM, respectively.
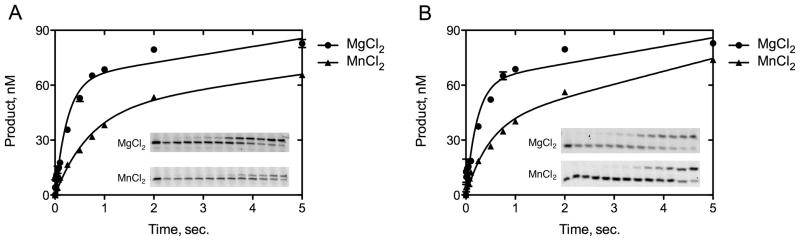
The rapid reaction analysis demonstrates that DNA pol κ catalyzes dCTP incorporation opposite the adduct N2-ethyl-Gua similarly to that opposite the unadducted Gua. The DNA pol κ rapidly incorporates dCTP opposite N2-ethyl-Gua (Fig. 3A) and Gua (Fig. 3B) with burst amplitudes of ~60 ± 4 nM using MgCl2 and ~40 ± 4 nM using MnCl2. Furthermore, DNA pol κ exhibits similar burst rates for dCTP incorporation opposite N2-ethyl-Gua (kobs = 4.0 ± 0.6 s−1) and Gua (kobs = 3.5 ± 0.3 s−1) in the presence of MgCl2 and just slightly lower rates of dCTP incorporation opposite N2-ethyl-Gua (kobs = 2.1 ± 0.3 s−1) and Gua (kobs = 1.4 ± 0.2 s−1) with MnCl2. These data indicate that the DNA pol κ active site accommodates the ethyl moiety of the N2-ethyl-Gua lesion with little or no effect on correct nucleotide incorporation.
Figure 3.
The kinetics of dCTP incorporation opposite Gua and N2-ethyl-Gua were measured in a rapid kinetic experiment under enzyme limiting conditions (50 nM DNA pol κ, 100 nM template-primer DNA, and 8 mM MgCl2 or MnCl2). The data were fit to the burst equation (see Materials and Methods) and the following kinetic parameters were determined: opposite template Gua (A) the kobs = 3.5 ± 0.3 s−1; kss = 4.4 ± 1.0 s−1; A = 63 ± 3 nM in the presence of MgCl2 and using MnCl2 the kobs = 1.4 ± 0.2 s−1; kss = 3.8 ± 1.0 s−1; A = 47 ± 5 nM; opposite template N2-ethyl-Gua (B) the kobs = 4.0 ± 0.6 s−1; kss = 4.7 ± 1.4 s−1; A = 62 ± 4 nM in the presence of MgCl2 and using MnCl2 the kobs = 2.1 ± 0.3 s−1; kss = 7 ± 1 s−1; A = 40 ± 4 nM.
3.5 Activating metal-dependent effects on DNA and dCTP binding in DNA Pol κ
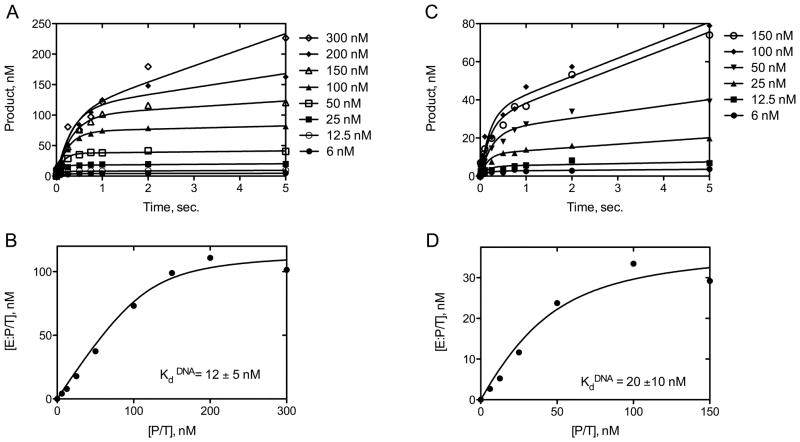
A rapid kinetic experiment was performed to directly measure the metal-dependent effects in DNA Pol κ on DNA binding (KdDNA), nucleotide binding (KddCTP) and on the first-order rate of polymerization (kpol). An active site titration analysis shows that Mg2+ is required to generate maximal levels of active DNA pol κ (Fig. 4). The burst amplitudes were measured using increased concentrations of DNA substrate in the presence of MgCl2 (Fig. 4A) and MnCl2 (Fig. 4C) as the activating metals. A plot of the burst amplitudes (E:P/T) relative to DNA concentrations (P/T) reveals about a 3-fold higher level of active DNA pol κ is generated in the presence of MgCl2 (Fig. 4B) compared to MnCl2 (Fig. 4D). These results show that the DNA pol κ preparation contains approximately 