Intramolecular O-N acyl migrations are well known reactions in organic chemistry[1-3]. This type of rearrangement has been the object of renewed interest in the field of chemical biology as it provides opportunities for the spatial and temporal control of peptide conformation and function.[4, 5] Incorporation of an O-acyl isomer of a peptide bond at a serine or threonine residue in a peptide sequence (herein referred to as an O-acyl linkage) introduces a kink into the main chain of the biopolymer and interrupts the normal backbone hydrogen-bonding characteristics, both of which can affect structure and function.[5-7] In the case where the Ser/Thr α-amino group is transiently protected, recovery of the normal amide backbone and, as a consequence, native structure and function, can be achieved by removal of this protecting group, thereby triggering the spontaneous O-N acyl shift. Accordingly, a variety of triggers of O-N acyl migrations have been developed that allow the restoration of native structure following the administration of light,[8] pH,[9] or enzymatic activity.[10]
Despite the successful use of O-N acyl migrations to control peptide conformation and function,[11-13] the strategy has yet to be applied to more complex processes such as the regulation of protein catalysis. Of particular interest to us, was whether a reversible O-acyl modification could be used to control protein splicing, a naturally occurring autocatalytic process in which an internal protein domain (intein) removes itself from a polypeptide sequence with concomitant ligation of its flanking regions (exteins).[14, 15] Engineered inteins have found widespread use in the biotechnology and chemical biology areas.[16] Of relevance to the current study are the conditional protein trans-splicing (CPS) systems which permit the activity of a given protein to be controlled by triggering the assembly of an artificially split intein.[17-19] In principle, use of the O-N acyl migration strategy would allow other types of chemical and biochemical triggers to be incorporated into these CPS systems. Moreover, it might be possible to generate conditional inteins based on the naturally split DnaE inteins,[20] which are incompatible with current CPS strategies due to their self-assembling nature.
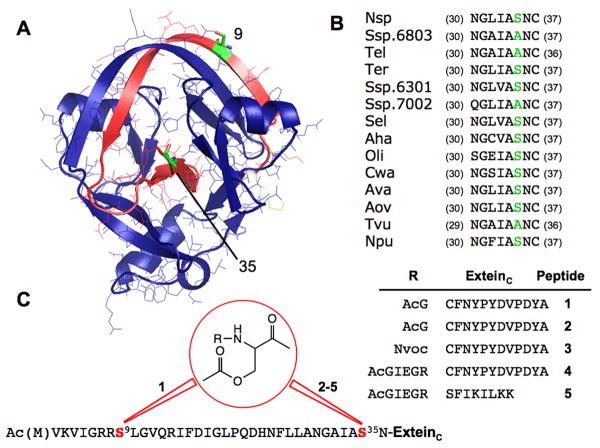
We hypothesized that the trans-splicing activity of the split Ssp DnaE intein could be controlled through the judicious introduction of an O-acyl linkage (Scheme 1). Analysis of the crystal structure of Ssp DnaE suggested two potential sites for the modification (Figure 1A). The first site, Ser-9, is the only native Ser/Thr residue in the C-terminal half of the split intein (referred to as DnaEC). This residue is located in the middle of a long β-strand that forms an anti-parallel β-sheet with the DnaEN fragment in the intein complex.[21] Introduction of a kink in this β-strand would be expected to disrupt this key element of secondary structure, destabilize the complex and prevent splicing. The second site was Ala-35, which is a serine in many other inteins of the DnaE family and is in the vicinity of the intein active site (Figure 1A, B). Introduction of a main chain kink at this position would be expected to alter the position of catalytic residues, which could lead to a loss of splicing activity. Importantly, both these sites are within the short DnaEC fragment, which we have previously shown is accessible to chemical synthesis.[22, 23] Accordingly, we synthesized two DnaEC analogues containing an O-acyl linkage at the β-hydroxyl of either Ser 9 (peptide 1) or 35 (peptide 2) and their α-amino groups irreversibly blocked by acetylglycine (Figure 1C; Supporting Information)
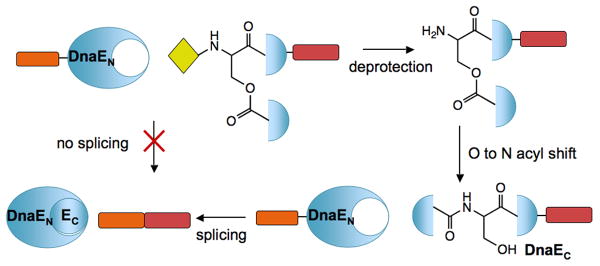
Scheme 1.
