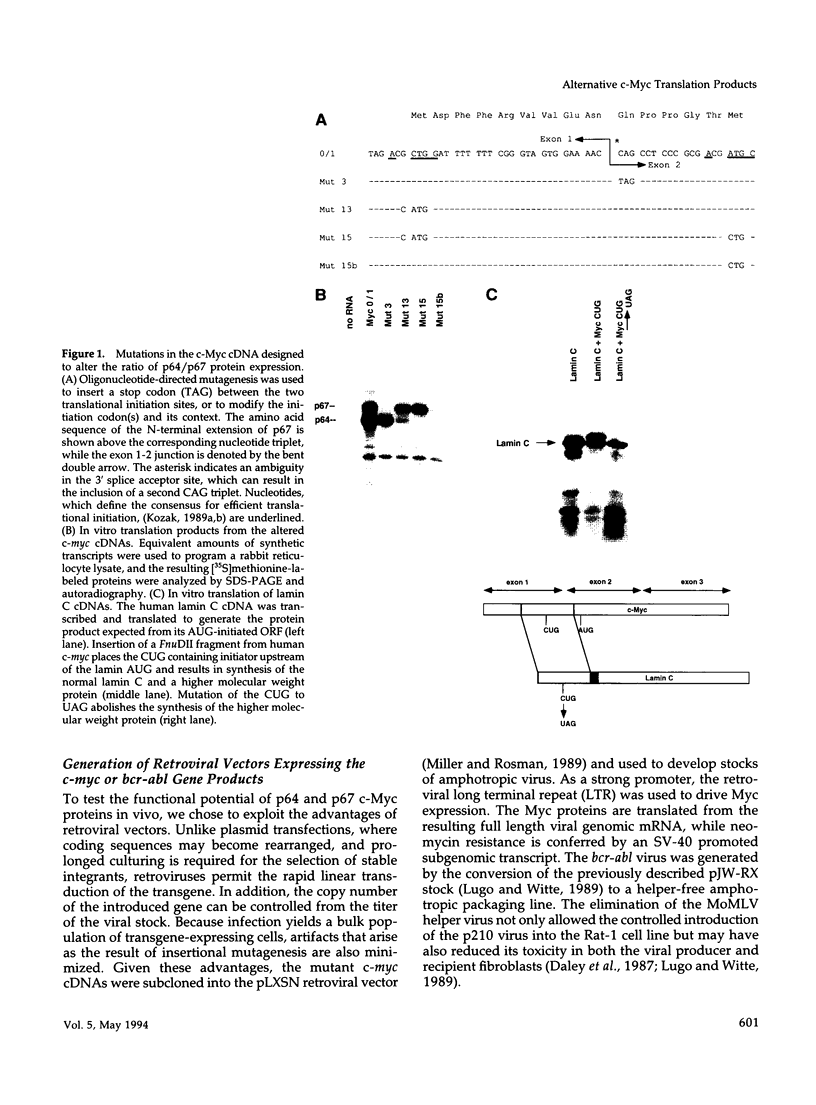
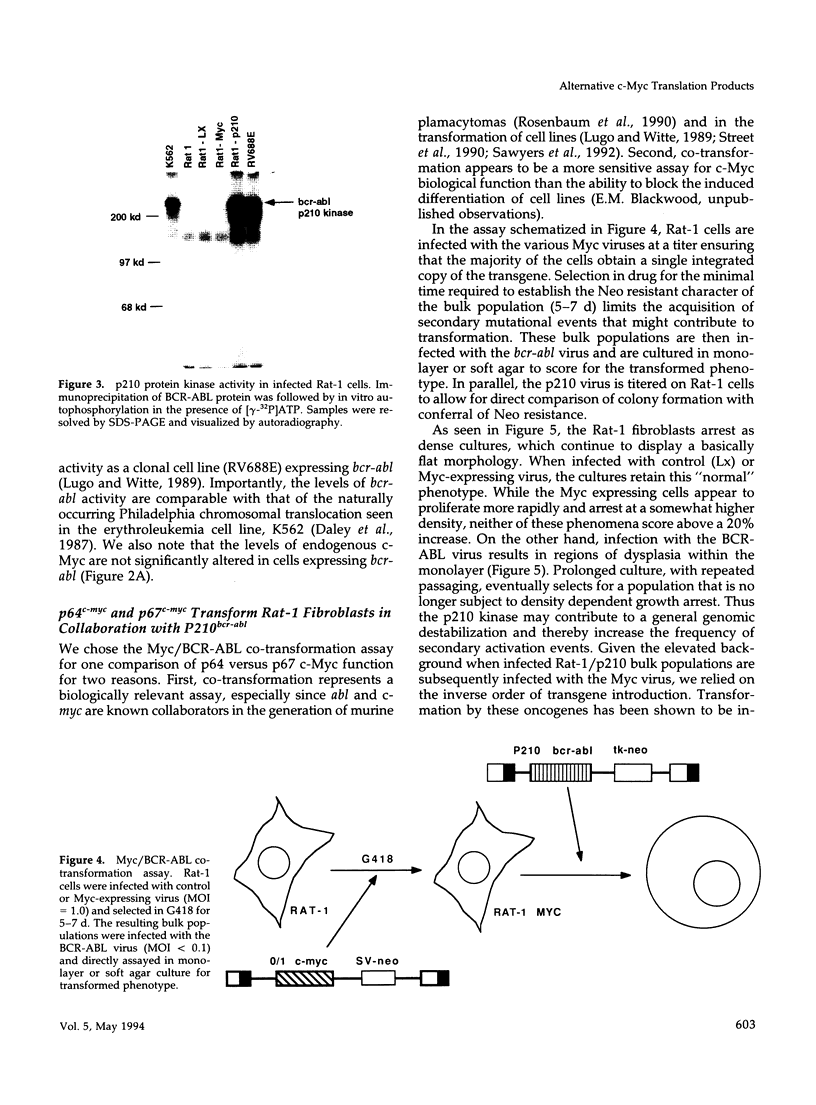
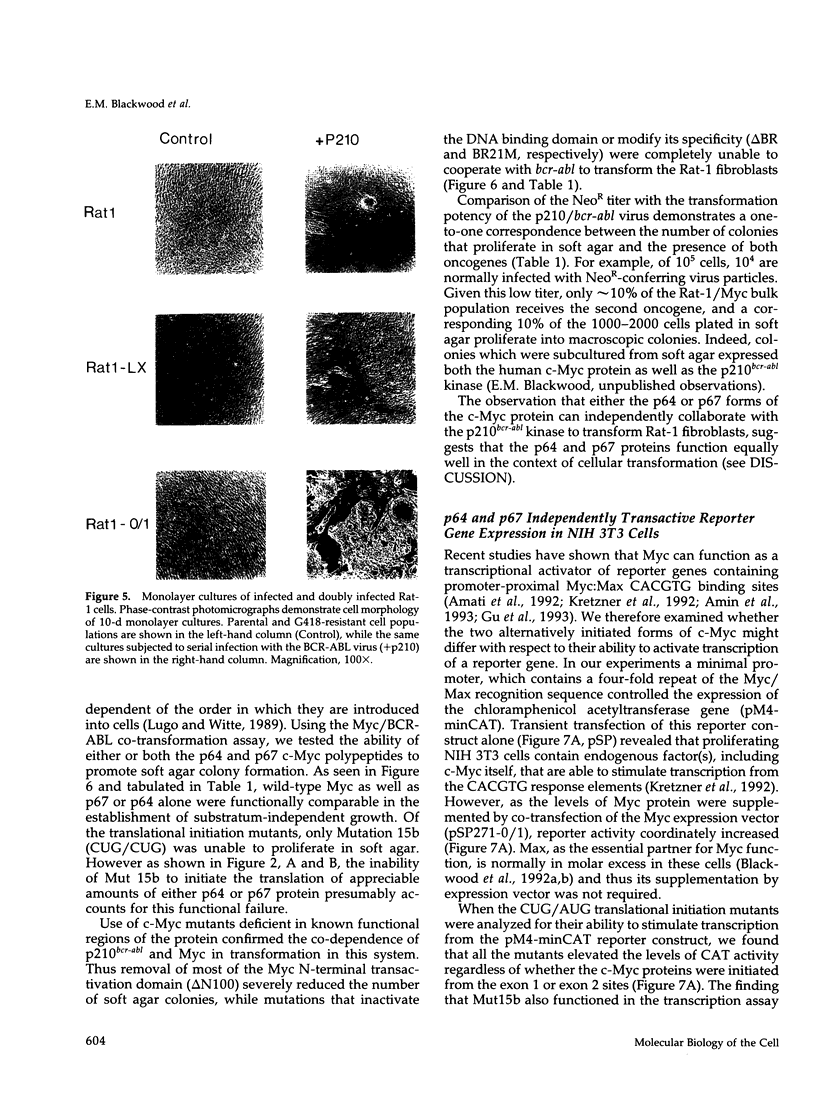
Abstract
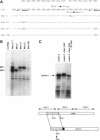
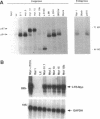
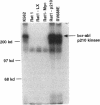
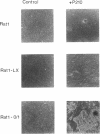
Activation of the c-myc proto-oncogene by chromosomal translocation or proviral insertion frequently results in the separation of the c-myc coding region from its normal regulatory elements. Such rearrangements are often accompanied by loss or mutation of c-myc exon 1 sequences. These genetic alterations do not affect synthesis of the major c-myc protein, p64, which is initiated from the first AUG codon in exon 2. However they can result in mutation or loss of the CUG codon located in exon 1 that normally serves as an alternative translational initiation codon for synthesis of an N-terminally extended form of c-Myc (p67). It has been hypothesized that p67 is a functionally distinct form of c-Myc whose specific loss during c-myc rearrangements confers a selective growth advantage. Here we describe experiments designed to test the functional properties of the two c-Myc protein forms. We introduced mutations within the translational initiation codons of a normal human c-myc cDNA that alter the pattern of Myc protein synthesis (p64 vs. p67). The functions of each of these proteins were experimentally addressed using co-transformation and transcriptional activation assays. Both the p64 and p67 c-Myc proteins were independently able to collaborate with bcr-abl in the transformation of Rat-1 fibroblasts. In addition, both the exon 1- and exon 2-initiated forms of the c-Myc protein stimulated transcription of a Myc/Max-responsive reporter construct to a similar level. Given the apparent absence of functional differences between p64 and p67, we conclude that the basis for c-Myc oncogenic activation lies primarily in the overall deregulation of its expression and not in alterations in the protein. The existence of the CUG translational initiator may reflect a mechanism for the continued synthesis of c-Myc protein under conditions where AUG initiation is inhibited.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Acland P., Dixon M., Peters G., Dickson C. Subcellular fate of the int-2 oncoprotein is determined by choice of initiation codon. Nature. 1990 Feb 15;343(6259):662–665. doi: 10.1038/343662a0. [DOI] [PubMed] [Google Scholar]
- Alitalo K., Koskinen P., Mäkelä T. P., Saksela K., Sistonen L., Winqvist R. myc oncogenes: activation and amplification. Biochim Biophys Acta. 1987 Apr 20;907(1):1–32. doi: 10.1016/0304-419x(87)90016-3. [DOI] [PubMed] [Google Scholar]
