Abstract
Lung cancer remains the leading cause of cancer death. Understanding lung tumors physiopathology should provide opportunity to prevent tumor development or/and improve their therapeutic management. Cancer stem cell (CSC) theory refers to a subpopulation of cancer cells, also named tumor-initiating cells, that can drive cancer development. Cells presenting these characteristics have been identified and isolated from lung cancer. Exploring cell markers and signaling pathways specific to lung CSCs may lead to progress in therapy and improve the prognosis of patients with lung cancer. Continuous efforts in developing in vitro and in vivo models may yield reliable tools to better understand CSC abilities and to test new therapeutic targets. Preclinical data on putative CSC targets are emerging by now. These preliminary studies are critical for the next generation of lung cancer therapies.
1. Introduction
Despite important improvements in therapies over the last decades, lung cancer prognosis remains very poor. After apparent successful initial therapy, development of secondary tumors often leads to a lethal relapse. Substantial growing experimental evidence has suggested that many cancers, including lung cancer, may be driven by a small subpopulation of self-renewing cells that could sustain malignant growth [1]. Innovative therapy could target this specific population of “cancer stem cells” (CSCs) or “tumor-initiating cells” in order to improve patients' outcome, with complete eradication of tumor growth. This review summarizes what is known about lung CSCs and focuses on potential clinical implications based on preclinical data obtained through new in vitro and in vivo models.
2. Cancer Stem Cells
Two major models have been described for tumor propagation: the clonal evolution model, involving a stochastic component, and the CSC model defined as hierarchical. According to the clonal evolution model, neoplasm arise from a single cell of origin, and tumor progression results from acquired genetic variability within the original clone allowing sequential selection of more aggressive sublines [2]. CSC model sustains that tumor cells are heterogeneous and only the CSC subset has the ability to proliferate extensively and form new tumors [1, 3]. These models are not mutually exclusive [4]. Recent data provide support for the CSC hypothesis, which suggests that a relatively rare subpopulation of tumor cells have the unique ability to initiate and perpetuate tumor growth [5]. CSCs have been identified and isolated in various malignancies including solid tumors [6–10] such as lung cancer [11]. These cells can be defined as cancer cells that specifically possess the ability to give rise to all cell types found in a particular cancer sample. CSCs share various characteristics with embryonic and somatic stem cells including self-renewal and multipotent differentiation [12, 13]. Self-renewal is defined as the ability to go through unlimited cycles of cell division while maintaining the undifferentiated state. Stem cells are characterized by the capacity to renew themselves through mitotic cell division and differentiate into a diverse range of specialized cell types. Indeed, self-renewal, which drives tumorigenesis, and differentiation albeit aberrant that contributes to cellular heterogeneity of the tumor, are considered the key properties of the CSC.
Stem cell populations are established in niches, which are defined as specific locations that regulate how these cells participate in tissue regeneration, maintenance, and repair [14]. The stem cell niche represents the microenvironment that interacts with stem cells to regulate stem cell self-renewal and differentiation. The dependence of normal stem cells on the niche limits their expansion. CSCs might arise from normal stem cells that have acquired mutations that enable them to escape from niche control [15]. There is increasing evidence of the importance of the tumor and the host microenvironment in conditioning the stem cell status itself [16]. The expansion of the niche cells may be driven by alterations in the CSCs or in the niche cells themselves. In either case, there is an expansion of the self-renewing CSC pool that gives rise to aberrantly differentiated cancer cells, which comprise the bulk of the tumor [17]. Alterations in CSCs may enable them to commandeer alternative niche cells to provide them with self-renewal signals. Vascular niche in brain tumors has been shown to contribute directly to the generation of CSCs and tumor growth [18]. Although some stem cells are perivascular, others may occupy hypoxic niches and be regulated by O2 gradients [19]. However, the underlying mechanisms remain unclear. O2 availability may have a direct role in stem cell regulation through HIF-1α modulation of Wnt/β-catenin signaling [20]. If CSCs depend upon aberrant niche microenvironments, then these niches might represent targets for treatments.
Some fundamental characteristics are needed to recognize CSCs. The term CSC is frequently referred to as “cancer initiating cells”, since serial transplantation of CSCs into animal models re-establishes part of the phenotypic heterogeneity of the primary tumor [21]. Xenotransplantation in immunocompromised mice, followed by serial transplantation, is now regarded as an essential criterion in defining CSCs. This preponderant characteristic might represent the mechanistic parallel of regenerating a tissue or an organ in normal stem cell. These cells can be grown in vitro as tumor spheres under nonadherent conditions [22]. Self-renewal capacity also has to be documented.
CSCs have been shown in in vitro models to be highly resistant to radiation [23] and chemotherapy [24], potentially resulting in residual disease that can lead to recurrence. However, radioresistance of CSCs remains controversial. Inconsistent experimental results could be related to difficulty in correctly isolating the CSC subpopulation within a tumor [25]. CSCs may be less vulnerable to treatments because of intrinsic properties of radio and chemoresistance [26]. One hypothesis is that CSCs could be more resistant to DNA damages induced by these treatments [27, 28]. They may have enhanced capacities of DNA repair [29].
The frequency of CSCs appears to be highly variable, reflecting biological diversity among cancer models as well as technical issues. This subpopulation of cells was identified utilizing flow-cytometer-based cell sorting, enabling isolation of a “side population” (SP) defined by Hoechst 33342 dye exclusion [30]. This test is based on ABCG2 transporter, which is the second member of the G subfamily of ATP binding cassette (ABC) transporters. ABCG2 is one of the most important multidrug-resistance transporters and its substrates include Hoeschst 33342 [31]. This transporter is highly expressed in the SP containing stem cells [32]. CSCs are capable of self-renewal and production of differentiated progenitors. Progenitors are engaged in a differentiation pathway that progressively generates the different mature cells. CSCs express tissue-specific cell surface markers, such as CD34 in several kinds of leukemia [33] and CD44 in breast cancer [6]. None of these markers are specific to CSCs: CD34 is expressed by endothelial and hematologic normal cells, while CD44 is expressed by endothelial and mesenchymal cells. CD133 has been observed in a wide spectrum of malignant tumors including lung cancer [11, 34]. The function of CD133 is still unknown. CD133 may have a role in stem cell activation/maintenance [35], and potential regulatory activity of cell-cell contacts [36]. The CSC phenotype may not necessarily be uniform between cancer subtypes or even tumors of the same subtype [37]. Aldehyde dehydrogenase (ALDH) metabolizes a wide variety of endogenous and exogenous aldehydes [38]. High ALDH activity has been detected in stem and progenitor cells in various lineages [39–41], suggesting that strong ALDH activity and/or antigen expression can be used as a marker for CSC in a variety of cancers, including lung cancer [42].