100% active enzyme (~116 ± 7 nM) in the presence of MgCl2 and approximately 40% active enzyme (~40 ± 5 nM) in the presence of MnCl2. The active site titration showed that metal ion activation affects the amount of catalytically competent DNA pol κ ternary complex and further indicates that Mg2+ is the preferred metal for maximal DNA pol κ activity. Interestingly, there was only a slight difference in the measured KdDNA for DNA pol κ whether Mg2+ (12 ± 5 nM) or Mn2+ (20 ± 10 nM) was used.
Figure 4.
Active site titration and determination of KdDNA for DNA Pol κ in the presence of MgCl2 or MnCl2. DNA pol κ (100 nM) was incubated with increasing concentrations of template-primer DNA (6–300 nM) and 500 μM dCTP at different reaction times in the presence of MgCl2 (A–B) or MnCl2 (C–D). The data were fit to the burst equation (A and C). The burst amplitudes ([E:P/T]) were plotted versus template-primer concentrations ([P/T]) (B and D) and fit to the quadratic equation (y = (0.5(Kd+Et+Dt))−((0.25(Kd+Et+Dt)2)−(EtDt))1/2). The active enzyme concentration was determined to be approximately 116 ± 7 nM.
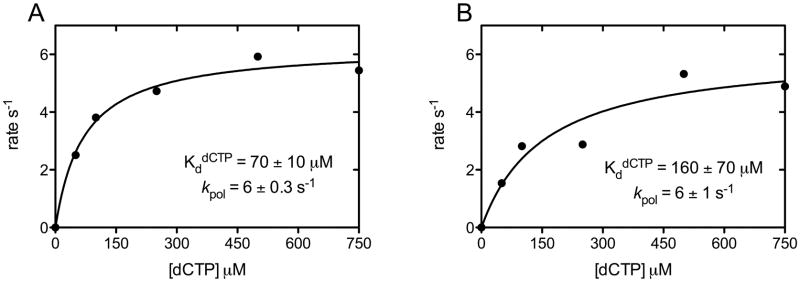
A rapid kinetic analysis further demonstrated that DNA pol κ binds nucleotide more efficiently in the presence of MgCl2 compared to MnCl2 as reflected in the lower nucleotide dissociation constant for dCTP (KddCTP) (Fig. 5). The DNA pol κ (50 nM) was incubated with increased concentrations of dCTP (50–750 μM) in the presence of 8 mM MgCl2 or MnCl2 and polymerization was measured at varied times from 5 msec to 5 s. The observed burst rates were fit to a hyperbolic equation to determine KddCTP and kpol. These data show that the KddCTP is ~70 ± 10 μM in the presence of Mg2+ (Fig. 5A) and ~160 ± 70 μM in the presence of Mn2+ (Fig. 5B). However, this 2-fold difference in the KddCTP does not apparently affect the polymerization rate (kpol) measured as ~6.0 ± 0.3 s−1 with Mg2+ and ~6.0 ± 1.0 s−1 with Mn2+. These rapid kinetic data indicating reduced levels of nucleotide binding by DNA pol κ upon Mn2+ activation relative to Mg2+ activation parrallel our findings of the inefficient incorporation of dCTP in the presence of MnCl2 in the steady-state analysis as reflected in the increased Km value for dCTP. The results of both steady-state and rapid kinetic analyses with DNA pol κ indicate that Mg2+ is preferentially utilized to achieve high efficiency and fidelity during DNA polymerization contrasting our previous findings with the DNA pol ι (9).