- Amati B., Brooks M. W., Levy N., Littlewood T. D., Evan G. I., Land H. Oncogenic activity of the c-Myc protein requires dimerization with Max. Cell. 1993 Jan 29;72(2):233–245. doi: 10.1016/0092-8674(93)90663-b. [DOI] [PubMed] [Google Scholar]
- Amati B., Dalton S., Brooks M. W., Littlewood T. D., Evan G. I., Land H. Transcriptional activation by the human c-Myc oncoprotein in yeast requires interaction with Max. Nature. 1992 Oct 1;359(6394):423–426. doi: 10.1038/359423a0. [DOI] [PubMed] [Google Scholar]
- Amin C., Wagner A. J., Hay N. Sequence-specific transcriptional activation by Myc and repression by Max. Mol Cell Biol. 1993 Jan;13(1):383–390. doi: 10.1128/mcb.13.1.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett J., Birrer M. J., Kato G. J., Dosaka-Akita H., Dang C. V. Activation domains of L-Myc and c-Myc determine their transforming potencies in rat embryo cells. Mol Cell Biol. 1992 Jul;12(7):3130–3137. doi: 10.1128/mcb.12.7.3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley D. L., Groudine M. Novel promoter upstream of the human c-myc gene and regulation of c-myc expression in B-cell lymphomas. Mol Cell Biol. 1986 Oct;6(10):3481–3489. doi: 10.1128/mcb.6.10.3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berberich S., Hyde-DeRuyscher N., Espenshade P., Cole M. max encodes a sequence-specific DNA-binding protein and is not regulated by serum growth factors. Oncogene. 1992 Apr;7(4):775–779. [PubMed] [Google Scholar]
- Bhatia K., Huppi K., Spangler G., Siwarski D., Iyer R., Magrath I. Point mutations in the c-Myc transactivation domain are common in Burkitt's lymphoma and mouse plasmacytomas. Nat Genet. 1993 Sep;5(1):56–61. doi: 10.1038/ng0993-56. [DOI] [PubMed] [Google Scholar]
- Bishop J. M. Molecular themes in oncogenesis. Cell. 1991 Jan 25;64(2):235–248. doi: 10.1016/0092-8674(91)90636-d. [DOI] [PubMed] [Google Scholar]
- Blackwell T. K., Kretzner L., Blackwood E. M., Eisenman R. N., Weintraub H. Sequence-specific DNA binding by the c-Myc protein. Science. 1990 Nov 23;250(4984):1149–1151. doi: 10.1126/science.2251503. [DOI] [PubMed] [Google Scholar]
- Blackwood E. M., Eisenman R. N. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991 Mar 8;251(4998):1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- Blackwood E. M., Kretzner L., Eisenman R. N. Myc and Max function as a nucleoprotein complex. Curr Opin Genet Dev. 1992 Apr;2(2):227–235. doi: 10.1016/s0959-437x(05)80278-3. [DOI] [PubMed] [Google Scholar]
