Abstract
Gene fusions prevalent in prostate cancer (CaP) lead to the elevated expression of the ERG proto-oncogene. ERG activation present in 50–70% of prostate tumors underscores one of the most common oncogenic alterations in CaP. Despite numerous reports of gene fusions and mRNA expression, ERG oncoprotein status in CaP still remains to be defined. Furthermore, development of ERG protein-based assays may provide a new dimension to evaluation of gene fusions involving diverse androgen-regulated promoters and the ERG protein-coding sequence. Through exhaustive evaluations of 132 whole-mount prostates (261 tumor foci and over 200 000 benign glands) for the ERG oncoprotein nuclear expression, we demonstrated 99.9% specificity for detecting prostate tumor cells using a highly specific anti-ERG monoclonal antibody. The ERG oncoprotein expression correlated well with fusion transcript or gene fusion in randomly selected specimens. Strong concordance of ERG-positive foci of prostatic intraepithelial neoplasia (PIN) with ERG-positive carcinoma (82 out of 85 sections with PIN, 96.5%) affirms the biological role of ERG in clonal selection of prostate tumors in 65% (86 out of 132) of patients. Conversely, ERG negative PINs were associated with ERG-negative carcinoma. Taken together, the homogeneous and strong ERG expression detected in individual tumors establishes the potential for ERG oncoprotein-based stratification of CaP.
Keywords: ERG, oncoprotein, prostatic intraepithelial neoplasia, clonal selection, patient stratification
Introduction
Prevalent gene fusions involving regulatory sequences of the androgen receptor (AR) regulated prostate-associated genes (predominantly TMPRSS2) and protein-coding sequences of nuclear transcription factors in the ETS gene family (primarily ERG), result in frequent overexpression of ERG in prostate tumors.1, 2, 3, 4, 5 Emerging studies suggest oncogenic functions of ERG and ETV1 in prostate cancer (CaP).1, 6, 7, 8, 9, 10, 11 Previous studies including our report have analyzed ERG gene fusions at genomic or mRNA levels in the context of multi-focal CaP and these data showed inter-tumoral heterogeneity within the same prostate.12, 13, 14, 15 Despite numerous reports of gene fusions and mRNA expression, ERG oncoprotein in CaP still remains to be defined. Using an anti-ERG monoclonal antibody (ERG–MAb) developed by our group, a global view of ERG oncoprotein expression has been established in the context of multi-focal CaP.
Materials and methods
Cell culture and androgen treatment
LNCaP (ATCC, no. CRL-1740) cells were grown in RPMI-1640 medium supplemented with 10% fetal bovine serum and 2 m glutamine. Cells (2 × 106) were seeded onto 10 cm dishes and maintained for 5 days in media containing 10% charcoal-stripped fetal bovine serum (c-FBS; no. 100119 Gemini Bio-Products, Calabasas, CA, USA). For androgen induction, fresh media was supplemented with 0.1 n R1881 or 1 n R1881 synthetic androgen for 48 h. VCaP cells (ATCC, no. CRL-2876) were grown in DMEM medium supplemented with 10% fetal bovine serum and 2 m glutamine. Cells (2 × 106) were seeded onto 10 cm dishes and maintained for 3 days in media containing 10% charcoal-stripped fetal bovine serum. For androgen induction, fresh media were supplemented with 0.1 n R1881 or 1 n R1881 for another 48 h. At the end of the incubation period, cells were harvested and analyzed by western blots and by microscopy.
ERG siRNA treatment of prostate cancer cells
VCaP cells were seeded onto 10 cm tissue culture dishes in DMEM medium containing 10% c-FBS for 3 days. Cells were transfected with ERG siRNA or non-targeting control RNA using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) as described before.7 Twelve hours after transfection with siRNAs, the cell culture medium was replaced with DMEM containing 10% charcoal-stripped serum and 0.1 n R1881 and maintained for 4 days before harvest and analysis by western blots and microscopy.
Immunoblot analysis
Cells were lysed in M-PER mammalian protein extraction reagent (Thermo, Rockford, IL,USA) containing protease and phosphatase inhibitor cocktails (Sigma, St Louis, MO, USA). Proteins were measured with Bradford Assay reagent (BioRad, Hercules, CA, USA) and lysates equivalent to 25 μg proteins were separated on NuPAGE Bis-Tris (4–12%) gels (Invitrogen, Carlsbad, CA, USA) and blotted onto PVDF membranes (Invitrogen). Immunoblot assays were performed with ERG–MAb (CPDR) mouse monoclonal antibody generated against immunizing polypeptide GQTSKMSPRVPQQDWLSQPPARVTI, anti-PSA (Cat # A056201–2, DAKO, Carpinteria, CA, USA) and anti--tubulin (Cat no. sc-5286, Santa Cruz, CA, USA) antibodies. Clustal W16 alignment did not reveal a significant homology of the ERG–MAb peptide antigen with 29 other protein sequences belonging to the human ETS family. Of note, FLI1 protein sequence, which showed 48% identity with the ERG-immunizing peptide was not recognized by the ERG–MAb (Supplementary Figure S1).