Figure 5.
Determination of KddCTP for DNA pol κ in the presence of MgCl2 (A) or MnCl2 (B). DNA pol κ (80 nM) was incubated with 100 nM template-primer DNA and mixed with increasing dCTP (50–750 μM) concentrations. A plot of “burst” rates (s−1) versus dCTP concentration was fit to a hyperbolic equation to determine the maximum rate (kpol) and the dissociation constant (Kd) for dCTP incorporation.
4. Discussion
The results presented here demonstrate that DNA pol κ is efficient at both the insertion and extension steps during bypass of N2-ethyl-Gua and indicate a role for DNA pol κ in translesion synthesis past bulky N2-alkyl-Gua lesions in vivo. Previous studies have shown that DNA pol κ is efficient at both the incorporation and extension steps when bypassing bulky N2-alkyl-Gua lesions such as N2-CH2(9-anthracenyl)-Gua, N2-furfuryl-Gua, and N2-BP-Gua [11,23–25]. Furthermore, mice that are deficient for DNA pol κ have been shown to have a spontaneous mutator phenotype with a broad mutation spectrum that includes Gua→Cyt transversion, and Gua→Ade and Gua→Thy transition mutations suggesting that in the absence of DNA pol κ error-prone translesion synthesis, past bulky Gua adducts, is carried out by other DNA pols. Also, the DNA pol κ −/− mouse cells are sensitive to treatment with benzo[a]pyrene-dihydrodiol epoxide suggesting that DNA pol κ functions to bypass N2-Gua lesions in vivo [40].
There is evidence that DNA pol κ is efficient at extending mis-matched primer termini and it has been hypothesized that DNA pol κ could act mainly at the extension step in a two-DNA polymerase-mechanism of lesion bypass [21,22,41]. Our data show the high efficiency of DNA pol κ-catalyzed incorporation of dCTP opposite N2-ethyl-Gua. Further, the DNA pol κ exhibits similar efficiencies extending from the Gua:Cyt or the Gua:Thy 3′-termini in the presence of MgCl2 and only a ~2.5 fold lower extension efficiency from the N2-ethyl-Gua:Cyt or N2-ethyl-Gua:Thy 3′-termini. Together, these data support a model where DNA pol κ participates at both the incorporation and extension steps during translesion synthesis of minor groove DNA adducts.
Y-family DNA pols bypass bulky DNA adducts that would otherwise be blocking to replicative DNA pols. The x-ray crystal structures of several Y-family DNA pols reveal that decreased flexibility in the nucleotide binding domain and an open active site contribute to the accommodation of bulky lesions and their efficient bypass during replication [42]. The size and shape of the DNA polymerase active site is not, however, the only factor in determining facile bypass of DNA adducts by Y-family DNA pols. This is evidenced by the fact that not all adducts are efficiently bypassed by all members of the Y-family. The Y-family DNA pols η, ι, and κ are able to bypass the N2-ethyl-Gua adduct but each recognize and bypass the lesion by distinct catalytic mechanisms [6,9–12]. DNA pol ι bypasses N2-ethylGua by rotating the adducted template base from the anti to syn configuration and forming Hoogsteen base pairs with the incoming nucleotides [9,43]. Biochemical data indicate that the DNA pols η and κ require Watson-Crick base pairing opposite undamaged DNA substrates [44,45]. Structural modeling of the N2-ethyl-Gua lesion into the active sites of DNA pols η and κ suggest that these DNA pols utilize wobble base pairing as a mechanism to avoid steric interference in forming the N2-ethyl-Gua:Cyt base pair during bypass of the adduct [9].
