Abstract
We previously demonstrated that IgG responses to a panel of 126 prostate tissue-associated antigens are common in patients with prostate cancer. In the current report we questioned whether changes in IgG responses to this panel might be used as a measure of immune response, and potentially antigen spread, following prostate cancer-directed immune-active therapies. Sera were obtained from prostate cancer patients prior to and three months following treatment with androgen deprivation therapy (n = 34), a poxviral vaccine (n = 31), and a DNA vaccine (n = 21). Changes in IgG responses to individual antigens were identified by phage immunoblot. Patterns of IgG recognition following three months of treatment were evaluated using a machine-learned Bayesian Belief Network (ML-BBN). We found that different antigens were recognized following androgen deprivation compared with vaccine therapies. While the number of clinical responders was low in the vaccine-treated populations, we demonstrate that ML-BBN can be used to develop potentially predictive models.
1. Introduction
Over the last two decades many new immunotherapy approaches to the treatment of cancer have entered clinical development due to the increased understanding of the mechanisms of antigen presentation, lymphocyte recognition, functions of the innate immune system, and the means of regulation of these responses and the means by which tumors can circumvent these responses. Many of these investigations have led to agents approved for standard clinical use, including infusional cytokine therapies for melanoma or renal cell cancer, intravesical BCG therapy for bladder cancer, and most recently an active cellular therapy targeting prostatic acid phosphatase (PAP, sipuleucel-T, Provenge, Dendreon) for patients with advanced metastatic prostate cancer. Many other agents have demonstrated benefit in large clinical trials, and approval is anticipated in the case of a monoclonal antibody targeting a T-cell checkpoint inhibitor targeting CTLA-4 (ipilimumab, Bristol-Myers Squibb) for advanced melanoma.
Ultimately, for these agents to be clinically approved there needs to be a demonstration that these treatments are relatively safe and patient care and outcome are positively affected. However, there is also an increasing recognition that some of these agents, while likely safe, may best be used in combination with other immune-activating or conventional therapies. This has presented challenges for evaluating these agents using traditional paradigms for clinical development. Consequently there is a need to identify markers of biological response, ideally associated with clinical outcome, but permitting an evaluation of biological effect of these agents used in combination. In the case of antigen-specific vaccines, it has been relatively straightforward to evaluate immune responses to the target antigen as a “biomarker” of immunological efficacy. Unfortunately, there are few instances in which target antigen immune response has been associated with clinical benefit. The situation is more difficult for broadly active immune modulating agents such as T-cell checkpoint inhibitors, including antibodies targeting CTLA-4 or PD-1, or TLR agonists, in which appropriate biomarkers of response have been more elusive. Studies with anti-CTLA-4 monoclonal antibodies, in particular, have sought to identify whether amplification of other T-cell costimulatory molecules [1], or antibodies to defined antigenic tumor-associated proteins [2, 3], might be useful as biomarkers. For whole cell tumor vaccines where there is not a specific, defined antigen being targeted, surrogate antigens known to be expressed by the tumor vaccine have been used as a means of monitoring immune responses from the vaccine [4]. The use of immunologically recognized surrogate antigens, including HER-2/neu, MUC1, and p53, has been possible in the case of breast cancer where T-cell and IgG responses to these antigens have been identified. However it is unknown whether responses to these antigens can be useful to study agents in combination or whether changes in responses to these antigens are associated with clinical outcome.
Over the last several years we have used SEREX- (serological analysis of recombinant cDNA expression libraries) based studies to identify immunologically recognized proteins expressed by normal and malignant prostate tissue that might serve as targets for anti-tumor vaccines [7]. In particular, we have evaluated the targets of IgG responses in patients with chronic prostatitis or autoimmune disorders [8, 9], patients with prostate cancer treated with immune-modulating therapies [10], and IgG responses to cancer-testis antigens in patients with prostate cancer [11, 12]. Over the course of these studies we have effectively identified hundreds of immunologically recognized proteins associated with prostate tissue and/or recognized by patients with prostate cancer. While the identification of hundreds of proteins presents challenges in prioritization for the development of antigen-specific vaccines, we previously questioned whether these antigens might also have diagnostic value with IgG responses being able to distinguish individuals with prostate cancer (or other inflammatory conditions of the prostate) from men without prostate disease. Other groups have similarly reported that IgG responses to tissue-associated antigens might have diagnostic value in identifying patients with prostate cancer [13] or nonsmall cell lung cancer [14]. We have previously reported that a subset of 23 of these antigens were recognized in patients with prostate cancer as well as individuals with symptomatic prostatitis, suggesting that such autoantibody signatures might be useful to identify inflammatory conditions of the prostate, and potentially in a premalignant setting [15].