Immunofluorescence assay
Cells were fixed in fresh 4% formaldehyde in phosphate-buffered saline (PBS) and permeabilized in PBS-T (PBS + 0.1% Triton X-100) and then centrifuged onto glass slides with a Cytospin 4 centrifuge. Cells were blocked in PBS-NT20 (PBS supplemented with 0.1% Tween-20 and 1% normal horse serum (Vector Laboratories, Burlingame, CA, USA). After incubation with a primary antibody, cells were rinsed and then treated with goat anti-mouse Alexa-594 (Cat no. A11302, Invitrogen) followed by DAPI staining. Images were captured using a × 40/0.65 N-Plan objective on a Leica DMIRE2 inverted microscope equipped with a QImaging Retiga-EX CCD camera (Burnaby, BC, Canada), operated by OpenLab software (Improvision, Lexington, MA, USA). Images were converted into color and merged by using Photoshop (Adobe, San Jose, CA, USA). For ERG peptide competition experiments, the ERG–MAb antibody was pre-incubated with 2000-fold molar excess of competing or non-competing peptide on ice for 30 min.
Prostate specimens
Under an Institutional Review Board-approved protocol, radical prostatectomy specimens from patients enrolled in the Center for Prostate Disease Research program were obtained by pathologists within 30 min after the surgical removal of the specimens. Prostates were processed as whole-mounts according to the Armed Forces Institute of Pathology (AFIP) protocol.15 From each of 132 patients, one whole-mount cross section containing one to four tumors (mostly two foci) was selected and tumors represented different grades and stages. Each tumor was separately diagnosed in the prostatectomy specimens and slices with more than one tumor focus represented separate tumors. The cohort includes 50 stage and grade matched patients of whom 25 developed metastasis and 25 had no recurrence with a mean follow up of 46.5 months with the intent to address possible prognostic features of ERG. To assess the relationship between mRNA and ERG oncoprotein data specimens from patients were included from previous studies investigating ERG transcripts by quantitative RT-PCR or by GeneChip (2, 5).
Immunohistochemistry (IHC) for ERG
Following deparaffinization, 4 μm sections were dehydrated and blocked in 0.6% hydrogen peroxide in methanol for 20 min. Sections were processed for antigen retrieval in EDTA (pH 9.0) for 30 min in a microwave followed by 30 min of cooling in EDTA buffer. Sections were then blocked in 1% horse serum for 40 min followed by incubation with the ERG–MAb mouse monoclonal antibody at a dilution of 1:1280 for 60 min at room temperature. Sections were incubated with the biotinylated horse anti-mouse antibody at a dilution of 1:200 (Vector Laboratories, Burlingame, CA, USA) for 30 min followed by treatment with the ABC Kit (Vector Laboratories) for 30 min. The color detection was achieved by treatment with VIP (Vector Laboratories) for 5 min. Sections were then counterstained in hematoxylin for 1 min, dehydrated, cleared and mounted. The ERG–MAb staining was determined according to percent of cells positive: up to 25% (1+), >25–50% (2+), >50–75% (3+) and >75% (4+). The intensity was scored as mild (1+), moderate (2+) and marked (3+). A combination of both measurements was calculated by multiplying the percent of positive cells with the degree of intensity, which resulted in a score. As, most of the tumors showed positivity in over 75% of cells and the intensity was uniform, we expressed the staining results as ERG positive or negative.
Analysis of ERG mRNA by branched-chain DNA (bDNA) signal amplification
One 4-μm thick section was selected from each of the 35 FFPE whole-mount prostate samples. Areas identified as tumors were marked, removed by scraping and were homogenized and processed as described previously17 and in the Supplementary Materials and Methods. The geometric mean of the expression of three housekeeping genes (ACTB, B2M, RPL19) was determined and only samples with this mean value of minimum three-fold over background signal were included in the analysis. The TMPRSS2–ERG expression data was normalized to the geometric mean of the three housekeeping genes in each sample. Samples with normalized TMPRSS2–ERG expression over an arbitrary cutoff of 0.5 were considered positive for the fusion. The TMPRSS2–ERG expression data tightly correlated with similarly normalized ERG mRNA expression in the same samples. The blinded transcript expression data were then compared with the protein expression data.
Statistical analysis
Sensitivity and specificity of ERG oncoprotein expression were analyzed for distinguishing all tumor foci from benign glands in whole-mount prostates (261 tumor foci and over 200 000 benign glands). Chi square test was used to test the association of ERG oncoprotein status with tumor differentiation and Gleason score for individual tumors. P-value of 0.05 was adopted as statistically significant. The SAS version 9.2 was used for all data analyses.