3. Does the Cancer Stem Cell Concept Remain Disputed?
In the stochastic model, transformation results from a random mutation and subsequent clonal selection. But this model may not be sufficient to fully explain biology of carcinogenesis. The conceptual gap left by this model could be filled in by CSC hypothesis which becomes very attractive in this context. Indeed carcinogenesis involves acquisition and accumulation of genetic mutations and epigenetic regulations that may take many years. Since the normal lung epithelium turns over fairly rapidly, there must be some very long-lived cell type that serves as the repository to safeguard all these accumulating mutations. This can fit with CSC hypothesis and therefore explain both heterogeneity of cancer cells and events of metastasis. This theory may bring fundamental bases for understanding resistance to chemotherapy or radiation treatment, tumor promotion and progression, as well as important clinical implications for detection, prevention, and treatment [3]. However, some controversies still remain [43–45]. Tumor growth may not need to be driven by rare cancer stem cells. Cancer stem cell theory may not be an absolute requisite to explain chemo-radio resistance. This characteristic may be related to multiple drug resistance (mdr) phenotype [46]. Metabolic flexibility may generate adaptive changes in cells of the residual disease that can lead to tumor growth [47] and relapse. Tumor progression and recurrence may be explained by a state of cancer dormancy [48]. CSC may not be a real entity. CSC theory could be the simple transposition of normal tissue development to cancer. Although the CSC hypothesis still has to be fully validated in lung cancer, its applicability would provide an improvement in our understanding of carcinogenesis.
4. Lung Cancer
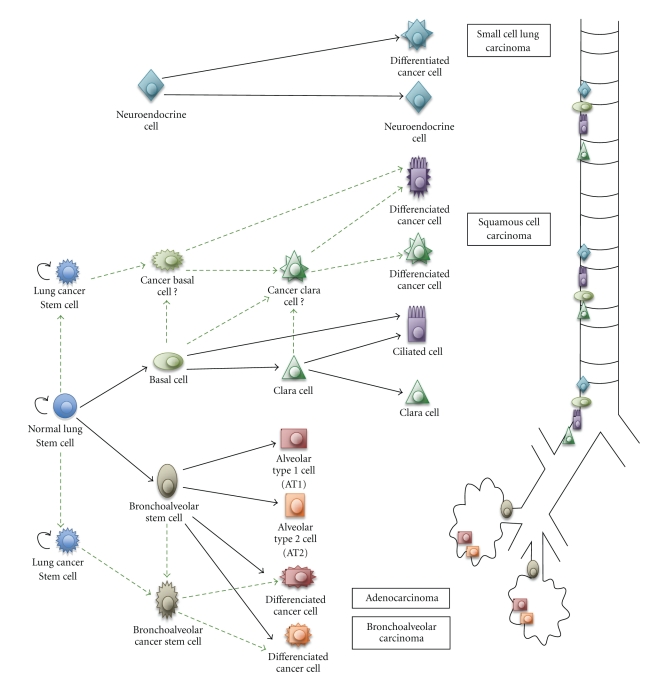
Lung cancer is one of the most intractable cancers with a five-year overall survival of around 15% all stages merged. Lung cancer has a poor prognosis, because of extra thoracic dissemination and frequent disease relapse. The main types of lung cancer are small cell lung carcinoma SCLC and nonsmall cell lung carcinoma NSCLC, which includes three major histological types: adenocarcinoma, squamous cell carcinoma (SCC), and large cell carcinoma [49]. About 15% of the tumors are SCLCs and arise in the larger airways, grow rapidly, and have a neuroendocrine component. Adenocarcinomas represent about 40% of NSCLCs and usually start in peripheral lung tissue. SCCs account for 25% and commonly originate near a central bronchus. Large cell carcinomas are believed to derive from neuroendocrine cells and may be observed in combination with other types of NSCLC. These neuroendocrine lung tumors appear to be epithelial tumors characterized by their preferential neuroendocrine differentiation as indicated by neuroendocrines granules, mucin granules, microvilli, and tonofilaments [50].
It is estimated that smoking causes 90% of lung cancers. However, different types of genetic modifications have been described in lung cancer cells such as recurrent chromosome abnormalities [51], upregulation of telomerase activity [52], and mutations of oncogenes or tumor suppressor genes (TP53, RB1, CDKN2A, KRAS, EGFR). Some may be markers of disease progression, others may have a direct role in lung cancer genesis in the context of gene-environment interactions [53]. The link between mutations and stem cell-ness is an important issue that needs to be explored. It is not clear whether mutations leading to CSCs occur in stem cells or in differentiated cells. These data strongly suggest that a deep knowledge of each tumor should improve patient outcome thanks to personalized therapy.
5. Normal Lung Stem Cells
The respiratory system arises from the ventral foregut endoderm with development of a tree-like system of epithelial tubules and vascular structures, which ultimately gives rise to the mature airways and alveoli. Precise sequence and pattern of branching events lead to highly ramified tubular networks possibly driven by a stereotyped hierarchical and modular branching program [54] which molecular regulation remains under study [55]. Lung mesenchymal development is crucially influenced by signals from the epithelium and the pleura that, in concert, appear to maintain a balance of differentiated and proliferating multipotent progenitors while the lung grows [56]. During lung branching morphogenesis, the transcription factor Sox2 is associated with the epithelium that is less morphogenetically active. Sox2 expression is lost at sites where nascent buds arise [57, 58]. As epithelial cells in branching airways continue to differentiate, it is crucial to maintain and expand a pool of uncommitted progenitor cells for continuous growth. It has been proposed that this pool resides in the distal lung, as a population of proliferating immature epithelial cells that expresses high levels of the proto-oncogene Mycn [59].
Mature respiratory epithelium consists of multiple cell types. Ciliated, neuroendocrine, and secretory cells are located in the proximal region of the respiratory system. Alveolar type 1 (AT1) and 2 (AT2) cells are typical of the distal alveolar region of the lung (Figure 1). Lineage analysis suggests that progenitor cells lining the trachea and proximal lung have a different origin from those lining the distal region of the lung [60]. The activity of the different pools of progenitor cells account for spatially restricted regional specificities in both proximal and distal cell lineages specifications during lung development and cellular composition of tracheobronchial and bronchiolar airways [61]. Basal cells have been proposed as a multipotent progenitor cell population of bronchial airways. They have the capacity for restoration of a fully differentiated epithelium [62, 63]. Lung CSCs and airway normal stem cells may use common pathways in development.
Figure 1.
Lung CSCs and progenitors which differentiation leads to heterogeneity of associated human carcinomas. Normal and tumor cellular hierarchy progressively generates progenitor cells and differentiated mature cells in both proximal and distal airways. Hypotheses are represented with discontinuous lines.