In the current report, we hypothesized that this same panel of previously identified prostate-associated antigens might be used as a monitoring tool to assess immune responses elicited following immune-modulating therapy. While B-cells or IgG production might not be an intended target of a particular therapeutic approach, IgG responses are often elicited with concurrent T-cell activation. We reasoned that IgG responses are easier to measure compared with antigen-specific T cells, and might be more stable over time in the peripheral blood compared with T-cell frequencies. Moreover, the identification of “off-target” IgG immune responses might further serve as an indication of “antigen spread” with secondary antigens recognized following immunological targeting and thus be more relevant to developing biomarkers associated with favorable clinical responses. To detect antibody responses to previously defined antigens, we applied a similar phage immunoblot approach evaluating IgG responses to multiple antigens simultaneously [15]. These types of complex biomarker data sets are historically very difficult to work with for two reasons: first is the complexity associated with biological networks; second is the challenge of infrequent observation of immune biomarkers in a complex system. As such, the identification of useful biomarkers in data sets such as this study can be very challenging. In this paper, we sought to evaluate the use of machine-learned Bayesian Belief Networks (ML-BBNs) as a method for identifying potentially promising biomarkers and potential biomarkers networks [16, 17]. We sought to train several ML-BBNs to identify promising biomarkers and then use these networks to select a subset of features to train a network of immune biomarkers as they related to observed declines in serum prostate-specific antigen (PSA). Our objective was to demonstrate the feasibility of this method to identify promising early biomarkers of immune response to vaccine therapies in our data.
For the current studies, sera samples were collected prior to treatment and after three months of treatment from three separate trials, one in which patients (n = 34) were treated with androgen deprivation (ADT) therapy only, a standard therapy known to elicit prostate-associated immune responses [18–20], a trial in which patients with castrate-resistant prostate cancer (n = 31) were treated with a viral vaccine encoding PSA (PSAV) [5], and one in which patients with early recurrent prostate cancer (n = 21) were treated with a plasmid DNA vaccine encoding PAP (PAPV) [6]. Patients treated with vaccines were subclassified as immunologic or clinical “responders” based on previously reported criteria to distinguish these groups. We report here that IgG immune responses could be detected to individual antigens, and as long as one year after therapy the recognition of specific antigens was associated with individual treatments. The evaluation of IgG responses to groups of antigens at three months suggests that predictive models might be developed with diagnostic potential. These findings support the concept of using measures of “antigen spread” as biomarkers of immunological efficacy for immune-active therapies, and IgG responses to panels of tissue-associated antigens as measures of this antigen spread.
2. Materials and Methods
2.1. Patient Populations
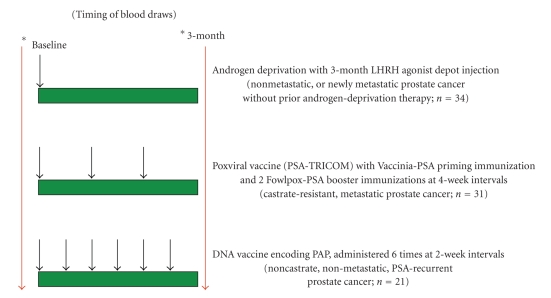
Sera used for the studies had been previously collected with IRB-approved, written consent as part of three separate clinical trials (Figure 1). All samples were stored at −80°C until used for analysis. These samples were all obtained prior to study treatment and after 3 months of treatment in the following settings: (1) a trial (ADT) in which patients (n = 34) with PSA-recurrent, or newly metastatic prostate cancer, who had never previously received androgen deprivation, received 22.5 mg leuprolide by intramuscular injection with or without daily oral bicalutamide; (2) a trial (PSAV) in which patients (n = 31) with castrate-resistant, metastatic prostate cancer were vaccinated at 2-week intervals with a poxviral vaccine (Prostvac, vaccinia virus encoding PSA priming immunization followed by fowlpox virus encoding PSA booster immunizations [5]; (3) a trial (PAPV) in which patients (n = 21) with PSA-recurrent nonmetastatic prostate cancer, not receiving androgen deprivation, were vaccinated at 2-week intervals with a plasmid DNA vaccine encoding PAP [6]. From the majority of patients treated with ADT (n = 24 of 34) or PAPV (n = 19 of 21), serum samples were also available 12 months after the baseline sample. From the vaccine studies, patients were grouped as clinical “responders” or “non-responders” as simply defined by a decrease in serum PSA level at the 3-month time point relative to the baseline value.