The rapid kinetic experiments with DNA pol κ indicate that the burst rates and burst amplitudes are the same for dCTP incorporation opposite Gua and N2-ethyl-Gua. These data suggest that DNA pol κ forms a wobble base pair (Supplemental Fig. 3) between the incoming dCTP and template N2-ethyl-Gua because there is no apparent interference by the adduct with formation of the active DNA pol κ:dNTP complex. The ethyl lesion at the N2 position of Gua might be predicted to sterically inhibit the formation of a Watson-Crick hydrogen bond with the O2 atom of Cyt. The N2-ethyl-Gua would therefore inhibit dCTP binding at the DNA pol κ active site via Watson-Crick hydrogen bonding resulting in a predicted decreased burst amplitude relative to that observed using a Gua containing DNA template. Alternatively, if a Watson-Crick hydrogen bond were formed between N2-ethyl-Gua and dCTP with the ethyl moiety rotated around the N2 atom in an anti conformation with respect to the N1 of Gua, lower burst amplitudes would be expected opposite N2-ethyl-Gua given the potential for the ethyl lesion to sample different rotational conformations during the reaction. Our data show that the burst amplitudes are the same whether Gua or N2-ethyl-Gua is used as the template base suggesting that a wobble base pair forms between N2-ethyl-Gua and incoming dCTP relieving the potential for steric overlap that would inhibit positioning of the incoming nucleotide for catalysis.
The activating metal ion affects the catalytic mechanism of Y-family DNA pols and is an important factor for maintaining high catalytic efficiency and maximum fidelity during translesion synthesis. The primer extension data demonstrate that DNA pol κ is fully active at low concentrations of Mg2+ or Mn2+ ions and the activity is metal ion concentration dependent. Steady state and rapid kinetic analyses with DNA pol κ demonstrate that MnCl2 activation results in decreased efficiency and lower fidelity during nucleotide incorporation opposite both adducted and unadducted DNA templates. These data contrast our findings with DNA pol ι that has higher activity in the presence of Mn2+ (9). The DNA pol κ preference for Mg2+ as the activating divalent metal ion for high fidelity DNA replication is more similar to replicative DNA pols than to DNA pol ι. This preference for Mg2+ by DNA pol κ may reflect the higher fidelity of DNA pol κ when compared to other Y-family DNA pols when replicating unadducted DNA templates [15,18,19].
A surprising finding in our work is detection of the increased Kd for dCTP binding at the DNA pol κ active site with Mn2+ ion relative to Mg2+ without a significant change to the rate of the chemical step. Together with our previous work these results suggest that differences in the nucleotide incorporation efficiencies of DNA pols κ and ι in the presence of MnCl2 likely reflect differences in shape and size of the active sites and the postions of DNA and incoming nucleotides for catalysis. DNA pol ι has a narrow binding pocket that makes tight contacts with both the template base and incoming nucleotide resulting in a shortened C1′-C1′ distance [46]. In contrast, DNA pol κ coordinates the DNA template more loosely and may thus rely more heavily on metal ion binding for coordination and correct positioning of the incoming dCTP. Differences in the inter-atomic distances between Mg2+ (3.197 Å) and Mn2+ (less than 3.0 Å) may affect the geometry of the DNA pol κ active site more than that of DNA pol ι [29]. The distance between two Mg2+ ions (usually 4 Å) is greater than that compared to two Mn2+ ions and would result in a tighter coordination of the incoming dCTP in the DNA pol κ active site than would Mn2+. The smaller distance between two Mn2+ ions would help to coordinate the incoming dNTP in the more confined active site of DNA pol ι [29]. The structures of DNA pol ι solved in the presence of Mg2+ show poor electron density around the proposed metal ion-binding sites [9,43,46]. A partial exclusion of Mg2+ from the DNA pol ι active site could be due to a preference for Mn2+ and its smaller inter-atomic distances that would help facilitate productive binding of the tri-phosphate nucleotide [32]. The structure of DNA pol ι reveals that the cysteine residue at position 196 lies in close proximity to the active site and may contribute to the coordination of Mn2+ that has a higher affinity for sulfur than Mg2+ [47]. These observations could explain the dramatic activation of DNA pol ι and less than efficient polymerization by DNA pol κ in the presence of Mn2+ [9,32].