6. Lung Cancer Stem Cells
Altered composition of airway epithelial cell that is associated with acute or chronic lung injury has the potential to significantly increase risk for developing lung cancer [64, 65]. It is currently unknown whether airway stem cells contribute significantly to normal epithelial maintenance and repair [66]. Clara cells participate in maintenance of both secretory and ciliated cell types after oxidant-mediated damage [67]. Meanwhile a population of pollutant-resistant stem cells localized in the bronchoalveolar duct junction (BADJ) contributes to restoration of a phenotypically diverse epithelium after clara cell depletion [68]. These cells are named bronchoalveolar stem cells (BASCs) (Figure 1). Both pulmonary neuroendocrine cells (PNEC) and Clara cells have been proposed as progenitors for the genesis of SCLC and NSCLC [69]. PNEC are specialized airway epithelial cells that produce specific neuropeptides and that are grouped in clusters termed neuroepithelial bodies (NEBs). Repair from airway injury is associated with PNEC hyperplasia [70]. Cells with Clara-like characteristics but reduced secretory protein expression have been identified in association with preneoplastic lesions [71]. These data suggest that cells engaged in differentiation could reactivate genes of immaturity. In this context, CSCs may arise from restricted progenitors or more differentiated cells that have reacquired self-renewing capacity [5]. This phenomenon could have direct bearing on the issue of stem cell plasticity, which is defined as the ability to cross lineage barriers and adopt cell-differentiated phenotypes [72]. It also may raise a parallel with transdifferentiation from differentiated cell to CSC that leads to cancer cell phenotypic diversification [73]. BASCs at the BADJ of adult airways have been proposed as an initiating cell source for lung adenocarcinoma [74]. Stem cells and progenitors could be considered ideal tumor initiating candidates, because dysregulation of proliferative capacity through mutation may rapidly cause dysplastic tumor-like growth. The deviation from a common endodermic stem cell could also be responsible for the multidirectional differentiation in lung tumors, including neuroendocrine phenotype in NSCLC and squamous or glandular signs of differentiation in authenticated neuroendocrine tumors [75].
Observed phenotypic heterogeneity between distinct tumor types suggests that the tumor's local pulmonary environment deeply impacts the fate of cancer cells [76]. Developmental programs of cells initially engaged in a specific lineage pathway can be altered by changing the type of signals in the local environment [77]. The different lung tumors could arise from regiospecific cancer stem cell niches that are still debated [78, 79].
7. Lung Cancer Stem Cell Models
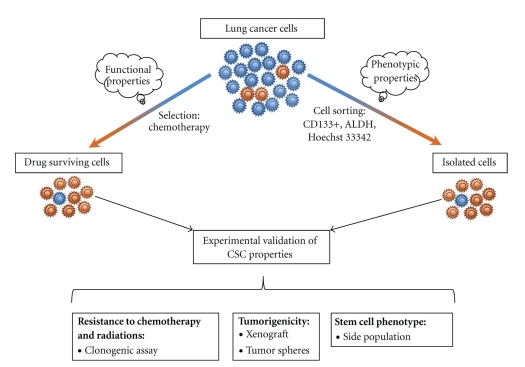
Tumorigenic human lung cancer cells have been isolated using different approaches from both cell lines and primary tumors. CSC models from lung cancer cell lines are based on the phenotype or on the functional characteristics of these cells (Figure 2). SP has showed repopulating ability and resistance to multiple chemotherapeutic drugs in lung cancer. In addition, human telomerase reverse transcriptase (hTERT) expression is higher in SP, suggesting that this fraction may represent an enriched source of lung tumor initiating cells with unlimited proliferative potential [80–83]. CSCs can be identified and isolated by flow-cytometry-based cell sorting using the cell surface marker CD133 [37] (Figure 2). The lack of early markers for lung progenitors represents a clear gap of knowledge. The known markers are not always ideal to sort for the CSC population [84]. In line with tumor heterogeneity, the phenotype of CSCs is not uniform, underlying the necessity to find more specific single markers or to define new marker combinations to potentially recognize the putative CSCs (Table 1). Isolation of lung CSCs based on increased ALDH activity was obtained using the Aldefluor assay followed by fluorescent-activated cell sorting (FACS) (Figure 2) [42, 85]. The second approach leading to the isolation of lung CSCs was based on their inherent functional resistance to chemotherapy (Figure 2). Drug-surviving cells were isolated after an in vitro treatment with cisplatin, doxorubicin, or etoposide. These cells present the phenotypic characteristics previously described [86]. These different CSC models developed on cancer cell lines can be grown in vitro as tumor spheres under nonadherent conditions using a serum-free medium that is supplemented with growth factors. They also exhibit high clonogenic potential, capacities for self-renewal, generation of differentiated progeny, and high in vivo tumorigenicity (Figure 3).
Figure 2.
In vitro models of lung CSCs. Experimental models of lung CSCs are based on the functional properties of chemoresistance of these cells or on their phenotypic characteristics. Populations enriched in CSCs obtained after treatment or cell sorting need to present all the stemcellness properties to be considered as lung CSC models.
Table 1.
Molecular characterization of putative human lung CSCs.
Figure 3.
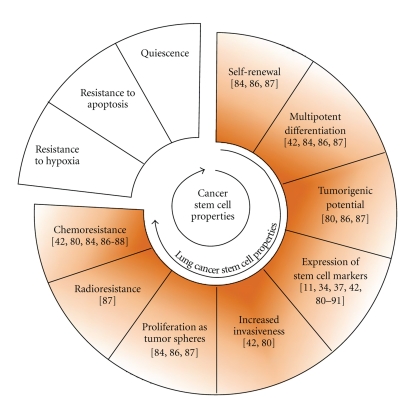
CSC and lung CSC properties. CSC characteristics verified on lung CSC models are presented with the references of the corresponding reports.
The first isolation and expansion of lung CSCs from primary patient tumors was based on their ability to survive under serum-free conditions and proliferate as cellular tumor spheres [11]. CD133 cells isolated from primary tumors displayed features of CSCs both in vitro and in vivo [87, 88]. The expression of CD133 in tumors was linked to shorter progression-free survival in patients treated with platinum-based chemotherapies, but this point remains controversial [88–91].
In vivo limiting dilution consists of injection of decreasing number of cells. This method used for subcutaneous tumor xenograft may prove enrichment of CSCs in an isolated subpopulation of cells [27, 92] (ratio to obtain a tumor from CSCs compared to tumor cells: 1/100). To date, there is no consistent data published on this method in lung CSC models that determine the minimal number of tumor-initiating cells necessary and sufficient to regenerate a tumor.
A few mouse models of lung cancer-initiating cells have been developed [74, 93]. However the frequency of CSCs in these models is highly variable. To date, there is no evidence of a subpopulation isolated from an animal model able to restore the tumor initial heterogeneity in secondary and tertiary hosts [84].