Figure 1.
Schema for sample collection. Sera were collected from men with prostate cancer undergoing treatment on three separate clinical trials. Shown are the timing of blood collection and basic schema for these studies. In one study, blood was collected immediately preceding, and at three months following, standard androgen deprivation therapy with a 3-month depot injection of an LHRH agonist. Patients were men (n = 34) with prostate cancer who had not previously received androgen depriving therapy, and had PSA-recurrent and/or metastatic prostate cancer. In the second study, blood was collected immediately preceding and three months following initiation of treatment with a poxviral vaccine encoding PSA (PSA-TRICOM) [5]. Patients were men (n = 31) with castrate-resistant metastatic prostate cancer. In the third study, blood was collected immediately preceding, and at three months following, biweekly treatment with a DNA vaccine encoding prostatic acid phosphatase (PAP) [6]. Patients were men (n = 21) with non-castrate, PSA-recurrent prostate cancer without evidence of metastatic disease.
2.2. High Throughput Immunoblot (HTI)
Phage immunoblot was performed as we have previously described [15]. In brief, 100,000 pfu lambda phage encoding 126 unique antigens were spotted manually in triplicate in a 16×24 array onto XL-1 blue E. coli. bacterial lawns in OmniTray plates using a Biomek FX liquid handling robot. These individual antigens included 29 cancer-testis antigens [21], 40 proteins identified in patients with chronic prostatitis [8], and 57 antigens identified in individual patients, some of whom were treated with androgen deprivation or other immunomodulatory therapies [9, 10, 20]. A listing of antigens and their GenBank Accession numbers is included in Supplemental Table 1 in supplementary material available online at doi:10.1155/2011/454861. Plates were allowed to air-dry after which 10-mM isopropyl ß-D-thiogalactopyranoside- (IPTG) suffused nitrocellulose membranes were overlain, and plates incubated at 37°C overnight to allow recombinant protein expression. Membranes were then washed, blocked, and probed with sera from patients pre- or post-treatment, diluted 1 : 100 in isotonic buffer. Human IgG was then detected with an IgG-specific secondary antibody conjugated to alkaline phosphatase and immunoreactivity detected by development with 0.3 mg/mL nitro blue tertazolium chloride (NBT) (Fisher Biotech) and 0.15 mg/mL 5-bromo 4-chloro 3-indoylphosphate (BCIP) (Fisher Biotech). Membranes were scanned and the digital format was assessed visually, with individual plaques scored positive or not by four independent observers, blinded to the treatment, timing of sample acquisition and membrane layout, as previously reported [8, 15]. All of the membranes for the entire study were evaluated by the same observers at the same time. Triplicate samples were evaluated for each antigen, and immunoreactivity to individual antigens was scored positive if there was concordance among 3 of 4 observers, and if immunoreactivity was scored positive in at least two of the three replicates. Heatmap Builder software (Version 1.1, Stanford University) was used to generate heatmaps displaying changes (gain, loss, or no change) of antibody immune responses following treatment.