Results
Characterization of ERG oncoprotein by ERG monoclonal antibody in cancer cell lines
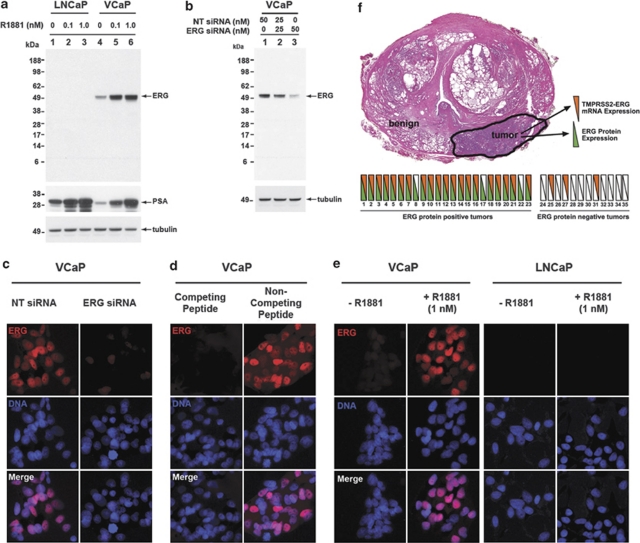
In TMPRRS2–ERG-positive VCaP cells, a mouse monoclonal anti-ERG antibody, ERG–MAb recognized predicted sizes of full length protein products (50–52 kDa) encoded by TMPRSS2–ERG2 and TMPRSS2–ERG3 fusion transcripts (Figure 1 and Supplementary Figure S2). As expected, ERG–MAb did not detect ERG oncoprotein in LNCaP cells, which do not harbor TMPRSS2–ERG fusion (Figure 1a and Supplementary Figure S2b). To further show the specificity of ERG–MAb, a significant inhibition of the endogenous ERG oncoprotein was noted in ERG siRNA7 transfected VCaP cells (Figure 1b). ERG protein was also detected in tumor cell lines (KG1, COLO 320, MOLT4) previously described to express ERG (Supplementary Figure 2b). Specificity of the ERG–MAb for ERG oncoprotein detection in VCaP cells was further validated by immunofluorescence (IF) assays (Figures 1c–e). These data together established the specificity of the ERG–MAb in detecting ERG oncoprotein in CaP cells.
Figure 1.
Detection of ERG oncoprotein by ERG–MAb in prostate cancer cells. (a) LNCaP cells treated with 0.1 n R1881 (lane 2) or 1 n R1881 (lane 3) and VCaP cells treated with 0.1 n R1881 (lane 5) or 1 n R1881 (lane 6) were analyzed for ERG oncoprotein by using ERG–MAb as described in Materials and Methods. LNCaP (lane 1) and VCaP cells (lane 4) were processed in parallel without R1881 served as controls. (b) ERG oncoprotein was analyzed in VCaP cells transfected with non-targeting (NT) or ERG siRNA oligonucleotides. Cell lysates were prepared 4 days after transfection with 50 n NT siRNA (lane 1), 25 n NT siRNA and 25 n ERG siRNA (lane 2) or 50 n ERG siRNA oligonucleotides (lane 3). Twenty-five micrograms of cell lysates were separated on NuPAGE Bis-Tris (4–12%) gels, transferred onto PVDF membrane and immunoblotted ERG–MAb. Identical samples were transferred onto PVDF membranes and probed with ERG–MAb, anti-PSA and anti--tubulin antibodies. The apparent size of the ERG protein products in the western blots correspond to predicted molecular weights of TMPRSS2 (exon 1)-ERG3 (exons from 8 to 16, GenBank accession number NM_001136154) or TMPRSS2 (exon 1)-ERG2 (exons from 8 to 16 lacking exon 12, GenBank accession number NM_004449). (c) VCaP cells transfected with either NT siRNA (left panel) or ERG siRNA (right panel) were immunostained with mouse ERG–MAb followed by goat anti-mouse Alexa-594 (red). (d) VCaP cells were grown in DMEM supplemented with 10% fetal bovine serum. Cells were incubated with ERG–MAb, pre-treated with competing or non-competing peptide. (e) VCaP or LNCaP cells treated with or without 1 n of R1881 were analyzed for ERG oncoprotein by ERG–MAb. (f) Schematic representation of the expression of ERG oncoprotein (IHC) and TMPRSS2–ERG fusion mRNA was determined in prostate tumors of 35 CaP patients treated with radical prostatectomy by using bDNA assay as described in Materials and Methods. Consecutive tissue slides from whole-mounted FFPE prostate specimens were used for the two assays in a blinded fashion. Green triangles represent positive ERG oncoprotein staining, orange triangles represent the detection of TMPRSS2–ERG fusion mRNA. Hollow triangles indicate specimens with undetectable ERG oncoprotein or TMPRSS2–ERG fusion transcript.
Relationship of ERG oncoprotein and TMPRSS2–ERG fusion status in prostate specimens
To determine the clinical utility of ERG–MAb, it was critical to establish the specificity of the ERG oncoprotein staining in tumor specimens in relation to TMPRSS2–ERG fusion status. A comparative analysis was performed on consecutive tissue sections of the ERG oncoprotein positive or negative FFPE specimens for the detection of TMPRSS2–ERG mRNA. Analysis of 35 evaluable specimens revealed a strong correlation between mRNA levels of TMPRSS2–ERG fusion type A transcript and ERG oncoprotein immunohistochemistry (Figure 1f). A concordance rate of 82.8% was noted between mRNA and protein data despite the expected differences in the sensitivity as well as read-outs of the two techniques. A comparative evaluation of TMPRSS2–ERG gene fusion analysis by fluorescence in situ hybridization (FISH) and ERG oncoprotein expression by ERG–MAb IHC in 10 specimens revealed no discrepancies (Supplementary Figure S3).