8. Preclinical Data on Lung Cancer Stem Cells
The surviving fraction of tumor cells after chemotherapy is low and may contain CSCs. These cells “left behind” may be responsible for recurrence. These cells need to be eradicated in order to provide long-term disease-free survival. Combinatorial treatments involving both cytotoxic and targeted therapies will probably be required to suppress all cancer cells [5]. Preclinical data on putative CSCs targets are emerging by now.
Oncogenes and tumor-suppressor genes are the 2 types of genes involved in carcinogenesis. Oncogenes are dominant genes [94] whereas tumor suppressor genes are recessive. Activating mutations or transcriptional deregulation can lead from proto-oncogenes to oncogenes. The Runx genes can present characteristics of both oncogenes and tumor suppressor genes. These genes encode transcription factors involved in normal development with tissue specific expression. Many chromosomal translocations involving Runx genes and leading to oncogenic fusion proteins have been reported [95]. These fusion proteins may have a tissue-restricted oncogenic spectrum [96] in accordance with the concept of lineage-specific oncogenes. This concept applied to CSCs could suggest that some specific oncogenes could select and maintain a particular malignant phenotype characteristic of a cell lineage. Runx3 is an essential transcription factor for the late phase of lung development. Runx3 is required for the control of differentiation and proliferation of bronchiolar epithelium [97]. The frequent silencing of Runx3 by promoter hypermethylation in the preneoplastic stage of the lung adenocarcinoma has been observed [98, 99]. Runx3 downregulation has been proposed as an early event in the development of the lung carcinoma interfering in the differentiation of progenitor cells [97]. Moreover, there are some data suggesting that in lung cancer TITF1 could be an adenocarcinoma lineage specific oncogene [100] and BRF2 a squamous cell lineage specific oncogene [101].
BASC possesses the potential to differentiate into Clara or AT2 cells, presenting coexpression of Clara cell secretory protein (CCSP) and surfactant protein C (SPC) (Figure 1) [74]. These cells also exhibit the capacity of self-renewal and the expression of Oct4. Oct4 is a transcriptional factor of the embryonic stem cells which expression was found to be associated with a poor prognosis [87, 102, 103]. In addition, knock down of Oct4 might lead to apoptosis of a CSC-like population of lung cancer cells [104].
Sox2 is a critical transcription regulator of embryonic stem cells [105]. Sox2 controls self-renewal and differentiation processes [106]. It is also implicated in lung branching morphogenesis during pulmonary development as previously evocated [57, 58]. Sox2 has been proposed to be an oncogene which expression is essential to lung SCC carcinogenesis and which is capable of transforming and conferring tumor initiating properties to human lung squamous cells [107]. Additionally, tumors with Sox2 overexpression seem to have a worse prognosis [108].
Oct4 and Sox2 are two of the four crucial transcription factors capable of cooperating to reprogram differentiated cells into an induced pluripotent stem cell-like phenotype [109, 110]. Nuclear transplantation of Oct4, Sox2, c-Myc, and Klf4 can reprogram a somatic genome back into an embryonic stem cell status. Is there a comparable way for cancer cells to reacquire an immature profile with properties of self-renewal? Oct4 and Sox2 may participate to a similar process leading to lung CSCs.
Stem Cell Factor (SCF) is a mitogenic and angiogenic factor involved in carcinogenesis. Human c-kit has been shown to operate as a SCF receptor promoting tumor growth [111–113]. Patients with either mutation or overexpression of c-kit have lower survival rates and show resistance to chemotherapy [114]. Blocking SCF/c-kit signaling pathway inhibits CSC proliferation and survival after chemotherapy exposure in human lung cancer cell lines [115]. Exposure to SCF/c-kit axis blockade by SCF-neutralizing antibodies or by imatinib (Gleevec), an inhibitor of c-kit, combined to cisplatin therapy might lead to inhibition of both non-CSCs and CSCs.
Differences between normal stem cells and CSCs may provide novel antigenic and molecular targets for therapy (Table 1). It is important to design new therapeutic approaches to selectively hit CSC-specific pathways, while sparing normal stem cells. Further identification of lung cancer stem cell-specifics target could allow for promising more effective combined therapies. In our view, the most important matter to address in a near future is to better characterize and isolate lung CSCs from patients' tumors combining marker studies and functional assays. Lung CSCs probably include a heterogeneous population of cells with different therapeutic resistance profiles. Identifying lineage specific oncogenes or their fusion proteins could bring new targets for specific therapeutic approaches selectively aimed at specific CSCs. In contrast to chemotherapy or radiotherapy on mature tumor cells, therapies against CSCs might have a slow but sustainable effect. Therefore experimental models will certainly have to be redrawn including longer observation periods and intermediate analysis.
9. Conclusion
CSC is central to cancer cell biology and cancer therapy. The knowledge of CSC signaling pathways may lead to new therapies able to eradicate lung CSC or induce differentiation of these cells. Promising models have recently been developed to isolate the subpopulation of CSCs within the lung-differentiated cells of the tumor mass. These models are useful tools to better characterize lung CSCs. Targeting the CSC or its microenvironmental niche could inhibit self-renewing and overcome chemo-radio resistance. Some consistent preclinical data are emerging on the role of transcriptional factors such as Runx3, Oct4, Sox2, and c-kit in lung tumor differentiation and treatment resistance. Further in vitro and in vivo studies are needed to design combined therapies in order to determine whether these candidates are potential targets for new therapeutic strategies against lung cancer. Moreover, the prognostic and predictive value of CSCs identification from tumor biopsies or among circulating tumor cells remains to be determined.