The different properties of the DNA pols κ and ι observed when Mn2+ is the activating divalent metal ion suggest distinct biological roles for these DNA pols dependent upon metal ion concentrations. It is possible that small changes to the nuclear environment that alter concentrations of divalent metal ions could contribute to the specific recruitment and activation of a particular Y-family DNA pol at a blocked replication fork to facilitate lesion bypass. The concentrations of metals in the cell are tightly regulated by efflux and influx of metal ions through membrane transporters [48,49]. A transient rise in the concentration of Mn2+ at the replication fork, for example, could act to inhibit DNA pol κ from participating in lesion bypass by decreasing its incorporation and extension efficiencies while stimulating the activity of DNA pol ι for efficient translesion synthesis. Transient regulation of DNA pol activity through changing metal ion concentrations at a blocked DNA replication fork would be beneficial for genomic integrity. A rapid and short-lived response would relieve a stalled replication fork quickly and eliminate unnecessary, prolonged DNA synthesis by the low-fidelity Y-family DNA pols thus permitting efficient bypass of the DNA adduct while preserving the overall genome integrity.
Supplementary Material
Acknowledgments
We thank Robert L. Eoff and F. Peter Guengerich for use and technical support on the KinTek Rapid Quench apparatus.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:
Footnotes
This work was supported by National Institutes of Health [GM069962 to F.W.P., CA52881 to J.C.F.].
Conflict of interest
The authors declare that there are no conflicts of interest.
Abbreviations: BSA, bovine serum albumin; DNA pol ι, DNA polymerase ι; N2-ethyl-Gua, N2-ethylguanine; pol, polymerase.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fang JL, Vaca CE. Development of a 32P-postlabelling method for the analysis of adducts arising through the reaction of acetaldehyde with 2′-deoxyguanosine-3′-monophosphate and DNA. Carcinogenesis. 1995;16:2177–2185. doi: 10.1093/carcin/16.9.2177. [DOI] [PubMed] [Google Scholar]
- 2.Fang JL, Vaca CE. Detection of DNA adducts of acetaldehyde in peripheral white blood cells of alcohol abusers. Carcinogenesis. 1997;18:627–632. doi: 10.1093/carcin/18.4.627. [DOI] [PubMed] [Google Scholar]
- 3.Baan R, Straif K, Grosse Y, Secretan B, El Ghissassi F, Bouvard V, Altieri A, Cogliano V. Carcinogenicity of alcoholic beverages. Lancet Oncol. 2007;8:292–293. doi: 10.1016/s1470-2045(07)70099-2. [DOI] [PubMed] [Google Scholar]
- 4.Terashima I, Matsuda T, Fang TW, Suzuki N, Kobayashi J, Kohda K, Shibutani S. Miscoding potential of the N2-ethyl-2′-deoxyguanosine DNA adduct by the exonuclease-free Klenow fragment of Escherichia coli DNA polymerase I. Biochemistry. 2001;40:4106–4114. doi: 10.1021/bi002719p. [DOI] [PubMed] [Google Scholar]
- 5.Seitz HK, Becker P. Alcohol metabolism and cancer risk. Alcohol Res Health. 2007;30:38–41. 44–37. [PMC free article] [PubMed] [Google Scholar]
- 6.Perrino FW, Blans P, Harvey S, Gelhaus SL, McGrath C, Akman SA, Jenkins GS, LaCourse WR, Fishbein JC. The N2-ethylguanine and the O6-ethyl- and O6-methylguanine lesions in DNA: contrasting responses from the “bypass” DNA polymerase eta and the replicative DNA polymerase alpha. Chem Res Toxicol. 2003;16:1616–1623. doi: 10.1021/tx034164f. [DOI] [PubMed] [Google Scholar]