Conditional protein splicing based on initiation of an O-N acyl migration. An O-acyl linkage judiciously placed within the sequence of DnaEC blocks protein trans-splicing activity, but upon rearrangement to an amide bond the intein regains normal activity.
Figure 1.
A) Crystal structure of Ssp DnaE intein.[21] DnaEN is shown in blue and DnaEC in red, with residues 9 and 35 highlighted in green. B) Sequence alignment of the conserved Block G of the DnaE intein family showing that both Ala and Ser are found at position 35. C) Structures of O-acyl peptide analogs of DnaEC used in this study. An O-acyl linkage was introduced at position 9 for peptide 1 and at position 35 for analogues 2-5.
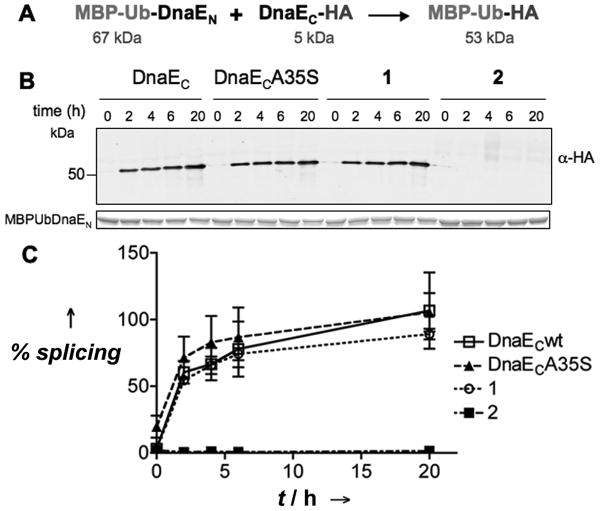
To evaluate the effect of each O-acyl linkage on trans-splicing activity, we compared splicing reactions between DnaEN and either wild-type DnaEC, an Ala35Ser mutant, O-acyl analog 1 or O-acyl analog 2. As model extein sequences, we fused maltose binding protein linked to ubiquitin (MBP-Ub) and a HA peptide tag to the N- and C-terminus of DnaEN and DnaEC, respectively (Figure 2A). The reactions were followed by western blot using an anti-HA antibody to detect formation of the MBP-Ub-HA splicing product (Figure 2). As predicted by the sequence analysis, the DnaECA35S mutant had identical activity to the wild-type DnaEC. More surprising was the splicing activity of O-acyl analog 1, which was also indistinguishable from that of the wild-type DnaEC. This indicates that the split intein can tolerate local disruption to the interfacial β-sheet caused by the O-acyl linkage. In contrast, no splicing product was observed for the reaction between O-acyl analog 2 and DnaEN. Moreover, HPLC and MS analysis of this reaction indicated that known splicing side-reactions, such as N-terminal cleavage,[24] also did not occur at detectable levels (Supporting Information). Thus, introduction of the O-acyl linkage at Ser35 blocks all trans-splicing activity of DnaEC. Steady-state and stopped-flow fluorescence experiments showed that O-acyl analog 2 associates with DnaEN with similar affinity and kinetics to wild-type DnaEC, and circular dichroism spectroscopy indicated the secondary and tertiary structure of the DnaEN/O-acyl analog 2 complex was not globally perturbed from that of the all-amide one (Supplemental Information). From this we conclude that the lack of trans-splicing activity associated with O-acyl analog 2 is due to local perturbations in the active site of the complex, rather than an inability of the fragment to associate with DnaEN and adopt the canonical intein fold.
Figure 2.
Effect of O-acyl linkages on DnaE mediated trans-splicing. A) Scheme of the trans-splicing reaction with the indicated model exteins. B) Western blot analysis with an anti-HA antibody of representative splicing reactions between DnaEN and DnaEC constructs at 20 and 12.5 μM, respectively, in splicing buffer (100 mM phosphate, 150 mM NaCl, 1mM EDTA, 1 mM DTT, pH 7.5). Bottom box: Coomassie stained loading control showing MBP-Ub-DnaEN. The splicing product (MBP-Ub-HA) can be detected as a band at just above 50 kDa in the western blot. C) Plot of the fraction splice product formed at different time-points relative to the maximum level of obtained for the wild-type intein (wt intein reaction went to ≈ 50 % completion, Supporting Information). Errors = s.e.m (n = 3).