References
- 1.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414(6859):105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 2.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194(4260):23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 3.Wicha MS, Liu S, Dontu G. Cancer stem cells: an old idea—a paradigm shift. Cancer Research. 2006;66(4):1883–1890. doi: 10.1158/0008-5472.CAN-05-3153. [DOI] [PubMed] [Google Scholar]
- 4.Campbell LL, Polyak K. Breast tumor heterogeneity: cancer stem cells or clonal evolution? Cell Cycle. 2007;6(19):2332–2338. doi: 10.4161/cc.6.19.4914. [DOI] [PubMed] [Google Scholar]
- 5.Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nature Reviews Cancer. 2008;8(10):755–768. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- 6.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(7):3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singh SK, Hawkins C, Clarke ID, et al. Identification of human brain tumour initiating cells. Nature. 2004;432(7015):396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 8.Collins AT, Maitland NJ. Prostate cancer stem cells. European Journal of Cancer. 2006;42(9):1213–1218. doi: 10.1016/j.ejca.2006.01.037. [DOI] [PubMed] [Google Scholar]
- 9.Hermann PC, Huber SL, Herrler T, et al. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1(3):313–323. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 10.Ricci-Vitiani L, Lombardi DG, Pilozzi E, et al. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445(7123):111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 11.Eramo A, Lotti F, Sette G, et al. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death and Differentiation. 2008;15(3):504–514. doi: 10.1038/sj.cdd.4402283. [DOI] [PubMed] [Google Scholar]
- 12.Jordan CT, Guzman ML, Noble M. Cancer stem cells. The New England Journal of Medicine. 2006;355(12):1253–1261. doi: 10.1056/NEJMra061808. [DOI] [PubMed] [Google Scholar]
- 13.Lobo NA, Shimono Y, Qian D, Clarke MF. The biology of cancer stem cells. Annual Review of Cell and Developmental Biology. 2007;23:675–699. doi: 10.1146/annurev.cellbio.22.010305.104154. [DOI] [PubMed] [Google Scholar]
- 14.Scadden DT. The stem-cell niche as an entity of action. Nature. 2006;441(7097):1075–1079. doi: 10.1038/nature04957. [DOI] [PubMed] [Google Scholar]
- 15.Zhu X, Zhou X, Lewis MT, Xia L, Wong S. Cancer stem cell, niche and EGFR decide tumor development and treatment response: a bio-computational simulation study. Journal of Theoretical Biology. 269(1):138–149. doi: 10.1016/j.jtbi.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cesana AE, Noonan DM. Cancer stem cells: soloists or choral singers within the tumor micro-environment? Current Pharmaceutical Biotechnology. Nov 2, 2010. PMID:21044010. [Google Scholar]
- 17.Clarke MF, Fuller M. Stem cells and cancer: two faces of eve. Cell. 2006;124(6):1111–1115. doi: 10.1016/j.cell.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 18.Calabrese C, Poppleton H, Kocak M, et al. A perivascular niche for brain tumor stem cells. Cancer Cell. 2007;11(1):69–82. doi: 10.1016/j.ccr.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 19.Parmar K, Mauch P, Vergilio JA, Sackstein R, Down JD. Distribution of hematopoietic stem cells in the bone marrow according to regional hypoxia. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(13):5431–5436. doi: 10.1073/pnas.0701152104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mazumdar J, O'Brien WT, Johnson RS, et al. O2 regulates stem cells through Wnt/β-catenin signalling. Nature Cell Biology. 2010;12(10):1007–1013. doi: 10.1038/ncb2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lapidot T, Sirard C, Vormoor J, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367(6464):645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 22.Reynolds BA, Rietze RL. Neural stem cells and neurospheres—re-evaluating the relationship. Nature Methods. 2005;2(5):333–336. doi: 10.1038/nmeth758. [DOI] [PubMed] [Google Scholar]
- 23.Debeb BG, Xu W, Woodward WA. Radiation resistance of breast cancer stem cells: understanding the clinical framework. Journal of Mammary Gland Biology and Neoplasia. 2009;14(1):11–17. doi: 10.1007/s10911-009-9114-z. [DOI] [PubMed] [Google Scholar]
- 24.Li X, Lewis MT, Huang J, et al. Intrinsic resistance of tumorigenic breast cancer cells to chemotherapy. Journal of the National Cancer Institute. 2008;100(9):672–679. doi: 10.1093/jnci/djn123. [DOI] [PubMed] [Google Scholar]
- 25.Al-Assar O, Muschel RJ, Mantoni TS, McKenna WG, Brunner TB. Radiation response of cancer stem-like cells from established human cell lines after sorting for surface markers. International Journal of Radiation Oncology Biology Physics. 2009;75(4):1216–1225. doi: 10.1016/j.ijrobp.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 26.Baumann M, Krause M, Hill R. Exploring the role of cancer stem cells in radioresistance. Nature Reviews Cancer. 2008;8(7):545–554. doi: 10.1038/nrc2419. [DOI] [PubMed] [Google Scholar]
- 27.Zhang M, Atkinson RL, Rosen JM. Selective targeting of radiation-resistant tumor-initiating cells. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(8):3522–3527. doi: 10.1073/pnas.0910179107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin LP, Hamilton TC, Schilder RJ. Platinum resistance: the role of DNA repair pathways. Clinical Cancer Research. 2008;14(5):1291–1295. doi: 10.1158/1078-0432.CCR-07-2238. [DOI] [PubMed] [Google Scholar]
- 29.Bao S, Wu Q, McLendon RE, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444(7120):756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 30.Goodell MA, Brose K, Paradis G, Conner AS, Mulligan RC. Isolation and functional properties of murine hematopoietic stem cells that are replicating in vivo. Journal of Experimental Medicine. 1996;183(4):1797–1806. doi: 10.1084/jem.183.4.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ding XIW, Wu JH, Jiang CP. ABCG2: a potential marker of stem cells and novel target in stem cell and cancer therapy. Life Sciences. 2010;86(17-18):631–637. doi: 10.1016/j.lfs.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 32.Zhou S, Schuetz JD, Bunting KD, et al. The ABC transporter Bcrp1/ABCG2 is expressed in a wide variety of stem cells and is a molecular determinant of the side-population phenotype. Nature Medicine. 2001;7(9):1028–1034. doi: 10.1038/nm0901-1028. [DOI] [PubMed] [Google Scholar]
- 33.Ryan DH, Chapple CW, Kossover SA, Sandberg AS, Cohen HJ. Phenotypic similarities and differences between CALLA-positive acute lymphoblastic leukemia cells and normal marrow CALLA-positive B cell precursors. Blood. 1987;70(3):814–821. [PubMed] [Google Scholar]