Expression map of the ERG oncoprotein in multi-focal prostate cancer
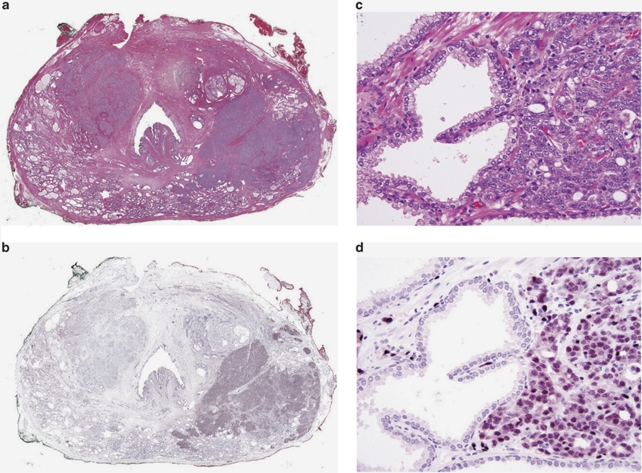
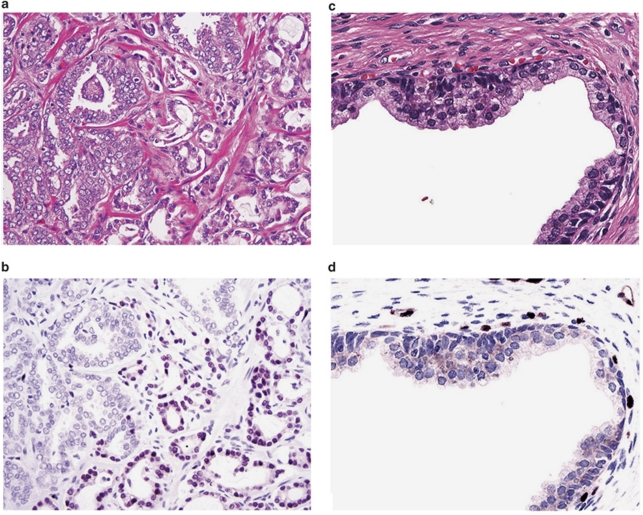
To delineate the expression map of ERG in benign glands, carcinoma and prostatic intraepithelial neoplasia (PIN), we utilized one entire cross section of each whole-mount radical prostatectomy from 132 patients with prostatic carcinoma. Each tumor was individually measured and graded. On average, one whole-mount section (3.5 × 2.5 cm or 4.0 × 3.5 cm) is equivalent to approximately 800–1400 tissue microarray cores of 1 mm diameter. In addition to index tumors, most of these cross sections contained benign prostatic tissue of the peripheral and the transition/periurethral zone as well as the urethra, utricle, ejaculatory ducts (Figures 2a and b), and seminal vesicles. A single tumor was present in 51 sections, and multiple individual tumors were present in 81 sections. Tumor grade, pathological stage, margin status and clinical data are summarized in Supplementary Table S1. In prostatic adenocarcinomas (Figures 2c and d) and in PIN (Figures 2e and f) the epithelial cells showed nuclear staining. ERG was positive in 117 of 261 (44.8%) individual tumors (Table 1a). ERG oncoprotein expression was highly specific (99.9%) in detecting carcinoma (Table 1a). Of 132 specimens only six specimens showed rare ERG-positive non-malignant cells. In three specimens, a single group of benign glands (average seven glands, raging from five to eight glands) each was positive for ERG in addition to carcinoma (Figures 2g and h). In three additional specimens, ERG was present in small aggregates of native glands (3–5 glands) with increased cellularity and nuclear enlargement and mild atypia, changes previously referred to as ‘low grade PIN'. Eight of the nine anterior/transition zone tumors were negative (Figures 2i and j). In all but five cases, over 85% of tumor cells showed moderate to strong nuclear staining with cytoplasmic blush (Figures 2c and d).
Figure 2.
Distribution of ERG oncoprotein in a patient's whole-mount cross section of the prostate. (a) Whole-mount cross section of one prostate with two tumors: left upper quadrant and right lower quadrant, H&E × 1. (b) Same section as (a). The tumor in the left upper quadrant is ERG negative, whereas the tumor in the right lower quadrant is ERG positive. Note that the entire tumor is positively outlined including the irregularly infiltrating borders, ERG–MAb × 1. (c) The tumor infiltrates as densely packed simple glands between benign glands, H&E × 20. (d) Same field as (c). Only tumor cells are positive for ERG. Note the strong reactivity in endothelial cells of the capillaries, some of which are in intimate proximity to benign glands, ERG–MAb × 20. (e) Native glands lined by secretory cells with nuclear anaplasia and recognizable basal cells are diagnostic of high-grade prostatic intraepithelial neoplasia. They are associated with infiltrating carcinoma, H&E × 20. (f) Same field as Figure (e). The nuclei of both the prostatic intraepithelial neoplasia and infiltrating carcinoma, are positive for ERG, but with variable intensity. Basal cells are negative. Note the uniformly strong nuclear staining in capillaries (arrows). (g) A cluster of benign glands appears to be prominent based on the dark staining cytoplasm, H&E × 20. (h) Same field as (g). Rare benign secretory cells show nuclear reactivity, ERG–MAb × 20. (i) Left upper quadrant tumor. Note benign glands in the lower left. H&E × 1. The inset shows infiltrating carcinoma at the left adjacent to a benign gland, H&E × 20. (j) Same field as (i). The tumor is negative for ERG. In the inset, both the benign gland and tumor are negative for ERG. ERG–MAb × 1 inset ERG–MAb × 20.
