Abstract
O-GlcNAcylation has now been added to the growing list of histone modifications making up the multifaceted ‘histone-code’ (Sakabe et al., 2010). The sites of O-GlcNAc-histone modification hint at a role in chromatin remodeling, thus adding to mounting evidence that O-GlcNAc cycling sits atop a robust regulatory network maintaining higher-order chromatin structure and epigenetic memory.
Preview
The hexosamine signaling pathway integrates cellular nutrient stores by regulating the synthesis of UDP-GlcNAc, a key metabolite required for the dynamic O-GlcNAc modification of nuclear and cytoplasmic proteins. Previous studies have directly linked O-GlcNAc cycling to signaling, transcription and translation, as well as to RNA and protein stability (Butkinaree et al., 2010; Hanover et al., 2010). Now, Sakabe et al., in an article published in the Proceedings of the National Academy of Sciences, demonstrate the GlcNAcylation of histones (Sakabe et al., 2010). This rigorous and compelling study provides a key element in a growing body of evidence directly linking O-GlcNAc to the ‘Histone Code’ now considered the ‘Holy Grail’ of modern epigenetics (Jenuwein and Allis, 2001).
The potential role of O-GlcNAc cycling in epigenetic regulation has been explored elsewhere (Butkinaree et al., 2010; Hanover et al., 2010; Love et al., 2010b). Here, a brief summary of previous studies linking O-GlcNAc to epigenetics may serve to highlight the importance of the Sakabe et al., contribution. O-GlcNAc cycling requires only two enzymes in higher metazoans the O-GlcNAc transferase (OGT), which catalyzes addition, and O-GlcNAcase (OGA; MGEA5), which mediates selective O-GlcNAc removal (Butkinaree et al., 2010; Hanover et al., 2010). Early studies demonstrated that O-GlcNAc was present on nuclear pores and many nuclear and chromatin associated proteins but largely absent from the transcriptionally active ‘puffs’ of Drosophila polytene chromosomes (Kelly and Hart, 1989). The molecular characterization of OGT in 1997 provided yet another essential clue, since OGT was identified as a tetratricopeptide repeat (TPR) protein with similarities to the yeast transcriptional repressor SSN6 (Kreppel et al., 1997; Lubas et al., 1997) (See also below). The link between O-GlcNAc and transcription was further solidified when RNA polymerase II was shown to be O-GlcNAc modified in its CTD domain. Studies also revealed that OGT was essential for stem cell survival in mice and associated with a number of transcriptional regulators mediating transcriptional repression, and stem cell pluripotency (Reviewed in (Love et al., 2010b). Recently, mutations in Drosophila OGT were shown to be the molecular defect underlying the homeotic mutant “super sex combs” genetically linked to the epigenetic modulators polycomb and trithorax (Gambetta et al., 2009; Sinclair et al., 2009). In parallel studies, mutants in O-GlcNAc cycling in C. elegans allowed a genome-wide glimpse of cycling of O-GlcNAc on promoters of genes involved in signaling, stress, immunity and aging (Love et al., 2010a). These sites of promoter occupancy by O-GlcNAc overlap significantly with sites occupied by the histone variant H2AZ resulting from histone exchange (Love et al., 2010a; Love et al., 2010b). In other studies, OGT was shown to interact with the methyltransferases MLL5, CARM1, and Whc1 (Reviewed in (Love et al., 2010b)). The O-GlcNAcase (Mgea5) has both a presumptive histone acetyltransferase domain (Butkinaree et al., 2010; Hanover et al., 2010) and is present in the NK cluster of homeobox genes in metazoans ranging from flies to mammals (Love et al., 2010b). In spite of these compelling findings linking O-GlcNAc cycling to epigenetics, evidence for direct modification of histones by O-GlcNAcylation was lacking until the Sakabe et al. study (Sakabe et al., 2010) previewed here.
O-GlcNAc has now been found on Histones H2A, H2B and H4 using a rigorous combination of techniques including immunoblotting, selective enzymatic labeling, chemoenzymatic detection, and mass spectrometry (Sakabe et al., 2010). The authors showed that acetylated histones are modified by O-GlcNAc and that O-GlcNAcylation increased upon heat stress. Heat shock and OGT over-expression also modestly increased chromatin condensation. Using a recently developed chemical enrichment procedure the authors then mapped three O-GlcNAc sites on histones: Thr101 of H2A, Ser36 of H2B and Ser47 of H4. These sites correspond to known histone phosphorylation sites potentially important for H2A-H2B dimerization and association with H3/H4 tetramers. Additionally, the authors also provide evidence that other sites must exist in the histone preparations including probable sites for modification of the remaining histone H3.
The identification of O-GlcNAc sites near known DNA interaction sites prompted Sakabe et al. to speculate that O-GlcNAcylation could induce major changes in chromatin structure. One particularly intriguing O-GlcNAcylated site discussed was the Ser47 site of H4. The authors point out that mutation in this site (Ser to Cys) induces a Swi/Snf independent (Sin) phenotype in yeast. Although not explicitly discussed, it should be noted that the histone remodeling complex Swi/Snf is normally antagonized by Ssn6-Tup1 controlling an extended stretch of chromatin structure around Ssn6-Tup1 repressed genes (Fleming and Pennings, 2007). As mentioned above, Ssn6 is a TPR protein with striking similarities to the extended TPR region of higher metazoan OGT but lacking the catalytic domain (Lubas et al., 1997). The findings suggest that O-GlcNAcylation of histones may represent a higher metazoan extension of a biochemical process conserved from yeast to man.