2.3. Statistical Analysis
Our statistical analysis consisted of using a commercially available machine-learning software package (FasterAnalytics, DecisionQ Corporation, Washington, DC). Machine learning is a field of computer science that uses intelligent algorithms to allow a computer to mimic the process of human learning. Machine learning algorithms allow the computer to learn dynamically from the data that resides in the training dataset, detecting associations between features without human supervision. The machine learning heuristics generate hypothetical models with different conditional independence assumptions. DecisionQ software generates several networks simultaneously and then continues to generate new hypotheses for each network. The software promotes the network with the best score as determined by goodness of fit relative to compactness. This allows for de novo exploration of associations in complex data sets.
In preprocessing our data, we compared the pre- and post-treatment status of biomarkers and encoded the change in each biomarker as a feature. We then used these encoded features and clinical response (PSA decline) to train models. The output of our machine-learning algorithms is a Bayesian Belief Network (BBN). A BBN encodes the joint probability distribution of all the variables in the domain by building a hierarchical network of conditional dependence. The graphical nature of the network allows the user to query the structure of conditional dependence to identify those features which provide the most information content in the network. In order to select a subset of features for inclusion in a final model, we used a stepwise process and trained a series of machine-learned (ML)BBNs for feature selection. We used this stepwise process as a means of identifying nodes with relatively high information content given our statistically challenging biomarker data sets. Because these data sets have a very high degree of dimensionality (features) relative to evidence (number of subjects), finding those features with the highest information content can be very challenging. To address this challenge, we trained multiple BBN-ML models and identified those features which recurred across multiple models as evidence of high information content. We modeled each of our study cohorts (ADT, PSAV, PAPV) and then compared to the model structures between individual cohorts to identify shared nodes. We also identified high-content nodes (greater than 10 associations) and combined these with the shared nodes to create a selected subset or training a final model to evaluate a network of biomarkers to evaluate clinical response (PSA decline). We used our selected markers to then train three additional models: (i) a final subset model including clinical response (PSA decline) on the vaccine cohorts, (ii) a model of subjects in the vaccine studies who were immune responders, and (iii) a model of subjects who were not immune responders in the vaccine studies. Finally, we performed tenfold cross-validation on our clinical response subset model and used receiver operating characteristic (ROC) curve analysis to calculate an area-under-the-curve (AUC) metric for the feature PSA decline, to determine if the subset model could robustly classify clinical response given immune biomarkers.
The frequencies of IgG responses to individual antigens were compared between treatment study populations using a chi-square test.
3. Results
3.1. IgG Responses to Prostate-Associated Antigens are Elicited Following Prostate Cancer-Directed Immune Therapies
We have previously reported that antibody responses to prostate antigens can be detected in patients with prostate cancer or other inflammatory conditions of the prostate [15]. Moreover, a subset of these prostate-associated antigens was commonly recognized in patients, relative to men without prostate disease, suggesting that the detection of IgG responses to specific prostate-associated antigens might have diagnostic value. In the current analysis, we wished to determine whether the detection of IgG responses to a panel of prostate-associated antigens might have utility in the evaluation of vaccine or other immunomodulatory therapies aimed specifically at eliciting immune responses to the prostate. For this, we obtained sera from men with prostate cancer prior to and following three months of therapy with standard androgen deprivation therapy (n = 34), and from men with prostate cancer (n = 52) prior to and following three months of therapy with one of two different antigen-specific vaccines (Figure 1). Sera from these individuals were used to screen for IgG responses to a panel of 126 antigens by immunoblot, as previously described [15]. Responses to all antigens were evaluated in blinded fashion at both time points, and in Figure 2, changes in IgG responses (gain or loss of response) after 3 months were determined. As demonstrated, androgen deprivation elicited immune responses to multiple antigens, and in particular to antigens previously identified as antigens recognized in patients with chronic prostatitis [8, 15]. Responses to these prostatitis antigens were uncommon over a similar 3-month period in patients treated with either of the vaccines. Gain or loss of IgG responses to some antigens appeared to be shared by these different treatments, while responses to some appeared more specific for individual treatments. Of note, gain or loss of IgG responses to PSA, while detected in one individual treated with ADT, were not detected in patients receiving the PSA-TRICOM vaccine. Similarly IgG responses to PAP were not detected in any of the patients, including those receiving the PAP-targeted vaccine, as previously reported [6].