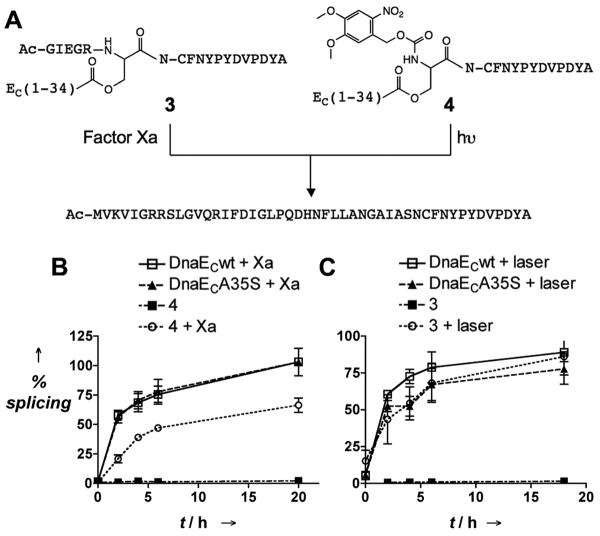
We next asked whether O-acyl analog 2 could be used as the basis of a CPS system. Triggering the O-N acyl shift at this site would generate the DnaECA35S mutant, which we have already shown supports trans-splicing (Figure 2). Thus, incorporation of a transient protecting group on the Ser35 α-amino group should lead to a conditional DnaE intein. Many α-amino protecting groups are known to be viable triggers for this kind of rearrangement.[8, 10, 13] We were especially attracted to protecting groups that are removed by proteases or light. Given that proteases are implicated in the pathology of many diseases,[25] the ability to activate splicing in response to their activities could lead to conditional inteins which sense the presence of these disease markers or generate polypeptide therapeutics (e.g. a toxin) in response to them. Photo-removable protecting groups have been widely used in the development of caged versions of bioactive molecules, including inteins,[26] Accordingly, we synthesized two protected versions of O-acyl peptide 2, one containing a photocleavable Nvoc group on the α-amino group of Ser35 (O-acyl analog 3) and one containing a Factor Xa protease recognition sequence at the same position (O-acyl analog 4) (Figure 3A). Initial studies demonstrated that peptides 3 and 4 could be de-protected by treatment with 325 nm laser light and Factor Xa protease, respectively, and that spontaneous O-N acyl migration then ensues (Supporting Information). As expected, in their protected forms, both 3 and 4 were inactive in our trans-splicing assay with DnaEN (Figure 3). However, exposure of the splicing reactions to the appropriate stimuli, either UV light or the protease, led to dramatic increases in the amount of splice product formed. The kinetics of the splicing reaction between 3 and DnaEN were indistinguishable from the wild-type reaction, whereas we observed a slight decrease in both the rate and extent of the reaction of 4 with DnaEN, compared to the controls. This difference in behavior likely reflects the significantly slower kinetics associated with the enzymatic deprotection compared to the photolysis reaction, and also the fact that a small amount of non-specific proteolysis of the intein occurs with prolonged incubations with Factor Xa (Supporting Information). Nonetheless, the key result is that we were able to trigger the DnaE trans-splicing reaction using either a photochemical or a biochemical stimulus.
Figure 3.
Light and protease triggered O-N acyl migration to activate protein trans-splicing. A) Scheme showing the basis of protease- and photo-mediated activation of O-acyl DnaEC analogs. B) Protease activated protein trans-splicing: MBP-Ub-DnaEN and indicated peptides were incubated at 20 and 12.5 mM, respectively, in buffer (20 mM Tris, 100 mM NaCl, 2 mM CaCl2, 1 mM DTT, pH 8.0) with or without Factor Xa (0.1 U / 50 μg) protease, errors = s.e.m. (n=3). C) Light activated protein trans-splicing: MBP-Ub-DnaEN and indicated peptides were incubated at 20 and 12.5 μM, respectively, in splicing buffer with or without 325 nm laser irradiation, errors = s.e.m. (n=3). The reactions were monitored as in Figure 2.