The findings of Sakabe et al., add yet another layer of complexity to the enormously multifaceted ‘histone code’ (Sakabe et al., 2010). O-GlcNAc also joins a vast array of post-translational modifications directly altering chromatin structure (Love et al., 2010b). Coupled with other recent observations summarized in Figure 1, the nutrient-driven, environmentally sensitive O-GlcNAc cycling enzymes appear to sit atop a cascade of biochemical regulators which including methyltransferases, acetyltransferases, and kinases already implicated in epigenetic programming. O-GlcNAc also directly modifies RNA polymerase II and could interact with the RNA pol II ‘CTD code’. Finally, the present findings are in accord with a complex interaction between O-GlcNAc cycling the histone remodeling complexes required for histone exchange, nucleosome stability, and the maintenance of higher order chromatin structure. As noted previously (Love et al., 2010b), these observations linking O-GlcNAc cycling to higher order chromatin structure provide new insights into how the environment may influence the epigenetic regulation of gene expression and developmental plasticity. Taken together, these findings make a compelling case that O-GlcNAcylation plays a pivotal role in the epigenetics of human disease.
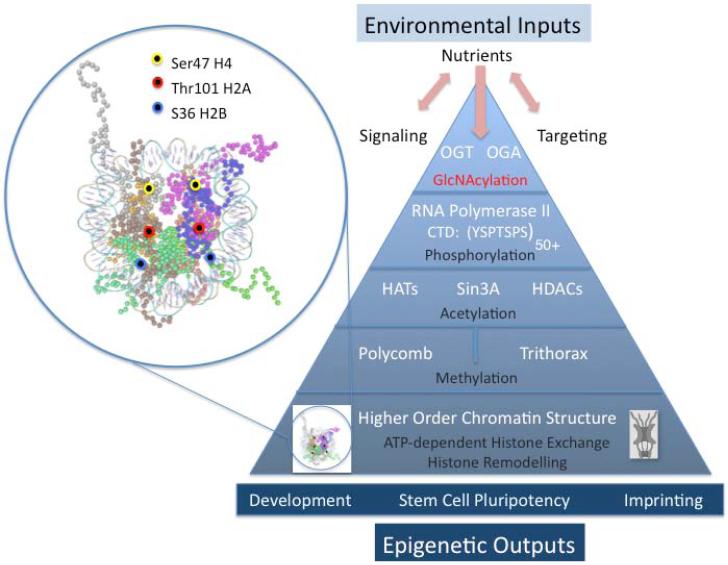
Figure 1. Environmentally responsive O-GlcNAc cycling sits atop a robust regulatory cascade regulating chromatin remodeling with potential epigenetic outcomes.
The expanded view in the circle at left shows the X-ray structure of the nucleosome core particle at 1.9 Å (pdb 1KX5, visualized with Cn3d). The mapped positions of the sites of O-GlcNAc modification on histones H2A, H2B and H4 are highlighted by the colored cirles as indicated in the associated legend.
The pyramid at right depicts the multilayered influence of O-GlcNAc cycling on chromatin structure. Nutrients and environmental inputs (stress, pathogens, aging) may alter signaling and targeting, ultimately resulting in modification of chromatin bound substrates by the enzymes of O-GlcNAc cycling. Chromatin associated substrates include RNA polymerase II, Sin3A: HDAC targets, members of the polycomb and trithorax group proteins involved in histone ubiquination and methylation, and nuclear pore complexes. In addition, as demonstrated by Sakabe, et al., (Sakabe et al., 2010) the histones are O-GlcNAcylated in regions thought to be involved in chromatin condensation, exchange and remodeling. Thus, changes in O-GlcNAc cycling may directly modify RNA polymerase II and histone cores while also influencing the diverse modifications occurring on histone tails. These hierarchical alterations in nucleosomes and chromatin structure have the potential to influence development, stem cell pluripotency, and genomic imprinting.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Butkinaree C, Park K, Hart GW. Biochim Biophys Acta. 2010;1800:96–106. doi: 10.1016/j.bbagen.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming AB, Pennings S. Nucleic Acids Res. 2007;35:5520–5531. doi: 10.1093/nar/gkm573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambetta MC, Oktaba K, Muller J. Science. 2009;325:93–96. doi: 10.1126/science.1169727. [DOI] [PubMed] [Google Scholar]
- Hanover JA, Krause MW, Love DC. Biochim Biophys Acta. 2010;1800:80–95. doi: 10.1016/j.bbagen.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Kelly WG, Hart GW. Cell. 1989;57:243–251. doi: 10.1016/0092-8674(89)90962-8. [DOI] [PubMed] [Google Scholar]
- Kreppel LK, Blomberg MA, Hart GW. J Biol Chem. 1997;272:9308–9315. doi: 10.1074/jbc.272.14.9308. [DOI] [PubMed] [Google Scholar]
- Love DC, Ghosh S, Mondoux MA, Fukushige T, Wang P, Wilson MA, Iser WB, Wolkow CA, Krause MW, Hanover JA. Proc Natl Acad Sci U S A. 2010a;107:7413–7418. doi: 10.1073/pnas.0911857107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love DC, Krause MW, Hanover JA. Semin Cell Dev Biol. 2010b;21:646–654. doi: 10.1016/j.semcdb.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubas WA, Frank DW, Krause M, Hanover JA. J Biol Chem. 1997;272:9316–9324. doi: 10.1074/jbc.272.14.9316. [DOI] [PubMed] [Google Scholar]
- Sakabe K, Wang Z, Hart GW. Proc Natl Acad Sci U S A. 2010 [Google Scholar]
- Sinclair DA, Syrzycka M, Macauley MS, Rastgardani T, Komljenovic I, Vocadlo DJ, Brock HW, Honda BM. Proc Natl Acad Sci U S A. 2009;106:13427–13432. doi: 10.1073/pnas.0904638106. [DOI] [PMC free article] [PubMed] [Google Scholar]