- Blackwood E. M., Lüscher B., Eisenman R. N. Myc and Max associate in vivo. Genes Dev. 1992 Jan;6(1):71–80. doi: 10.1101/gad.6.1.71. [DOI] [PubMed] [Google Scholar]
- Bugler B., Amalric F., Prats H. Alternative initiation of translation determines cytoplasmic or nuclear localization of basic fibroblast growth factor. Mol Cell Biol. 1991 Jan;11(1):573–577. doi: 10.1128/mcb.11.1.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celenza J. L., Carlson M. Mutational analysis of the Saccharomyces cerevisiae SNF1 protein kinase and evidence for functional interaction with the SNF4 protein. Mol Cell Biol. 1989 Nov;9(11):5034–5044. doi: 10.1128/mcb.9.11.5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987 Apr;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Clark S. S., Crist W. M., Witte O. N. Molecular pathogenesis of Ph-positive leukemias. Annu Rev Med. 1989;40:113–122. doi: 10.1146/annurev.me.40.020189.000553. [DOI] [PubMed] [Google Scholar]
- Daley G. Q., McLaughlin J., Witte O. N., Baltimore D. The CML-specific P210 bcr/abl protein, unlike v-abl, does not transform NIH/3T3 fibroblasts. Science. 1987 Jul 31;237(4814):532–535. doi: 10.1126/science.2440107. [DOI] [PubMed] [Google Scholar]
- Davis R. L., Cheng P. F., Lassar A. B., Weintraub H. The MyoD DNA binding domain contains a recognition code for muscle-specific gene activation. Cell. 1990 Mar 9;60(5):733–746. doi: 10.1016/0092-8674(90)90088-v. [DOI] [PubMed] [Google Scholar]
- DePinho R. A., Schreiber-Agus N., Alt F. W. myc family oncogenes in the development of normal and neoplastic cells. Adv Cancer Res. 1991;57:1–46. doi: 10.1016/s0065-230x(08)60994-x. [DOI] [PubMed] [Google Scholar]
- Dean M., Levine R. A., Ran W., Kindy M. S., Sonenshein G. E., Campisi J. Regulation of c-myc transcription and mRNA abundance by serum growth factors and cell contact. J Biol Chem. 1986 Jul 15;261(20):9161–9166. [PubMed] [Google Scholar]
- Eilers M., Schirm S., Bishop J. M. The MYC protein activates transcription of the alpha-prothymosin gene. EMBO J. 1991 Jan;10(1):133–141. doi: 10.1002/j.1460-2075.1991.tb07929.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari S., Tagliafico E., D'Incá M., Ceccherelli G., Manfredini R., Selleri L., Donelli A., Sacchi S., Torelli G., Torelli U. Ratios between the abundance of messenger RNA and the corresponding protein of two growth-related genes, c-myc and vimentin, in leukemia blast cells. Cancer Res. 1990 Apr 1;50(7):1988–1991. [PubMed] [Google Scholar]