- 34.Tirino V, Camerlingo R, Franco R, et al. The role of CD133 in the identification and characterisation of tumour-initiating cells in non-small-cell lung cancer. European Journal of Cardio-Thoracic Surgery. 2009;36(3):446–453. doi: 10.1016/j.ejcts.2009.03.063. [DOI] [PubMed] [Google Scholar]
- 35.Yu Y, Flint A, Dvorin EL, Bischoff J. AC133-2, a novel isoform of human AC133 stem cell antigen. Journal of Biological Chemistry. 2002;277(23):20711–20716. doi: 10.1074/jbc.M202349200. [DOI] [PubMed] [Google Scholar]
- 36.Taïeb N, Maresca M, Guo XJ, Garmy N, Fantini J, Yahi N. The first extracellular domain of the tumour stem cell marker CD133 contains an antigenic ganglioside-binding motif. Cancer Letters. 2009;278(2):164–173. doi: 10.1016/j.canlet.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 37.Klonisch T, Wiechec E, Hombach-Klonisch S, et al. Cancer stem cell markers in common cancers—therapeutic implications. Trends in Molecular Medicine. 2008;14(10):450–460. doi: 10.1016/j.molmed.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 38.Yoshida A, Rzhetsky A, Hsu LC, Chang C. Human aldehyde dehydrogenase gene family. European Journal of Biochemistry. 1998;251(3):549–557. doi: 10.1046/j.1432-1327.1998.2510549.x. [DOI] [PubMed] [Google Scholar]
- 39.Jones RJ, Barber JP, Vala MS, et al. Assessment of aldehyde dehydrogenase in viable cells. Blood. 1995;85(10):2742–2746. [PubMed] [Google Scholar]
- 40.Ginestier C, Hur MH, Charafe-Jauffret E, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1(5):555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang EH, Hynes MJ, Zhang T, et al. Aldehyde dehydrogenase 1 is a marker for normal and malignant human colonic stem cells (SC) and tracks SC overpopulation during colon tumorigenesis. Cancer Research. 2009;69(8):3382–3389. doi: 10.1158/0008-5472.CAN-08-4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiang F, Qiu Q, Khanna A, et al. Aldehyde dehydrogenase 1 is a tumor stem cell-associated marker in lung cancer. Molecular Cancer Research. 2009;7(3):330–338. doi: 10.1158/1541-7786.MCR-08-0393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kelly PN, Dakic A, Adams JM, Nutt SL, Strasser A. Tumor growth need not be driven by rare cancer stem cells. Science. 2007;317(5836):p. 337. doi: 10.1126/science.1142596. [DOI] [PubMed] [Google Scholar]
- 44.Yoo MH, Hatfield DL. The cancer stem cell theory: is it correct? Molecules and Cells. 2008;26(5):514–516. [PMC free article] [PubMed] [Google Scholar]
- 45.Rowan K. Are cancer stem cells real? After four decades, debate still simmers. Journal of the National Cancer Institute. 2009;101(8):546–547. doi: 10.1093/jnci/djp083. [DOI] [PubMed] [Google Scholar]
- 46.Baguley BC. Multiple drug resistance mechanisms in cancer. Molecular Biotechnology. 2010;46(3):308–316. doi: 10.1007/s12033-010-9321-2. [DOI] [PubMed] [Google Scholar]
- 47.Berridge MV, Herst PM, Tan AS. Metabolic flexibility and cell hierarchy in metastatic cancer. Mitochondrion. 2010;10(6):584–588. doi: 10.1016/j.mito.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 48.Aguirre-Ghiso JA. The problem of cancer dormancy: understanding the basic mechanisms and identifying therapeutic opportunities. Cell Cycle. 2006;5(16):1740–1743. doi: 10.4161/cc.5.16.3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Collins LG, Haines C, Perkel R, Enck RE. Lung cancer: diagnosis and management. American Family Physician. 2007;75(1):56–63. [PubMed] [Google Scholar]
- 50.Brambilla E, Veale D, Moro D, Morel F, Dubois F, Brambilla C. Neuroendocrine phenotype in lung cancers: comparison of immunohistochemistry with biochemical determination of enolase isoenzymes. American Journal of Clinical Pathology. 1992;98(1):88–97. doi: 10.1093/ajcp/98.1.88. [DOI] [PubMed] [Google Scholar]
- 51.Yanada M, Yaoi T, Shimada J, et al. Frequent hemizygous deletion at 1p36 and hypermethylation downregulate RUNX3 expression in human lung cancer cell lines. Oncology Reports. 2005;14(4):817–822. [PubMed] [Google Scholar]
- 52.Hiyama K, Hiyama E, Ishioka S, et al. Telomerase activity in small-cell and non-small-cell lung cancers. Journal of the National Cancer Institute. 1995;87(12):895–902. doi: 10.1093/jnci/87.12.895. [DOI] [PubMed] [Google Scholar]
- 53.Amos CI, Xu W, Spitz MR. Is there a genetic basis for lung cancer susceptibility? Recent Results in Cancer Research. 1999;151:3–12. doi: 10.1007/978-3-642-59945-3_1. [DOI] [PubMed] [Google Scholar]
- 54.Metzger RJ, Klein OD, Martin GR, Krasnow MA. The branching programme of mouse lung development. Nature. 2008;453(7196):745–750. doi: 10.1038/nature07005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cardoso WV, Lü J. Regulation of early lung morphogenesis: questions, facts and controversies. Development. 2006;133(9):1611–1624. doi: 10.1242/dev.02310. [DOI] [PubMed] [Google Scholar]
- 56.Weaver M, Batts L, Hogan BLM. Tissue interactions pattern the mesenchyme of the embryonic mouse lung. Developmental Biology. 2003;258(1):169–184. doi: 10.1016/s0012-1606(03)00117-9. [DOI] [PubMed] [Google Scholar]
- 57.Ishii Y, Rex M, Scotting PJ, Yasugi S. Region-specific expression of chicken Sox2 in the developing gut and lung epithelium: regulation by epithelial-mesenchymal interactions. Developmental Dynamics. 1998;213(4):464–475. doi: 10.1002/(SICI)1097-0177(199812)213:4<464::AID-AJA11>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 58.Gontan C, de Munck A, Vermeij M, Grosveld F, Tibboel D, Rottier R. Sox2 is important for two crucial processes in lung development: branching morphogenesis and epithelial cell differentiation. Developmental Biology. 2008;317(1):296–309. doi: 10.1016/j.ydbio.2008.02.035. [DOI] [PubMed] [Google Scholar]
- 59.Okubo T, Knoepfler PS, Eisenman RN, Hogan BLM. Nmyc plays an essential role during lung development as a dosage-sensitive regulator of progenitor cell proliferation and differentiation. Development. 2005;132(6):1363–1374. doi: 10.1242/dev.01678. [DOI] [PubMed] [Google Scholar]
- 60.Perl AKT, Wert SE, Nagy A, Lobe CG, Whitsett JA. Early restriction of peripheral and proximal cell lineages during formation of the lung. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(16):10482–10487. doi: 10.1073/pnas.152238499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pack RJ, Al-Ugaily LH, Morris G, Widdicombe JG. The distribution and structure of cells in the tracheal epithelium of the mouse. Cell and Tissue Research. 1980;208(1):65–84. doi: 10.1007/BF00234174. [DOI] [PubMed] [Google Scholar]