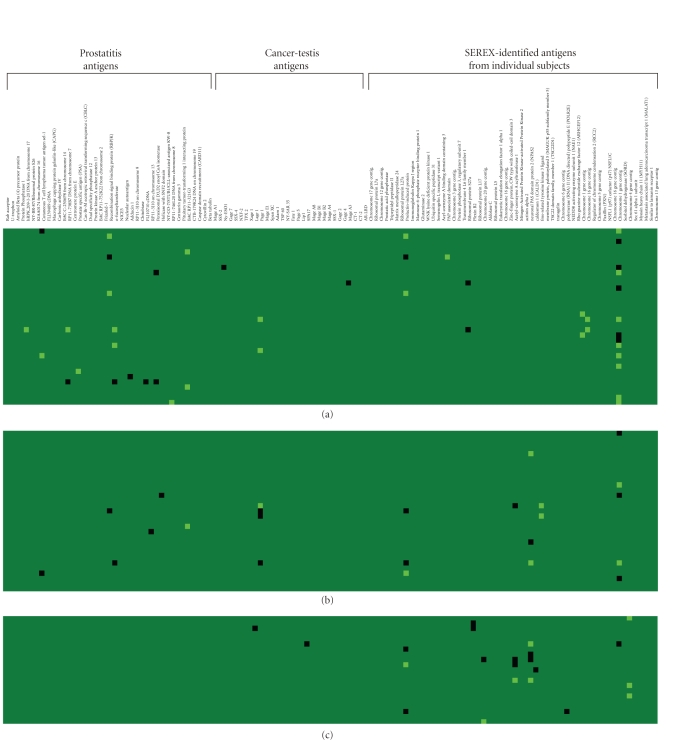
Figure 2.
IgG responses to prostate-associated antigens are elicited following prostate cancer-directed therapies. Sera from patients pretreatment and three months following treatment with androgen deprivation ((a), n = 34), a PSA-targeted viral vaccine ((b), n = 31), or a PAP-targeted DNA vaccine ((c), n = 21) were evaluated for IgG responses to 126 prostate-associated antigens. Antigens (detailed in Supplemental table 1) are grouped according to the original studies from which they were derived (prostatitis antigens, cancer-testis antigens, or antigens detected by SEREX from individual subjects), and IgG responses were scored as previously described [15]. Shown is a heatmap representing gain of response pretreatment to posttreatment (light green), loss of response following treatment (black), or no change in response (dark green) for all subjects (in rows) and all antigens (in columns).
3.2. IgG Responses to Individual Antigens are Specific for the Type of Prostate-Directed Therapy
We next wanted to determine whether IgG responses observed were generally stable, or increased over time, and also identify more specifically whether responses to some antigens were more generally associated with different therapies. In the majority of patients treated with ADT and PAPV, sera samples were also available 12 months later. Evaluation of IgG responses gained or lost after 12 months of ADT, or 12 months after PAP vaccine treatment compared with baseline demonstrated overall an increased number of antigens recognized (Figure 3). Interestingly, responses to individual antigens were observed to be highly specific for the treatment. For example, IgG responses were elicited to the ribosomal L5 protein in 8/24 patients receiving ADT, and 0/19 patients after receiving the PAPV (P = .005, chi-square test). Similarly, IgG responses elicited to the neuronal PAS domain protein 2 (NPAS2) antigen were observed more frequently in patients receiving the PAP vaccine (5/19) compared with patients receiving androgen deprivation (0/24, P = .0075, chi-square test). Even after one year, responses gained or lost to antigens previously identified as prostatitis antigens were not detected in patients treated with the vaccine. While we did not have access to a control population of sera from untreated men, given that these represented populations of subjects with nearly identical stage of disease, collected at the same institution, the differences in IgG response patterns to individual antigens appears most related to the difference in treatment. Moreover, these findings suggest that IgG responses are elicited to “off-target” antigens by means of prostate-directed therapies, and the patterns of IgG responses differ with respect to therapy.