- Florkiewicz R. Z., Sommer A. Human basic fibroblast growth factor gene encodes four polypeptides: three initiate translation from non-AUG codons. Proc Natl Acad Sci U S A. 1989 Jun;86(11):3978–3981. doi: 10.1073/pnas.86.11.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg M. E., Ziff E. B. Stimulation of 3T3 cells induces transcription of the c-fos proto-oncogene. Nature. 1984 Oct 4;311(5985):433–438. doi: 10.1038/311433a0. [DOI] [PubMed] [Google Scholar]
- Gu W., Cechova K., Tassi V., Dalla-Favera R. Opposite regulation of gene transcription and cell proliferation by c-Myc and Max. Proc Natl Acad Sci U S A. 1993 Apr 1;90(7):2935–2939. doi: 10.1073/pnas.90.7.2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hann S. R., Abrams H. D., Rohrschneider L. R., Eisenman R. N. Proteins encoded by v-myc and c-myc oncogenes: identification and localization in acute leukemia virus transformants and bursal lymphoma cell lines. Cell. 1983 Oct;34(3):789–798. doi: 10.1016/0092-8674(83)90535-4. [DOI] [PubMed] [Google Scholar]
- Hann S. R., Eisenman R. N. Proteins encoded by the human c-myc oncogene: differential expression in neoplastic cells. Mol Cell Biol. 1984 Nov;4(11):2486–2497. doi: 10.1128/mcb.4.11.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hann S. R., King M. W., Bentley D. L., Anderson C. W., Eisenman R. N. A non-AUG translational initiation in c-myc exon 1 generates an N-terminally distinct protein whose synthesis is disrupted in Burkitt's lymphomas. Cell. 1988 Jan 29;52(2):185–195. doi: 10.1016/0092-8674(88)90507-7. [DOI] [PubMed] [Google Scholar]
- Hann S. R., Sloan-Brown K., Spotts G. D. Translational activation of the non-AUG-initiated c-myc 1 protein at high cell densities due to methionine deprivation. Genes Dev. 1992 Jul;6(7):1229–1240. doi: 10.1101/gad.6.7.1229. [DOI] [PubMed] [Google Scholar]
- Hann S. R., Thompson C. B., Eisenman R. N. c-myc oncogene protein synthesis is independent of the cell cycle in human and avian cells. 1985 Mar 28-Apr 3Nature. 314(6009):366–369. doi: 10.1038/314366a0. [DOI] [PubMed] [Google Scholar]
- Holt J. T., Redner R. L., Nienhuis A. W. An oligomer complementary to c-myc mRNA inhibits proliferation of HL-60 promyelocytic cells and induces differentiation. Mol Cell Biol. 1988 Feb;8(2):963–973. doi: 10.1128/mcb.8.2.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karn J., Watson J. V., Lowe A. D., Green S. M., Vedeckis W. Regulation of cell cycle duration by c-myc levels. Oncogene. 1989 Jun;4(6):773–787. [PubMed] [Google Scholar]
- Kato G. J., Barrett J., Villa-Garcia M., Dang C. V. An amino-terminal c-myc domain required for neoplastic transformation activates transcription. Mol Cell Biol. 1990 Nov;10(11):5914–5920. doi: 10.1128/mcb.10.11.5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato G. J., Lee W. M., Chen L. L., Dang C. V. Max: functional domains and interaction with c-Myc. Genes Dev. 1992 Jan;6(1):81–92. doi: 10.1101/gad.6.1.81. [DOI] [PubMed] [Google Scholar]