- 62.Hong KU, Reynolds SD, Watkins S, Fuchs E, Stripp BR. In vivo differentiation potential of tracheal basal cells: evidence for multipotent and unipotent subpopulations. American Journal of Physiology. 2004;286(4):L643–L649. doi: 10.1152/ajplung.00155.2003. [DOI] [PubMed] [Google Scholar]
- 63.Hong KU, Reynolds SD, Watkins S, Fuchs E, Stripp BR. Basal cells are a multipotent progenitor capable of renewing the bronchial epithelium. American Journal of Pathology. 2004;164(2):577–588. doi: 10.1016/S0002-9440(10)63147-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nakanishi K, Hiroi S, Kawai T, Suzuki M, Torikata C. Argyrophilic nucleolar-organizer region counts and DNA status in bronchioloalveolar epithelial hyperplasia and adenocarcinoma of the lung. Human Pathology. 1998;29(3):235–239. doi: 10.1016/s0046-8177(98)90041-7. [DOI] [PubMed] [Google Scholar]
- 65.Trichopoulos D, Mollo F, Tomatis L, et al. Active and passive smoking and pathological indicators of lung cancer risk in an autopsy study. Journal of the American Medical Association. 1992;268(13):1697–1701. [PubMed] [Google Scholar]
- 66.Giangreco A, Arwert EN, Rosewell IR, Snyder J, Watt FM, Stripp BR. Stem cells are dispensable for lung homeostasis but restore airways after injury. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(23):9286–9291. doi: 10.1073/pnas.0900668106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Evans MJ, Cabral Anderson LJ, Freeman G. Role of the clara cell in renewal of the bronchiolar epithelium. Laboratory Investigation. 1978;38(6):648–655. [PubMed] [Google Scholar]
- 68.Giangreco A, Reynolds SD, Stripp BR. Terminal bronchioles harbor a unique airway stem cell population that localizes to the bronchoalveolar duct junction. American Journal of Pathology. 2002;161(1):173–182. doi: 10.1016/S0002-9440(10)64169-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Reynolds SD, Giangreco A, Power JHT, Stripp BR. Neuroepithelial bodies of pulmonary airways serve as a reservoir of progenitor cells capable of epithelial regeneration. American Journal of Pathology. 2000;156(1):269–278. doi: 10.1016/S0002-9440(10)64727-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stevens TP, Mcbride JT, Peake JL, Pinkerton KE, Stripp BR. Cell proliferation contributes to PNEC hyperplasia after acute airway injury. American Journal of Physiology. 1997;272(3):L486–L493. doi: 10.1152/ajplung.1997.272.3.L486. [DOI] [PubMed] [Google Scholar]
- 71.Linnoila RI, Jensen SM, Steinberg SM, Mulshine JL, Eggleston JC, Gazdar AF. Peripheral airway cell marker expression in non-small cell lung carcinoma. Association with distinct clinicopathologic features. American Journal of Clinical Pathology. 1992;97(2):233–243. doi: 10.1093/ajcp/97.2.233. [DOI] [PubMed] [Google Scholar]
- 72.Eisenberg LM, Eisenberg CA. Stem cell plasticity, cell fusion, and transdifferentiation. Birth Defects Research Part C. 2003;69(3):209–218. doi: 10.1002/bdrc.10017. [DOI] [PubMed] [Google Scholar]
- 73.Forbes SJ, Vig P, Poulsom R, Wright NA, Alison MR. Adult stem cell plasticity: new pathways of tissue regeneration become visible. Clinical Science. 2002;103(4):355–369. doi: 10.1042/cs1030355. [DOI] [PubMed] [Google Scholar]
- 74.Kim CF, Jackson EL, Woolfenden AE, et al. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell. 2005;121(6):823–835. doi: 10.1016/j.cell.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 75.Onuki N, Wistuba II, Travis WD, et al. Genetic changes in the spectrum of neuroendocrine lung tumors. Cancer. 1999;85(3):600–607. doi: 10.1002/(sici)1097-0142(19990201)85:3<600::aid-cncr10>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 76.Meuwissen R, Berns A. Mouse models for human lung cancer. Genes and Development. 2005;19(6):643–664. doi: 10.1101/gad.1284505. [DOI] [PubMed] [Google Scholar]
- 77.Ohtsuka N, Urase K, Momoi T, Nogawa H. Induction of bud formation of embryonic mouse tracheal epithelium by fibroblast growth factor plus transferrin in mesenchyme-free culture. Developmental Dynamics. 2001;222(2):263–272. doi: 10.1002/dvdy.1206. [DOI] [PubMed] [Google Scholar]
- 78.Giangreco A, Groot KR, Janes SM. Lung cancer and lung stem cells: strange bedfellows? American Journal of Respiratory and Critical Care Medicine. 2007;175(6):547–553. doi: 10.1164/rccm.200607-984PP. [DOI] [PubMed] [Google Scholar]
- 79.Li L, Neaves WB. Normal stem cells and cancer stem cells: the niche matters. Cancer Research. 2006;66(9):4553–4557. doi: 10.1158/0008-5472.CAN-05-3986. [DOI] [PubMed] [Google Scholar]
- 80.Ho MM, Ng AV, Lam S, Hung JY. Side population in human lung cancer cell lines and tumors is enriched with stem-like cancer cells. Cancer Research. 2007;67(10):4827–4833. doi: 10.1158/0008-5472.CAN-06-3557. [DOI] [PubMed] [Google Scholar]
- 81.Summer R, Kotton DN, Sun X, Ma B, Fitzsimmons K, Fine A. Side population cells and Bcrp1 expression in lung. American Journal of Physiology. 2003;285(1):L97–L104. doi: 10.1152/ajplung.00009.2003. [DOI] [PubMed] [Google Scholar]
- 82.Sung JM, Cho HJ, Yi H, et al. Characterization of a stem cell population in lung cancer A549 cells. Biochemical and Biophysical Research Communications. 2008;371(1):163–167. doi: 10.1016/j.bbrc.2008.04.038. [DOI] [PubMed] [Google Scholar]
- 83.Salcido CD, Larochelle A, Taylor BJ, Dunbar CE, Varticovski L. Molecular characterisation of side population cells with cancer stem cell-like characteristics in small-cell lung cancer. British Journal of Cancer. 2010;102(11):1636–1644. doi: 10.1038/sj.bjc.6605668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Eramo A, Haas TL, De Maria R. Lung cancer stem cells: tools and targets to fight lung cancer. Oncogene. 2010 doi: 10.1038/onc.2010.207. [DOI] [PubMed] [Google Scholar]