Table 1a. Frequency of ERG oncoprotein expression in whole-mount prostatectomy specimens.
| ERG | Individual tumors | Benign glands |
|---|---|---|
| Positive | 117 | 22 |
| Negative | 144 | 200 000 |
| Specificity=99.99% Sensitivity=44.83% PPV 84.17% and NPV=99.93% | ||
Sensitivity and specificity of the ERG oncoprotein nuclear staining distinguishing tumor foci from benign glands in 132 whole-mounted prostate sections (261 tumor foci and about 200,000 benign glands). Number of benign glands represents an estimate based on counting of the number of benign glands in three average size sections of this cohort (average 1550 benign glands/section) multiplied by 132 sections.
Association of the ERG oncoprotein status was evaluated with various clinico-pathological features (Supplementary Table S1a and b). Although, ERG expression did not show correlation with most clinico-pathological features, when all of the tumor foci in a given whole-mount section were taken into account, higher Gleason sum and less-differentiated tumors showed significant correlation with ERG-positive immunostaining (Table 1b).
Table 1b. Association of ERG oncoprotein status with tumor differentiation and Gleason pattern of individual tumors (N=261).
| Tumor grade | ERG status | P-value | |
|---|---|---|---|
| Negative (N=144) | Positive (N=117) | ||
| Tumor differentiation | |||
| Gleason pattern 3 (Well differentiated) | 100 (62.5%) | 60 (37.5%) | 0.0027 |
| Gleason pattern 4/5 (moderate/poorly differentiated) | 44 (43.6%) | 57 (56.4%) | |
| Tumor Gleason sum | 0.0094 | ||
| 6 | 100 (62.5%) | 60 (37.5%) | |
| 7 | 26 (41.3%) | 37 (58.7%) | |
| 8–10 | 18 (47.4%) | 20 (52.6%) | |
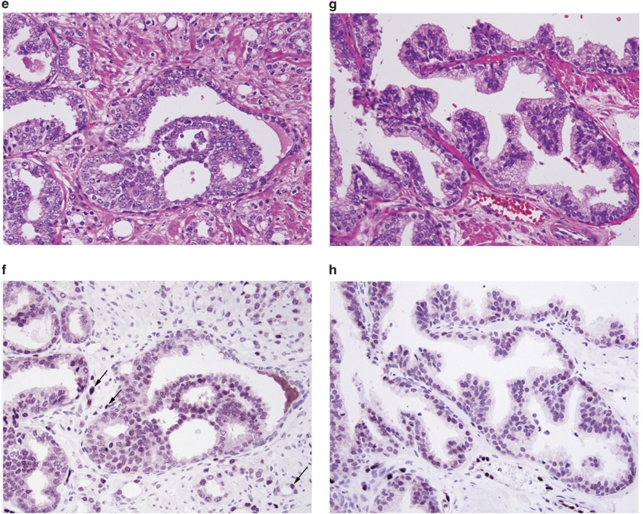
Eighty-two of eighty-five (96.5%) evaluable specimens with ERG-positive tumor foci contained ERG-positive PIN lesions, and all of the ERG-positive PIN foci were co-located with ERG-positive tumors (Table 2). Eighty-one sections contained multiple tumors; in 15 of these all tumors were positive; in 31 all tumor foci were negative and in 35 some tumors were diffusely positive and others completely negative. Thus, in a multi-focal tumor context, 50 of 81 sections (61.7%) had one or more ERG-positive tumors. In the 51 sections containing only one tumor, 36 (70.6%) were ERG positive, and two of these contained clones of completely ERG-negative tumor cells embedded in the positive areas (Figures 3a and b). A weak non-discriminatory cytoplasmic staining was observed in all epithelial cell types (prostatic and non-prostatic), which was consistent with the cell line data (Figures 3c and d).
Table 2. Summary of the ERG oncoprotein status in individual tumors of whole-mount prostate sections.
Figure 3.
ERG oncoprotein in carcinoma with heterogeneous expression and non-discriminatory staining of benign glands. (a) Tumor shows a ‘diverse' ERG expression pattern with ERG positive alternating with ERG negative clones, H&E × 20. (b) Same field as (a). Although, the tumor cells appear similar in the H&E-stained section, they differ in their ERG oncoprotein distribution, ERG–MAb × 20. (c) Benign gland with basal and secretory cells, H&E × 20. (d) Same field as (c). The nuclei of the secretory and basal cells are negative for ERG. Note weak cytoplasmic reactivity in the secretory cells. The endothelial cells show strong nuclear positivity for ERG, ERG–MAb × 20.
The ERG–MAb consistently detected ERG in the nuclei of all endothelial cells (lympho/vascular), which served as intrinsic positive control for the ERG IHC assay. ERG expression in endothelial cells has also been noted previously in other contexts; however, its significance remains to be defined.18, 19, 20 Endothelial cells can be easily identified by ERG-positive nuclei in cells with very little discernible cytoplasm in contrast to carcinoma, in which most of the tumor cells have ERG-positive nuclei and easily identifiable cytoplasm (Figures 2c and d). In ERG negative poorly differentiated/Gleason pattern 4 or 5 carcinomas, positive nuclei of endothelial cells often have a linear narrow distribution (Supplementary Figure S4a and b).