- Kelly K., Cochran B. H., Stiles C. D., Leder P. Cell-specific regulation of the c-myc gene by lymphocyte mitogens and platelet-derived growth factor. Cell. 1983 Dec;35(3 Pt 2):603–610. doi: 10.1016/0092-8674(83)90092-2. [DOI] [PubMed] [Google Scholar]
- Kozak M. Context effects and inefficient initiation at non-AUG codons in eucaryotic cell-free translation systems. Mol Cell Biol. 1989 Nov;9(11):5073–5080. doi: 10.1128/mcb.9.11.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzner L., Blackwood E. M., Eisenman R. N. Myc and Max proteins possess distinct transcriptional activities. Nature. 1992 Oct 1;359(6394):426–429. doi: 10.1038/359426a0. [DOI] [PubMed] [Google Scholar]
- Lock P., Ralph S., Stanley E., Boulet I., Ramsay R., Dunn A. R. Two isoforms of murine hck, generated by utilization of alternative translational initiation codons, exhibit different patterns of subcellular localization. Mol Cell Biol. 1991 Sep;11(9):4363–4370. doi: 10.1128/mcb.11.9.4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugo T. G., Witte O. N. The BCR-ABL oncogene transforms Rat-1 cells and cooperates with v-myc. Mol Cell Biol. 1989 Mar;9(3):1263–1270. doi: 10.1128/mcb.9.3.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüscher B., Eisenman R. N. c-myc and c-myb protein degradation: effect of metabolic inhibitors and heat shock. Mol Cell Biol. 1988 Jun;8(6):2504–2512. doi: 10.1128/mcb.8.6.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüscher B., Kuenzel E. A., Krebs E. G., Eisenman R. N. Myc oncoproteins are phosphorylated by casein kinase II. EMBO J. 1989 Apr;8(4):1111–1119. doi: 10.1002/j.1460-2075.1989.tb03481.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcu K. B., Bossone S. A., Patel A. J. myc function and regulation. Annu Rev Biochem. 1992;61:809–860. doi: 10.1146/annurev.bi.61.070192.004113. [DOI] [PubMed] [Google Scholar]
- McCarthy D. M., Rassool F. V., Goldman J. M., Graham S. V., Birnie G. D. Genomic alterations involving the c-myc proto-oncogene locus during the evolution of a case of chronic granulocytic leukaemia. Lancet. 1984 Dec 15;2(8416):1362–1365. doi: 10.1016/s0140-6736(84)92058-0. [DOI] [PubMed] [Google Scholar]
- McKeon F. D., Kirschner M. W., Caput D. Homologies in both primary and secondary structure between nuclear envelope and intermediate filament proteins. Nature. 1986 Feb 6;319(6053):463–468. doi: 10.1038/319463a0. [DOI] [PubMed] [Google Scholar]