- 7.Choi JY, Guengerich FP. Analysis of the effect of bulk at N2-alkylguanine DNA adducts on catalytic efficiency and fidelity of the processive DNA polymerases bacteriophage T7 exonuclease- and HIV-1 reverse transcriptase. J Biol Chem. 2004;279:19217–19229. doi: 10.1074/jbc.M313759200. [DOI] [PubMed] [Google Scholar]
- 8.Upton DC, Wang X, Blans P, Perrino FW, Fishbein JC, Akman SA. Replication of N2-ethyldeoxyguanosine DNA adducts in the human embryonic kidney cell line 293. Chem Res Toxicol. 2006;19:960–967. doi: 10.1021/tx060084a. [DOI] [PubMed] [Google Scholar]
- 9.Pence MG, Blans P, Zink CN, Hollis T, Fishbein JC, Perrino FW. Lesion bypass of N2-ethylguanine by human DNA polymerase iota. J Biol Chem. 2009;284:1732–1740. doi: 10.1074/jbc.M807296200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi JY, Guengerich FP. Kinetic evidence for inefficient and error-prone bypass across bulky N2-guanine DNA adducts by human DNA polymerase iota. J Biol Chem. 2006;281:12315–12324. doi: 10.1074/jbc.M600112200. [DOI] [PubMed] [Google Scholar]
- 11.Choi JY, Angel KC, Guengerich FP. Translesion synthesis across bulky N2-alkyl guanine DNA adducts by human DNA polymerase kappa. J Biol Chem. 2006;281:21062–21072. doi: 10.1074/jbc.M602246200. [DOI] [PubMed] [Google Scholar]
- 12.Choi JY, Guengerich FP. Adduct size limits efficient and error-free bypass across bulky N2-guanine DNA lesions by human DNA polymerase eta. J Mol Biol. 2005;352:72–90. doi: 10.1016/j.jmb.2005.06.079. [DOI] [PubMed] [Google Scholar]
- 13.Swan MK, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Structural basis of high-fidelity DNA synthesis by yeast DNA polymerase delta. Nat Struct Mol Biol. 2009;16:979–986. doi: 10.1038/nsmb.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franklin MC, Wang J, Steitz TA. Structure of the replicating complex of a pol alpha family DNA polymerase. Cell. 2001;105:657–667. doi: 10.1016/s0092-8674(01)00367-1. [DOI] [PubMed] [Google Scholar]
- 15.Ohashi E, Bebenek K, Matsuda T, Feaver WJ, Gerlach VL, Friedberg EC, Ohmori H, Kunkel TA. Fidelity and processivity of DNA synthesis by DNA polymerase kappa, the product of the human DINB1 gene. J Biol Chem. 2000;275:39678–39684. doi: 10.1074/jbc.M005309200. [DOI] [PubMed] [Google Scholar]
- 16.Gerlach VL, Feaver WJ, Fischhaber PL, Friedberg EC. Purification and characterization of pol kappa, a DNA polymerase encoded by the human DINB1 gene. J Biol Chem. 2001;276:92–98. doi: 10.1074/jbc.M004413200. [DOI] [PubMed] [Google Scholar]
- 17.Gerlach VL, Aravind L, Gotway G, Schultz RA, Koonin EV, Friedberg EC. Human and mouse homologs of Escherichia coli DinB (DNA polymerase IV), members of the UmuC/DinB superfamily. Proc Natl Acad Sci USA. 1999;96:11922–11927. doi: 10.1073/pnas.96.21.11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y, Yuan F, Xin H, Wu X, Rajpal DK, Yang D, Wang Z. Human DNA polymerase kappa synthesizes DNA with extraordinarily low fidelity. Nucleic Acids Res. 2000;28:4147–4156. doi: 10.1093/nar/28.21.4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y, Yuan F, Wu X, Wang M, Rechkoblit O, Taylor JS, Geacintov NE, Wang Z. Error-free and error-prone lesion bypass by human DNA polymerase kappa in vitro. Nucleic Acids Res. 2000;28:4138–4146. doi: 10.1093/nar/28.21.4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carlson KD, Johnson RE, Prakash L, Prakash S, Washington MT. Human DNA polymerase kappa forms nonproductive complexes with matched primer termini but