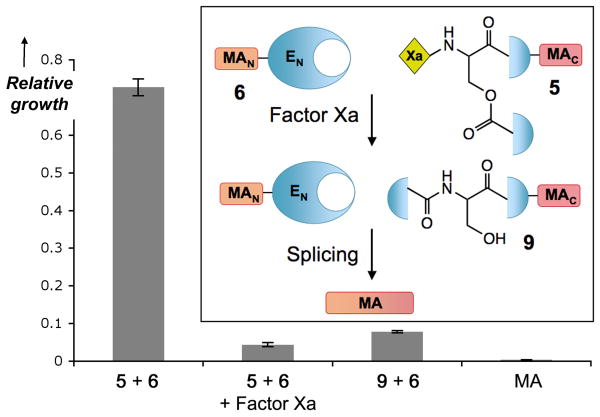
Finally, to illustrate the generality and potential applications of our CPS system we asked whether the approach could be used to proteolytically trigger the synthesis of a toxin. Among the many extein options possible, we focused on the antimicrobial peptides,[27] as the vast structure-activity relationship literature available on these molecules helped with the design of functionally silent mutations that would allow the use of DnaE trans-splicing for the assembly of the peptide. Accordingly, we designed an analog of magainin[28] that had native like activity and could be split and regenerated upon DnaE-mediated trans-splicing leading to an antimicrobial response (Supporting Information and Figure 4). We then fused the C-terminus of this magainin peptide to O-acyl analog 4, which contains the Factor Xa trigger, to give peptide 5. Consistent with the initial model studies, we could not detect any trans-splicing between 5 and the corresponding DnaEN fused to the N-terminus of magainin (protein 6) in the absence of the protease. Treatment of 5 with Factor Xa in the presence 6 resulted in the formation of magainin and the recovery of antimicrobial activity against both E. coli and S. aureus strains, thus demonstrating the potential of the approach (Figure 4 and Supporting Information). By exporting this system to more active toxins under the control of pathogenesis related proteases, potent and specific antimicrobial agents could be developed.
Figure 4.
Protease activation of intein mediated antimicrobial activity. Growth of E. coli incubated with constructs 5 and 6 at 30 μM in the presence and absence of Factor Xa protease. Magainin (MA) and the constitutively active wild type intein (9 + 6) at 30 μM are also shown as positive controls. Bacterial growth (OD650 after 18 h incubation) of each sample relative to a culture control is shown, errors = s.d. (n=3). Inset) Scheme of the DnaE mediated splicing of magainin.
Our results demonstrate that an O-acyl linkage conveniently placed at the Ser35 side chain of the C-terminal fragment of the DnaE intein abolishes protein splicing. The modification does not have a significant effect on either the association of N- and C-terminal intein fragments or the global conformation of the complex between them, which indicates that inhibition is accomplished by a localized perturbation of the intein active site. More importantly, intein activity can be recovered by an O-N acyl migration triggered by proteolytic or photochemical removal of specific protecting groups at the switch site. This result is, to the best of our knowledge, the first application of the O-N acyl shift method for the control of protein activity. In principle, this type of reversible modification could be used to control the activity of other proteins accessible to chemical synthesis.
Supplementary Material
Footnotes
We thank Prof. D. Raleigh and H. Taskent for access to stop-flow and circular dichroism instruments and to them and E. George for helpful discussions. This work was supported by NIH grants EB001991 and GM55843 to T.W.M. M.V.P. and M.R. were supported by the Spanish Ministry of Science and Education.
Supporting information for this article is available on the WWW under http://www.angewandte.org or from the author.