- 85.Deng S, Yang X, Lassus H, et al. Distinct expression levels and patterns of stem cell marker, aldehyde dehydrogenase isoform 1 (ALDH1), in human epithelial cancers. PLoS One. 2010;5(4) doi: 10.1371/journal.pone.0010277. Article ID e10277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Levina V, Marrangoni AM, DeMarco R, Gorelik E, Lokshin AE. Drug-selected human lung cancer stem cells: cytokine network, tumorigenic and metastatic properties. PLoS One. 2008;3(8) doi: 10.1371/journal.pone.0003077. Article ID e3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chen YC, Hsu HS, Chen YW, et al. Oct-4 expression maintained cancer stem-like properties in lung cancer-derived CD133-positive cells. PLoS One. 2008;3(7) doi: 10.1371/journal.pone.0002637. Article ID e2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bertolini G, Roz L, Perego P, et al. Highly tumorigenic lung cancer CD133 cells display stem-like features and are spared by cisplatin treatment. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(38):16281–16286. doi: 10.1073/pnas.0905653106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tirino V, Desiderio V, D’Aquino R, et al. Detection and characterization of CD133 cancer stem cells in human solid tumours. PLoS One. 2008;3(10) doi: 10.1371/journal.pone.0003469. Article ID e3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Salnikov AV, Gladkich J, Moldenhauer G, Volm M, Mattern J, Herr I. CD133 is indicative for a resistance phenotype but does not represent a prognostic marker for survival of non-small cell lung cancer patients. International Journal of Cancer. 2010;126(4):950–958. doi: 10.1002/ijc.24822. [DOI] [PubMed] [Google Scholar]
- 91.Li F, Zeng H, Ying K. The combination of stem cell markers CD133 and ABCG2 predicts relapse in stage I non-small cell lung carcinomas. doi: 10.1007/s12032-010-9646-5. Medical Oncology. Aug 18, 2010. PMID:20717756. [DOI] [PubMed] [Google Scholar]
- 92.Bonnefoix T, Bonnefoix P, Verdiel P, Sotto JJ. Fitting limiting dilution experiments with generalized linear models results in a test of the single-hit Poisson assumption. Journal of Immunological Methods. 1996;194(2):113–119. doi: 10.1016/0022-1759(96)00077-4. [DOI] [PubMed] [Google Scholar]
- 93.Jackson EL, Willis N, Mercer K, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes and Development. 2001;15(24):3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bishop JM. The molecular genetics of cancer. Science. 1987;235(4786):305–311. doi: 10.1126/science.3541204. [DOI] [PubMed] [Google Scholar]
- 95.Cameron ER, Neil JC. The Runx genes: lineage-specific oncogenes and tumor suppressors. Oncogene. 2004;23(24):4308–4314. doi: 10.1038/sj.onc.1207130. [DOI] [PubMed] [Google Scholar]
- 96.de Guzman CG, Warren AJ, Zhang Z, et al. Hematopoietic stem cell expansion and distinct myeloid developmental abnormalities in a murine model of the AML1-ETO translocation. Molecular and Cellular Biology. 2002;22(15):5506–5517. doi: 10.1128/MCB.22.15.5506-5517.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lee KS, Lee YS, Lee JM, et al. Runx3 is required for the differentiation of lung epithelial cells and suppression of lung cancer. Oncogene. 2010;29(23):3349–3361. doi: 10.1038/onc.2010.79. [DOI] [PubMed] [Google Scholar]
- 98.Li QL, Kim HR, Kim WJ, et al. Transcriptional silencing of the RUNX3 gene by CpG hypermethylation is associated with lung cancer. Biochemical and Biophysical Research Communications. 2004;314(1):223–228. doi: 10.1016/j.bbrc.2003.12.079. [DOI] [PubMed] [Google Scholar]
- 99.Licchesi JDF, Westra WH, Hooker CM, Machida EO, Baylin SB, Herman JG. Epigenetic alteration of wnt pathway antagonists in progressive glandular neoplasia of the lung. Carcinogenesis. 2008;29(5):895–904. doi: 10.1093/carcin/bgn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kwei KA, Kim YH, Girard L, et al. Genomic profiling identifies TITF1 as a lineage-specific oncogene amplified in lung cancer. Oncogene. 2008;27(25):3635–3640. doi: 10.1038/sj.onc.1211012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lockwood WW, Chari R, Coe BP, et al. Integrative genomic analyses identify BRF2 as a novel lineage-specific oncogene in lung squamous cell carcinoma. PLoS Medicine. 2010;7(7) doi: 10.1371/journal.pmed.1000315. Article ID e1000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhang X, Han B, Huang J, et al. Prognostic significance of OCT4 expression in adenocarcinoma of the lung. Japanese Journal of Clinical Oncology. 2010;40(10):961–966. doi: 10.1093/jjco/hyq066. [DOI] [PubMed] [Google Scholar]
- 103.Karoubi G, Gugger M, Schmid R, Dutly A. OCT4 expression in human non-small cell lung cancer: implications for therapeutic intervention. Interactive Cardiovascular and Thoracic Surgery. 2009;8(4):p. 397. doi: 10.1510/icvts.2008.193995. [DOI] [PubMed] [Google Scholar]
- 104.Hu T, Liu S, Breiter DR, Wang F, Tang Y, Sun S. Octamer 4 small interfering RNA results in cancer stem cell-like cell apoptosis. Cancer Research. 2008;68(16):6533–6540. doi: 10.1158/0008-5472.CAN-07-6642. [DOI] [PubMed] [Google Scholar]
- 105.Boyer LA, Tong IL, Cole MF, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122(6):947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ivanova N, Dobrin R, Lu R, et al. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006;442(7102):533–538. doi: 10.1038/nature04915. [DOI] [PubMed] [Google Scholar]
- 107.Hussenet T, Dali S, Exinger J, et al. SOX2 is an oncogene activated by recurrent 3q26.3 amplifications in human lung squamous cell carcinomas. PLoS One. 2010;5:15 pages. doi: 10.1371/journal.pone.0008960. Article ID e8960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lu Y, Futtner C, Rock JR, et al. Evidence that SOX2 overexpression is oncogenic in the lung. PLoS One. 2010;5(6) doi: 10.1371/journal.pone.0011022. Article ID e11022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 110.Wernig M, Meissner A, Foreman R, et al. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448(7151):318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 111.Krystal GW, Hines SJ, Organ CP. Autocrine growth of small cell lung cancer mediated by coexpression of c-kit and stem cell factor. Cancer Research. 1996;56(2):370–376. [PubMed] [Google Scholar]
- 112.Hines SJ, Organ C, Kornstein MJ, Krystal GW. Coexpression of the c-kit and stem cell factor genes in breast carcinomas. Cell Growth and Differentiation. 1995;6(6):769–779. [PubMed] [Google Scholar]
- 113.Yasuda A, Sawai H, Takahashi H, et al. Stem cell factor/c-kit receptor signaling enhances the proliferation and invasion of colorectal cancer cells through the PI3K/Akt Pathway. Digestive Diseases and Sciences. 2007;52(9):2292–2300. doi: 10.1007/s10620-007-9759-7. [DOI] [PubMed] [Google Scholar]
- 114.Hassan HT. c-Kit expression in human normal and malignant stem cells prognostic and therapeutic implications. Leukemia Research. 2009;33(1):5–10. doi: 10.1016/j.leukres.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 115.Levina V, Marrangoni A, Wang T, et al. Elimination of human lung cancer stem cells through targeting of the stem cell factor-c-kit autocrine signaling loop. Cancer Research. 2010;70(1):338–346. doi: 10.1158/0008-5472.CAN-09-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]