not with mismatched primer termini. Proc Natl Acad Sci USA. 2006;103:15776–15781. doi: 10.1073/pnas.0605785103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Washington MT, Johnson RE, Prakash L, Prakash S. Human DINB1-encoded DNA polymerase kappa is a promiscuous extender of mispaired primer termini. Proc Natl Acad Sci USA. 2002;99:1910–1914. doi: 10.1073/pnas.032594399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haracska L, Prakash L, Prakash S. Role of human DNA polymerase kappa as an extender in translesion synthesis. Proc Natl Acad Sci USA. 2002;99:16000–16005. doi: 10.1073/pnas.252524999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rechkoblit O, Zhang Y, Guo D, Wang Z, Amin S, Krzeminsky J, Louneva N, Geacintov NE. trans-Lesion synthesis past bulky benzo[a]pyrene diol epoxide N2-dG and N6-dA lesions catalyzed by DNA bypass polymerases. J Biol Chem. 2002;277:30488–30494. doi: 10.1074/jbc.M201167200. [DOI] [PubMed] [Google Scholar]
- 24.Jarosz DF, Godoy VG, Delaney JC, Essigmann JM, Walker GC. A single amino acid governs enhanced activity of DinB DNA polymerases on damaged templates. Nature. 2006;439:225–228. doi: 10.1038/nature04318. [DOI] [PubMed] [Google Scholar]
- 25.Jia L, Geacintov NE, Broyde S. The N-clasp of human DNA polymerase kappa promotes blockage or error-free bypass of adenine- or guanine-benzo[a]pyrenyl lesions. Nucleic Acids Res. 2008;36:6571–6584. doi: 10.1093/nar/gkn719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lone S, Townson SA, Uljon SN, Johnson RE, Brahma A, Nair DT, Prakash S, Prakash L, Aggarwal AK. Human DNA polymerase kappa encircles DNA: implications for mismatch extension and lesion bypass. Mol Cell. 2007;25:601–614. doi: 10.1016/j.molcel.2007.01.018. [DOI] [PubMed] [Google Scholar]
- 27.Castro C, Smidansky ED, Arnold JJ, Maksimchuk KR, Moustafa I, Uchida A, Gotte M, Konigsberg W, Cameron CE. Nucleic acid polymerases use a general acid for nucleotidyl transfer. Nat Struct Mol Biol. 2009;16:212–218. doi: 10.1038/nsmb.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steitz TA. A mechanism for all polymerases. Nature. 1998;391:231–232. doi: 10.1038/34542. [DOI] [PubMed] [Google Scholar]
- 29.Yang W, Lee JY, Nowotny M. Making and breaking nucleic acids: two-Mg2+-ion catalysis and substrate specificity. Mol Cell. 2006;22:5–13. doi: 10.1016/j.molcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 30.Sirover MA, Loeb LA. Metal activation of DNA synthesis. Biochem Biophys Res Commun. 1976;70:812–817. doi: 10.1016/0006-291x(76)90664-1. [DOI] [PubMed] [Google Scholar]
- 31.Sirover MA, Loeb LA. Metal-induced infidelity during DNA synthesis. Proc Natl Acad Sci USA. 1976;73:2331–2335. doi: 10.1073/pnas.73.7.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frank EG, Woodgate R. Increased catalytic activity and altered fidelity of human DNA polymerase iota in the presence of manganese. J Biol Chem. 2007;282:24689–24696. doi: 10.1074/jbc.M702159200. [DOI] [PubMed] [Google Scholar]
- 33.Boosalis MS, Petruska J, Goodman MF. DNA polymerase insertion fidelity. Gel assay for site-specific kinetics. J Biol Chem. 1987;262:14689–14696. [PubMed] [Google Scholar]
- 34.Sirover MA, Loeb LA. Infidelity of DNA synthesis in vitro: screening for potential metal mutagens or carcinogens. Science. 1976;194:1434–1436. doi: 10.1126/science.1006310. [DOI] [PubMed] [Google Scholar]