References
- 1.Moore JA, Dice JR, Nicolaides ED, Westland RD. J Am Chem Soc. 1954;76:2884–2887. [Google Scholar]
- 2.Sakakibara S, Shin KH, Hess GP. J Am Chem Soc. 1962;84:4921–4928. [Google Scholar]
- 3.Kemp DS, Kerkman DJ, Leung SL, Hanson G. J Org Chem. 1981;46:490–498. [Google Scholar]
- 4.Sohma Y, Sasaki M, Hayashi Y, Kimura T, Kiso Y. Chem Commun (Camb) 2004:124–125. doi: 10.1039/b312129a. [DOI] [PubMed] [Google Scholar]
- 5.Mutter M, et al. Angew Chem. 2004;116:4267–4273. [Google Scholar]; Angew Chem Int Ed. 2004;43:4172–4178. doi: 10.1002/anie.200454045. [DOI] [PubMed] [Google Scholar]
- 6.Anfinsen CB. Science. 1973;181:223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- 7.Muir TW, Williams MJ, Kent SB. Anal Biochem. 1995;224:100–109. doi: 10.1006/abio.1995.1013. [DOI] [PubMed] [Google Scholar]
- 8.Taniguchi A, et al. J Am Chem Soc. 2006;128:696–697. doi: 10.1021/ja057100v. [DOI] [PubMed] [Google Scholar]
- 9.Coin I, Dolling R, Krause E, Bienert M, Beyermann M, Sferdean CD, Carpino LA. J Org Chem. 2006;71:6171–6177. doi: 10.1021/jo060914p. [DOI] [PubMed] [Google Scholar]
- 10.Tuchscherer G, et al. Biopolymers. 2007;88:239–252. doi: 10.1002/bip.20663. [DOI] [PubMed] [Google Scholar]
- 11.Sohma Y, Hayashi Y, Skwarczynski M, Hamada Y, Sasaki M, Kimura T, Kiso Y. Biopolymers. 2004;76:344–356. doi: 10.1002/bip.20136. [DOI] [PubMed] [Google Scholar]
- 12.Mimna R, Camus MS, Schmid A, Tuchscherer G, Lashuel HA, Mutter M. Angew Chem Int Ed. 2007;46:2681–2684. doi: 10.1002/anie.200603681. [DOI] [PubMed] [Google Scholar]
- 13.Shigenaga A, Tsuji D, Nishioka N, Tsuda S, Itoh K, Otaka A. Chembiochem. 2007;8:1929–1931. doi: 10.1002/cbic.200700442. [DOI] [PubMed] [Google Scholar]
- 14.Hirata R, Ohsumk Y, Nakano A, Kawasaki H, Suzuki K, Anraku Y. J Biol Chem. 1990;265:6726–6733. [PubMed] [Google Scholar]
- 15.Kane PM, Yamashiro CT, Wolczyk DF, Neff N, Goebl M, Stevens TH. Science. 1990;250:651–657. doi: 10.1126/science.2146742. [DOI] [PubMed] [Google Scholar]
- 16.Muralidharan V, Muir TW. Nat Methods. 2006;3:429–438. doi: 10.1038/nmeth886. [DOI] [PubMed] [Google Scholar]
- 17.Mootz HD, Blum ES, Tyszkiewicz AB, Muir TW. J Am Chem Soc. 2003;125:10561–10569. doi: 10.1021/ja0362813. [DOI] [PubMed] [Google Scholar]
- 18.Schwartz EC, Saez L, Young MW, Muir TW. Nat Chem Biol. 2007;3:50–54. doi: 10.1038/nchembio832. [DOI] [PubMed] [Google Scholar]
- 19.Tyszkiewicz AB, Muir TW. Nat Methods. 2008;5:303–305. doi: 10.1038/nmeth.1189. [DOI] [PubMed] [Google Scholar]
- 20.Wu H, Xu MQ, Liu XQ. Biochim Biophys Acta. 1998;1387:422–423. doi: 10.1016/s0167-4838(98)00157-5. [DOI] [PubMed] [Google Scholar]
- 21.Sun P, Ye S, Ferrandon S, Evans TC, Xu MQ, Rao Z. J Mol Biol. 2005;353:1093–1105. doi: 10.1016/j.jmb.2005.09.039. [DOI] [PubMed] [Google Scholar]
- 22.Giriat I, Muir TW. J Am Chem Soc. 2003;125:7180–7181. doi: 10.1021/ja034736i. [DOI] [PubMed] [Google Scholar]
- 23.Shi J, Muir TW. J Am Chem Soc. 2005;127:6198–6206. doi: 10.1021/ja042287w. [DOI] [PubMed] [Google Scholar]
- 24.Nichols NM, Benner JS, Martin DD, Evans TC. Biochemistry. 2003;42:5301–5311. doi: 10.1021/bi020679e. [DOI] [PubMed] [Google Scholar]
- 25.Turk B. Nat Rev Drug Discov. 2006;5:785–799. doi: 10.1038/nrd2092. [DOI] [PubMed] [Google Scholar]
- 26.Cook SN, Jack WE, Xiong X, Danley LE, Ellman JA, Schultz PG, Noren CJ. Angew Chem. 1995;107:1736–1737. [Google Scholar]; Angew Chem Int Ed. 1995;34:1629–1630. [Google Scholar]
- 27.Zasloff M. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 28.Maloy WL, Kari UP. Biopolymers. 1995;37:105–122. doi: 10.1002/bip.360370206. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.