Tumor cells with amphophilic cytoplasm were more strongly positive than those with pale or foamy cytoplasm (Supplementary Figure S5a and b). Three of the four mucinous carcinomas were positive for ERG (Supplementary Figure S5c and d). Only two of the five tumors with a ductal component were positive for ERG (Supplementary Figure S5e–h). One tumor with vacuolated/signet ring-like appearance was positive for ERG. The focus with lymphoepithelioma-like features was negative. In seven patients with lymph node metastases at the time of prostatectomy, the ERG expression mirrored the expression status of the index tumor. Four ERG-positive primary tumors had ERG-positive metastases, and conversely, three ERG-negative primary tumors had ERG-negative metastases (Supplementary Figure S3). By FISH assay, ERG-positive primary tumors (Supplementary Figure S3a) and the corresponding metastases (Supplementary Figure S3b) showed identical fusion patterns.
Basal cells, urothelial cells of the prostatic urethra and periurethral prostatic ducts were non-reactive. Ejaculatory ducts, seminal vesicles, nerve bundles, fibromuscular stroma, variants of glandular hyperplasia including microacinar hyperplasia (synonyms: adenosis, atypical adenomatous hyperplasia), sclerosing adenosis and basal cell hyperplasia were all negative for ERG. Different patterns of atrophy including proliferative inflammatory atrophy and evolving or partial atrophy were also negative for ERG.
Discussion
As the gene fusion events in CaP commonly involve regulatory sequences of AR-regulated prostate-associated genes, for example, TMPRSS2, SLC45A3 or NDRG1 along with protein coding sequences of the nuclear transcription factors in the ETS gene family (ERG, ETV1, ETV4–6 and ELK4), the resultant protein products are ETS-related oncogenic transcription factors with ERG being the most common.1 The ERG–MAb described herein exhibits a high degree of specificity and sensitivity in recognizing ERG oncoprotein. Positive nuclear staining for the ERG oncoprotein is highly specific (99.9%) in identifying tumor cells in 65% of patients. Nuclear ERG staining is virtually absent in benign epithelial cells. Overall 44.8% of all 261 individual tumors were ERG positive in this cohort, whereas 70.6% of 51 specimens with single tumor were ERG positive and 62% of 81 specimens with more than one tumor were ERG positive. Overall frequencies of ERG expression in CaP specimens noted here are similar to the reported rate of gene fusions involving ERG locus reviewed in Kumar Sinha et al., and Clark and Cooper.1, 21 Furthermore, this study points to the potential contribution of sample bias in assessing frequency of ERG alterations in CaP. In previous studies specificity and sensitivity of ERG protein detection was not addressed due to limited number of specimens examined.6, 22 In general, tumors are either homogeneously positive or negative for ERG expression. This study highlights the association (96.5%) of ERG-positive PINs with ERG-positive tumors (Table 2). Although other studies1, 23, 24 have shown lower frequency of ERG fusion-positive PIN (15–20%), this study of whole-mount prostate sections allows more comprehensive evaluation of PIN and tumors in the context of ERG oncoprotein expression (Figure 4).
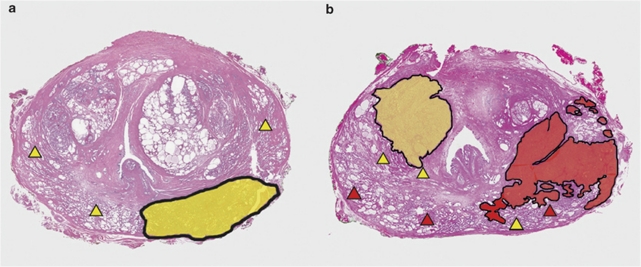
Figure 4.
Schematic representation of the concordance of ERG status between PIN and carcinoma in whole-mount prostates. Two scenarios of whole-mount radical prostatectomy specimens are represented with carcinoma and PIN areas are marked. (a) Specimens with ERG (−) carcinoma (yellow areas) and ERG (−) PIN foci (yellow triangles) in the same prostates. (b) Specimens with at least one ERG (+) carcinoma (red areas) and PIN foci (red triangles) in the same prostates.
The rare ERG-positive benign glands and the rare atypical native glands, referred to as low-grade PIN, may harbor sub-morphological molecular alterations, particularly in view of their topographical relationship to PIN and/or carcinoma. This finding is in agreement with previous studies reporting the presence of TMPRSS2–ERG fusion transcripts in rare instances of benign prostatic glands.12, 15 The confirmation of TMPRSS2–ERG fusions in these foci is challenging due to their small size. When considering the high concordance rate between ERG oncoprotein expression and TMPRSS2–ERG gene fusion transcript status, one could employ the ERG IHC as an excellent surrogate marker for gene fusions leading to ERG overexpression. Thus, in addition to complementing genomic and mRNA-based assays ERG oncoprotein detection provides a significant advance in assessing ERG alterations in CaP. For example, translational products resulting from genomic fusion events of ERG protein-coding sequence and regulatory sequence of any 5′ fusion partners (TMPRSS2, SLC45A3 and NDRG1)3, 25, 26 can be detected by ERG–MAb. On the practical side, evaluation of ERG protein by IHC will be more rapid and informative for morphological assessment of ERG oncogenic activation in ‘front-end' pathology setting.