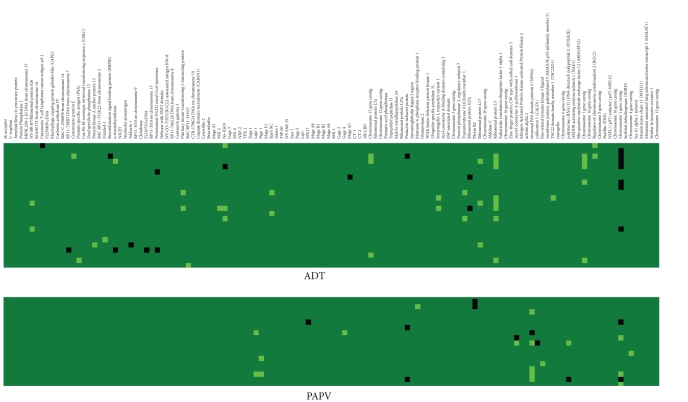
Figure 3.
IgG responses to specific prostate-associated antigens are detectable several months after initiation of treatment with ADT or a PAP vaccine. Immunoblot analysis was performed with the same panel of antigens using sera from individual subjects for whom sera was available 12 months after beginning treatment with ADT (24 of the original 34) or the PAP vaccine (19 of the original 21). The heatmap similarly shows gain of response pretreatment to 12 months posttreatment (light green), loss of response following treatment (black), or no change in response (dark green) for all subjects (in rows) and all antigens (in columns).
3.3. Machine-Based Learning Algorithms can be Designed to Detect Early IgG Response Changes that Might have Predictive Value
The results above demonstrated that, indeed, IgG immune responses are elicited as a result of prostate-directed immune-active therapies. Moreover, antigen-specific IgG immune responses were highly associated with specific treatments, suggesting that the generation of immune responses to these “off-target” antigens might be associated with other measures of immune response or clinical outcome. These responses, however, were most detectable at 12 months after therapy initiation, a time relatively late to be useful in most circumstances as a predictive biomarker. Responses detectable at three months would be more useful. However, the sample sizes for each individual trial were small, and multiple comparisons made by evaluating IgG responses to multiple antigens present difficulties in statistically assessing the importance of an individual marker. Consequently, we evaluated IgG responses to multiple antigens by training a ML-BBN model to determine whether we could identify groups of IgG responses that are associated with clinical response, using data obtained at three months. Because the vaccine trials were conducted in different patient populations where different definitions of clinical response were used, we defined it simply for this purpose as a serum PSA value at the 3-month time point lower than the baseline time point (n = 1 for the PAPV trial, and n = 4 for the PSAV trial subjects).
We trained classifiers on each cohort and compared classifier structure between cohorts. We identified nine (9) biomarkers that were shared between one or more model structures, as described in Table 1, as well as two high-content nodes in the all-cohort model. This resulted in a final subset of biomarkers to include in the final ML-BBN model: chromosome 20 gene contig CHANGE, RP11-738B7 DNA on chromosome 7 CHANGE, chromosome 1 gene contig CHANGE1, prolactin-induced protein CHANGE, acetyl-coenzyme A acyltransferase 1 CHANGE, BAC RP11-321G3 CHANGE, cutaneous T-cell lymphoma tumor antigen sel-1 CHANGE, neuronal PAS domain protein 2 CHANGE, o-fucosyltansferase CHANGE, PAGE 1 CHANGE, and recombination signal binding protein (RBPJK) CHANGE. The structure of the final subset model is displayed in Figure 4. This indicates that there are two first-degree associates of PSA decline, IgG responses to chromosome 1 gene contig 1 and BAC RP11-321G3, and three immune biomarkers features which can be used to estimate PSA decline: IgG responses to chromosome 1 gene contig 1, BAC RP11-321G3, and RP11-738B7 DNA on chromosome 7. Further, these biomarkers are associated with IgG responses to chromosome 20 gene contig, o-fucosyltansferase, PAGE 1, and cutaneous T-cell lymphoma tumor antigen sel-1. To evaluate the robustness of this model, we performed tenfold cross-validation and calculated an AUC for clinical response (PSA decline) of 0.357. This indicates that our first model is not a robust classifier, but is rather an exploratory model.