- Meichle A., Philipp A., Eilers M. The functions of Myc proteins. Biochim Biophys Acta. 1992 Dec 16;1114(2-3):129–146. doi: 10.1016/0304-419x(92)90011-m. [DOI] [PubMed] [Google Scholar]
- Mellentin J. D., Smith S. D., Cleary M. L. lyl-1, a novel gene altered by chromosomal translocation in T cell leukemia, codes for a protein with a helix-loop-helix DNA binding motif. Cell. 1989 Jul 14;58(1):77–83. doi: 10.1016/0092-8674(89)90404-2. [DOI] [PubMed] [Google Scholar]
- Miller A. D., Rosman G. J. Improved retroviral vectors for gene transfer and expression. Biotechniques. 1989 Oct;7(9):980-2, 984-6, 989-90. [PMC free article] [PubMed] [Google Scholar]
- Miller J. B. Myoblast diversity in skeletal myogenesis: how much and to what end? Cell. 1992 Apr 3;69(1):1–3. doi: 10.1016/0092-8674(92)90111-o. [DOI] [PubMed] [Google Scholar]
- Moberg K. H., Logan T. J., Tyndall W. A., Hall D. J. Three distinct elements within the murine c-myc promoter are required for transcription. Oncogene. 1992 Mar;7(3):411–421. [PubMed] [Google Scholar]
- Moerschell R. P., Hosokawa Y., Tsunasawa S., Sherman F. The specificities of yeast methionine aminopeptidase and acetylation of amino-terminal methionine in vivo. Processing of altered iso-1-cytochromes c created by oligonucleotide transformation. J Biol Chem. 1990 Nov 15;265(32):19638–19643. [PubMed] [Google Scholar]
- Mukherjee B., Morgenbesser S. D., DePinho R. A. Myc family oncoproteins function through a common pathway to transform normal cells in culture: cross-interference by Max and trans-acting dominant mutants. Genes Dev. 1992 Aug;6(8):1480–1492. doi: 10.1101/gad.6.8.1480. [DOI] [PubMed] [Google Scholar]
- Murre C., McCaw P. S., Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989 Mar 10;56(5):777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- Nepveu A., Marcu K. B., Skoultchi A. I., Lachman H. M. Contributions of transcriptional and post-transcriptional mechanisms to the regulation of c-myc expression in mouse erythroleukemia cells. Genes Dev. 1987 Nov;1(9):938–945. doi: 10.1101/gad.1.9.938. [DOI] [PubMed] [Google Scholar]
- Nowell P., Finan J., Dalla-Favera R., Gallo R. C., ar-Rushdi A., Romanczuk H., Selden J. R., Emanuel B. S., Rovera G., Croce C. M. Association of amplified oncogene c-myc with an abnormally banded chromosome 8 in a human leukaemia cell line. Nature. 1983 Dec 1;306(5942):494–497. doi: 10.1038/306494a0. [DOI] [PubMed] [Google Scholar]