- 35.Kunkel TA, Loeb LA. On the fidelity of DNA replication. Effect of divalent metal ion activators and deoxyrionucleoside triphosphate pools on in vitro mutagenesis. J Biol Chem. 1979;254:5718–5725. [PubMed] [Google Scholar]
- 36.Rabkin SD, Moore PD, Strauss BS. In vitro bypass of UV-induced lesions by Escherichia coli DNA polymerase I: specificity of nucleotide incorporation. Proc Natl Acad Sci USA. 1983;80:1541–1545. doi: 10.1073/pnas.80.6.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goodman MF, Keener S, Guidotti S, Branscomb EW. On the enzymatic basis for mutagenesis by manganese. J Biol Chem. 1983;258:3469–3475. [PubMed] [Google Scholar]
- 38.Johnson KA. Rapid quench kinetic analysis of polymerases, adenosinetriphosphatases, and enzyme intermediates. Methods Enzymol. 1995;249:38–61. doi: 10.1016/0076-6879(95)49030-2. [DOI] [PubMed] [Google Scholar]
- 39.Patel SS, Wong I, Johnson KA. Pre-steady-state kinetic analysis of processive DNA replication including complete characterization of an exonuclease-deficient mutant. Biochemistry. 1991;30:511–525. doi: 10.1021/bi00216a029. [DOI] [PubMed] [Google Scholar]
- 40.Stancel JN, McDaniel LD, Velasco S, Richardson J, Guo C, Friedberg EC. Polk mutant mice have a spontaneous mutator phenotype. DNA Repair. 2009;8:1355–1362. doi: 10.1016/j.dnarep.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Washington MT, Minko IG, Johnson RE, Wolfle WT, Harris TM, Lloyd RS, Prakash S, Prakash L. Efficient and error-free replication past a minor-groove DNA adduct by the sequential action of human DNA polymerases iota and kappa. Mol Cell Biol. 2004;24:5687–5693. doi: 10.1128/MCB.24.13.5687-5693.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu Rev Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 43.Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Human DNA polymerase iota incorporates dCTP opposite template G via a G.C + Hoogsteen base pair. Structure. 2005;13:1569–1577. doi: 10.1016/j.str.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 44.Washington MT, Helquist SA, Kool ET, Prakash L, Prakash S. Requirement of Watson-Crick hydrogen bonding for DNA synthesis by yeast DNA polymerase eta. Mol Cell Biol. 2003;23:5107–5112. doi: 10.1128/MCB.23.14.5107-5112.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wolfle WT, Washington MT, Kool ET, Spratt TE, Helquist SA, Prakash L, Prakash S. Evidence for a Watson-Crick hydrogen bonding requirement in DNA synthesis by human DNA polymerase kappa. Mol Cell Biol. 2005;25:7137–7143. doi: 10.1128/MCB.25.16.7137-7143.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nair DT, Johnson RE, Prakash S, Prakash L, Aggarwal AK. Replication by human DNA polymerase-iota occurs by Hoogsteen base-pairing. Nature. 2004;430:377–380. doi: 10.1038/nature02692. [DOI] [PubMed] [Google Scholar]
- 47.Tari LW, Matte A, Goldie H, Delbaere LTJ. Mg2+-Mn2+ clusters in enzyme-catalyzed phosphoryl-transfer reactions. Nat Struct Biol. 1997;4:990–994. doi: 10.1038/nsb1297-990. [DOI] [PubMed] [Google Scholar]
- 48.Romani A. Regulation of magnesium homeostasis and transport in mammalian cells. Arch Biochem Biophys. 2007;458:90–102. doi: 10.1016/j.abb.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 49.Kehres DG, Zaharik ML, Finlay BB, Maguire ME. The NRAMP proteins of Salmonella typhimurium and Escherichia coli are selective manganese transporters involved in the response to reactive oxygen. Mol Microbiol. 2000;36:1085–1100. doi: 10.1046/j.1365-2958.2000.01922.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.