Among the currently used diagnostic markers, α-methylacyl-CoA racemase detects approximately 80% of prostatic carcinomas and a variety of other carcinomas.27 However, the specificity of α-methylacyl-CoA racemase is lower than that of the ERG, because 25–30% of benign prostatic glands may stain for α-methylacyl-CoA racemase. Thus, inclusion of ERG–MAb in a diagnostic IHC panel may increase the specificity for tumor detection. The strong positive reaction of ERG-MAb in endothelial cells observed highlights many more capillaries in the prostate than were previously appreciated using conventional endothelial cell markers (CD 31, CD 34 and Factor VIII-related antigens). However, this feature of ERG expression may cause some difficulties in the interpretation of the ERG IHC staining. For example, capillaries in intimate contact with glands may suggest basal cell staining, or dilated capillaries with reactive endothelium may mimic small tumor glands or atrophy. This initial limitation can be overcome by gaining experience recognizing ERG-positive vascular patterns (Supplementary Figure S4a and b).
Although prognostic features of ERG alterations in CaP remain to be better understood, both positive and negative associations have been reported and reviewed in Kumar Sinha et al. and Clark and Cooper.1, 21 In this evaluation of ERG oncoprotein, when all of the tumor foci in a given whole-mount section were taken into account, higher Gleason sum and less-differentiated tumors showed correlation with ERG immunostaining (Table 1b). However, there was no significant correlation with progression (Supplementary Figure S6). Considering the ERG expression in the multi-focal tumor context, further independent evaluations in larger and better-defined cohorts are warranted.
In summary, among the currently known CaP protein biomarkers the detection of the homogeneous, strong and highly specific ERG oncoprotein offers unprecedented opportunities in CaP diagnostic setting. These findings substantiate the role of ERG activation in clonal selection and expansion of ERG-positive tumor cells during the transition from pre-invasive to invasive CaP in two-thirds of patients. Finally, with a better understanding of ERG functions in prostate tumor biology, ERG–MAb-based stratification of prostate tumors in the future may be used in the context of imaging, targeted therapy or monitoring efficacy of androgen ablation therapy.
Acknowledgments
We thank Ms Amina Ali, Mr Zhe Chang and Ms Lakshmi Ravindranath for outstanding medical informatics, biospecimen banking and technical assistance. This research was supported by Grants RO1 DK065977 to SS and GP; DoD, CDMRP, Grant PC073614 to SS, TS and AD; and Center for Prostate Disease Research Program HU001-04-C-1502 to DGM.
The Henry M Jackson Foundation for the Advancement of Military Medicine filed a patent on ERG–MAb, in which AD, ST and SS are co-inventors. GM is an employee of the Affymetrix.
Footnotes
Supplementary Information accompanies the paper on the Prostate Cancer and Prostatic Diseases website (http://www.nature.com/pcan)
Disclaimer
The views expressed in this article are those of the authors and do not reflect the official policy of the Department of the Army, Department of Defense or the US Government.
Supplementary Material
References
- Kumar-Sinha C, Tomlins SA, Chinnaiyan AM. Recurrent gene fusions in prostate cancer. Nat Rev Cancer. 2008;8:497–511. doi: 10.1038/nrc2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovics G, Liu A, Shaheduzzaman S, Furusato B, Sun C, Chen Y, et al. Frequent overexpression of ETS-related gene-1 (ERG1) in prostate cancer transcriptome. Oncogene. 2005;24:3847–3852. doi: 10.1038/sj.onc.1208518. [DOI] [PubMed] [Google Scholar]
- Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- Gopalan A, Leversha MA, Satagopan JM, Zhou Q, Al-Ahmadie HA, Fine SW, et al. TMPRSS2-ERG gene fusion is not associated with outcome in patients treated by prostatectomy. Cancer Res. 2009;69:1400–1406. doi: 10.1158/0008-5472.CAN-08-2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Dobi A, Sreenath T, Cook C, Tadase AY, Ravindranath L, et al. Delineation of TMPRSS2-ERG splice variants in prostate cancer. Clin Cancer Res. 2008;14:4719–4725. doi: 10.1158/1078-0432.CCR-08-0531. [DOI] [PubMed] [Google Scholar]