- Ohno S., Migita S., Wiener F., Babonits M., Klein G., Mushinski J. F., Potter M. Chromosomal translocations activating myc sequences and transduction of v-abl are critical events in the rapid induction of plasmacytomas by pristane and abelson virus. J Exp Med. 1984 Jun 1;159(6):1762–1777. doi: 10.1084/jem.159.6.1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peabody D. S. Translation initiation at non-AUG triplets in mammalian cells. J Biol Chem. 1989 Mar 25;264(9):5031–5035. [PubMed] [Google Scholar]
- Penn L. J., Brooks M. W., Laufer E. M., Land H. Negative autoregulation of c-myc transcription. EMBO J. 1990 Apr;9(4):1113–1121. doi: 10.1002/j.1460-2075.1990.tb08217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn L. J., Brooks M. W., Laufer E. M., Littlewood T. D., Morgenstern J. P., Evan G. I., Lee W. M., Land H. Domains of human c-myc protein required for autosuppression and cooperation with ras oncogenes are overlapping. Mol Cell Biol. 1990 Sep;10(9):4961–4966. doi: 10.1128/mcb.10.9.4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prats A. C., De Billy G., Wang P., Darlix J. L. CUG initiation codon used for the synthesis of a cell surface antigen coded by the murine leukemia virus. J Mol Biol. 1989 Jan 20;205(2):363–372. doi: 10.1016/0022-2836(89)90347-1. [DOI] [PubMed] [Google Scholar]
- Prats H., Kaghad M., Prats A. C., Klagsbrun M., Lélias J. M., Liauzun P., Chalon P., Tauber J. P., Amalric F., Smith J. A. High molecular mass forms of basic fibroblast growth factor are initiated by alternative CUG codons. Proc Natl Acad Sci U S A. 1989 Mar;86(6):1836–1840. doi: 10.1073/pnas.86.6.1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preisler H. D., Sato H., Yang P. M., Wilson M., Kaufman C., Watt R. Assessment of c-myc expression in individual leukemic cells. Leuk Res. 1988;12(6):507–516. doi: 10.1016/0145-2126(88)90118-x. [DOI] [PubMed] [Google Scholar]
- Prendergast G. C., Lawe D., Ziff E. B. Association of Myn, the murine homolog of max, with c-Myc stimulates methylation-sensitive DNA binding and ras cotransformation. Cell. 1991 May 3;65(3):395–407. doi: 10.1016/0092-8674(91)90457-a. [DOI] [PubMed] [Google Scholar]
- Prendergast G. C., Ziff E. B. Methylation-sensitive sequence-specific DNA binding by the c-Myc basic region. Science. 1991 Jan 11;251(4990):186–189. doi: 10.1126/science.1987636. [DOI] [PubMed] [Google Scholar]
- Rosenbaum H., Harris A. W., Bath M. L., McNeall J., Webb E., Adams J. M., Cory S. An E mu-v-abl transgene elicits plasmacytomas in concert with an activated myc gene. EMBO J. 1990 Mar;9(3):897–905. doi: 10.1002/j.1460-2075.1990.tb08187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussel M. F., Cleveland J. L., Shurtleff S. A., Sherr C. J. Myc rescue of a mutant CSF-1 receptor impaired in mitogenic signalling. Nature. 1991 Sep 26;353(6342):361–363. doi: 10.1038/353361a0. [DOI] [PubMed] [Google Scholar]
- Saris C. J., Domen J., Berns A. The pim-1 oncogene encodes two related protein-serine/threonine kinases by alternative initiation at AUG and CUG. EMBO J. 1991 Mar;10(3):655–664. doi: 10.1002/j.1460-2075.1991.tb07994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyers C. L., Callahan W., Witte O. N. Dominant negative MYC blocks transformation by ABL oncogenes. Cell. 1992 Sep 18;70(6):901–910. doi: 10.1016/0092-8674(92)90241-4. [DOI] [PubMed] [Google Scholar]
- Spencer C. A., Groudine M. Control of c-myc regulation in normal and neoplastic cells. Adv Cancer Res. 1991;56:1–48. doi: 10.1016/s0065-230x(08)60476-5. [DOI] [PubMed] [Google Scholar]
- Stone J., de Lange T., Ramsay G., Jakobovits E., Bishop J. M., Varmus H., Lee W. Definition of regions in human c-myc that are involved in transformation and nuclear localization. Mol Cell Biol. 1987 May;7(5):1697–1709. doi: 10.1128/mcb.7.5.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Street A. J., Blackwood E., Lüscher B., Eisenman R. N. Mutational analysis of the carboxy-terminal casein kinase II phosphorylation site in human c-myc. Curr Top Microbiol Immunol. 1990;166:251–258. doi: 10.1007/978-3-642-75889-8_31. [DOI] [PubMed] [Google Scholar]
- Thompson C. B., Challoner P. B., Neiman P. E., Groudine M. Levels of c-myc oncogene mRNA are invariant throughout the cell cycle. 1985 Mar 28-Apr 3Nature. 314(6009):363–366. doi: 10.1038/314363a0. [DOI] [PubMed] [Google Scholar]
- Vogelstein B., Kinzler K. W. The multistep nature of cancer. Trends Genet. 1993 Apr;9(4):138–141. doi: 10.1016/0168-9525(93)90209-z. [DOI] [PubMed] [Google Scholar]
- Waters C. M., Littlewood T. D., Hancock D. C., Moore J. P., Evan G. I. c-myc protein expression in untransformed fibroblasts. Oncogene. 1991 May;6(5):797–805. [PubMed] [Google Scholar]
- Webb E., Barri G., Cory S., Adams J. M. Lymphomagenesis in Emu-myc transgenic mice does not require transgene rearrangement or mutation of myc exon 1. Mol Biol Med. 1989 Dec;6(6):475–480. [PubMed] [Google Scholar]
- Wisdom R., Lee W. The protein-coding region of c-myc mRNA contains a sequence that specifies rapid mRNA turnover and induction by protein synthesis inhibitors. Genes Dev. 1991 Feb;5(2):232–243. doi: 10.1101/gad.5.2.232. [DOI] [PubMed] [Google Scholar]
- Yano T., Sander C. A., Clark H. M., Dolezal M. V., Jaffe E. S., Raffeld M. Clustered mutations in the second exon of the MYC gene in sporadic Burkitt's lymphoma. Oncogene. 1993 Oct;8(10):2741–2748. [PubMed] [Google Scholar]