- Klezovitch O, Risk M, Coleman I, Lucas JM, Null M, True LD, et al. A causal role for ERG in neoplastic transformation of prostate epithelium. Proc Natl Acad Sci USA. 2008;105:2105–2110. doi: 10.1073/pnas.0711711105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Dobi A, Mohamed A, Li H, Thangapazham RL, Furusato B, et al. TMPRSS2-ERG fusion, a common genomic alteration in prostate cancer activates C-MYC and abrogates prostate epithelial differentiation. Oncogene. 2008;27:5348–5353. doi: 10.1038/onc.2008.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlins SA, Laxman B, Varambally S, Cao X, Yu J, Helgeson BE, et al. Role of the TMPRSS2-ERG gene fusion in prostate cancer. Neoplasia. 2008;10:177–188. doi: 10.1593/neo.07822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver BS, Tran J, Gopalan A, Chen Z, Shaikh S, Carracedo A, et al. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat Genet. 2009;41:619–624. doi: 10.1038/ng.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King JC, Xu J, Wongvipat J, Hieronymus H, Carver BS, Leung DH, et al. Cooperativity of TMPRSS2-ERG with PI3-kinase pathway activation in prostate oncogenesis. Nat Genet. 2009;41:524–526. doi: 10.1038/ng.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong Y, Xin L, Goldstein AS, Lawson DA, Teitell MA, Witte ON. ETS family transcription factors collaborate with alternative signaling pathways to induce carcinoma from adult murine prostate cells. Proc Natl Acad Sci USA. 2009;106:12465–12470. doi: 10.1073/pnas.0905931106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark J, Merson S, Jhavar S, Flohr P, Edwards S, Foster CS, et al. Diversity of TMPRSS2-ERG fusion transcripts in the human prostate. Oncogene. 2007;26:2667–2673. doi: 10.1038/sj.onc.1210070. [DOI] [PubMed] [Google Scholar]
- Barry M, Perner S, Demichelis F, Rubin MA. TMPRSS2-ERG fusion heterogeneity in multifocal prostate cancer: clinical and biologic implications. Urology. 2007;70:630–633. doi: 10.1016/j.urology.2007.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehra R, Han B, Tomlins SA, Wang L, Menon A, Wasco MJ, et al. Heterogeneity of TMPRSS2 gene rearrangements in multifocal prostate adenocarcinoma: molecular evidence for an independent group of diseases. Cancer Res. 2007;67:7991–7995. doi: 10.1158/0008-5472.CAN-07-2043. [DOI] [PubMed] [Google Scholar]
- Furusato B, Gao CL, Ravindranath L, Chen Y, Cullen J, McLeod DG, et al. Mapping of TMPRSS2-ERG fusions in the context of multi-focal prostate cancer. Mod Pathol. 2008;21:67–75. doi: 10.1038/modpathol.3800981. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Knudsen BS, Allen AN, McLerran DF, Vessella RL, Karademos J, Davies JE, et al. Evaluation of the branched-chain DNA assay for measurement of RNA in formalin-fixed tissues. J Mol Diagn. 2008;10:169–176. doi: 10.2353/jmoldx.2008.070127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltzinger M, Mager-Heckel AM, Remy P. Xl erg: expression pattern and overexpression during development plead for a role in endothelial cell differentiation. Dev Dyn. 1999;216:420–433. doi: 10.1002/(SICI)1097-0177(199912)216:4/5<420::AID-DVDY10>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Birdsey GM, Dryden NH, Amsellem V, Gebhardt F, Sahnan K, Haskard DO, et al. Transcription factor Erg regulates angiogenesis and endothelial apoptosis through VE-cadherin. Blood. 2008;111:3498–3506. doi: 10.1182/blood-2007-08-105346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellett F, Kile BT, Lieschke GJ. The role of the ETS factor erg in zebrafish vasculogenesis. Mech Dev. 2009;126:220–229. doi: 10.1016/j.mod.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark JP, Cooper CS. ETS gene fusions in prostate cancer. Nat Rev Urol. 2009;6:429–439. doi: 10.1038/nrurol.2009.127. [DOI] [PubMed] [Google Scholar]
- Bonaccorsi L, Nesi G, Nuti F, Paglierani M, Krausz C, Masieri L, et al. Persistence of expression of the TMPRSS2: ERG fusion gene after pre-surgery androgen ablation may be associated with early prostate specific antigen relapse of prostate cancer: preliminary results. J Endocrinol Invest. 2009;32:590–596. doi: 10.1007/BF03346514. [DOI] [PubMed] [Google Scholar]
- Cerveira N, Ribeiro FR, Peixoto A, Costa V, Henrique R, Jeronimo C, et al. TMPRSS2-ERG gene fusion causing ERG overexpression precedes chromosome copy number changes in prostate carcinomas and paired HGPIN lesions. Neoplasia. 2006;8:826–832. doi: 10.1593/neo.06427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perner S, Mosquera JM, Demichelis F, Hofer MD, Paris PL, Simko J, et al. TMPRSS2-ERG fusion prostate cancer: an early molecular event associated with invasion. Am J Surg Pathol. 2007;31:882–888. doi: 10.1097/01.pas.0000213424.38503.aa. [DOI] [PubMed] [Google Scholar]
- Han B, Mehra R, Dhanasekaran SM, Yu J, Menon A, Lonigro RJ, et al. A fluorescence in situ hybridization screen for E26 transformation-specific aberrations: identification of DDX5-ETV4 fusion protein in prostate cancer. Cancer Res. 2008;68:7629–7637. doi: 10.1158/0008-5472.CAN-08-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflueger D, Rickman DS, Sboner A, Perner S, LaFargue CJ, Svensson MA, et al. N-myc downstream regulated gene 1 (NDRG1) is fused to ERG in prostate cancer. Neoplasia. 2009;11:804–811. doi: 10.1593/neo.09572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hameed O, Sublett J, Humphrey PA. Immunohistochemical stains for p63 and alpha-methylacyl-CoA racemase, versus a cocktail comprising both, in the diagnosis of prostatic carcinoma: a comparison of the immunohistochemical staining of 430 foci in radical prostatectomy and needle biopsy tissues. Am J Surg Pathol. 2005;29:579–587. doi: 10.1097/01.pas.0000157936.93999.18. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.