Table 1.
Biomarker co-occurrence among models. Feature comparison analysis describing which biomarkers have in population specific ML-BBNs. An IgG response change to Chromosome 1 gene contig CHANGE1, for example, has associations in all three population-specific ML-BBNs. Conversely, an IgG response change to Adducin 1 CHANGE only has an association in the ADT population ML-BBN.
| ADT | PAPV | PSAV | Total | |
|---|---|---|---|---|
| Chromosome 1 gene contig CHANGE1 | Yes | Yes | Yes | 3 |
| Prolactin-induced protein CHANGE | Yes | Yes | Yes | 3 |
| Acetyl-coenzyme A acyltransferase 1 CHANGE | No | Yes | Yes | 2 |
| BAC RP11-321G3 CHANGE | Yes | No | Yes | 2 |
| Cutaneous T cell CHANGE | Yes | No | Yes | 2 |
| neuronal PAS domain protein 2 CHANGE | No | Yes | Yes | 2 |
| o-fucosyltansferase CHANGE | Yes | No | Yes | 2 |
| Page 1 CHANGE | Yes | No | Yes | 2 |
| Recombination signal CHANGE | Yes | No | Yes | 2 |
| Adducin 1 CHANGE | Yes | No | No | 1 |
| caldesmon 1 (CALD1) CHANGE | No | Yes | No | 1 |
| carcimona-associated antigen 64 CHANGE | Yes | No | No | 1 |
| Chromosome 1 gene contig CHANGE | Yes | No | No | 1 |
| Chromosome 16 gene contig CHANGE | Yes | No | No | 1 |
| chromosome 17 CHANGE | Yes | No | No | 1 |
| Chromosome 20 gene CHANGE | No | Yes | No | 1 |
| Chromosome 4 gene contig CHANGE | No | Yes | No | 1 |
| FLJ10710 cDNA CHANGE | Yes | No | No | 1 |
| fms-related tyrosine kinase 3 ligand CHANGE | No | No | Yes | 1 |
| Helicase with SNF2 domain CHANGE | No | No | Yes | 1 |
| Lage 1 CHANGE | No | Yes | No | 1 |
| Mage A3 CHANGE | Yes | No | No | 1 |
| Ny-ESO1 CHANGE | Yes | No | No | 1 |
| PAP associated domain CHANGE | Yes | No | No | 1 |
| PAP ELISPOT 12 months | No | Yes | No | 1 |
| Plexin B2 CHANGE | No | Yes | No | 1 |
| polypeptide E (POLR2E) CHANGE | No | Yes | No | 1 |
| Prostate specific antigen (PSA) CHANGE | Yes | No | No | 1 |
| PSA ELISPOT 3m | No | No | Yes | 1 |
| Ribosomal protein S27a CHANGE | Yes | No | No | 1 |
| RP11-3J10 on chromosome 13 CHANGE | No | No | Yes | 1 |
| RP11-738B7 DNA on chromosome 7 CHANGE | Yes | No | No | 1 |
| RP11-746L20 DNA on chromosome 8 CHANGE | Yes | No | No | 1 |
| SPA17 CHANGE | No | Yes | No | 1 |
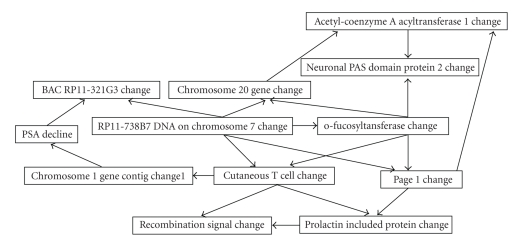
Figure 4.
Structure of Bayesian Belief Network representing selected subset of biomarkers. The structure of the network represents the hierarchy of conditional dependence between features, hence we can identify that the two first degree associates of clinical response are change in IgG responses to BACRP11-321G3 and chromosome 1 gene contig 1. Further, because IgG response to BAC RP11-321G3 is a shared child between PSA decline and IgG response to RP11-738B7 DNA on chromosome 7, IgG response to RP11-738B7 DNA on chromosome 7 still influences the estimate of PSA decline even when IgG response to BAC RP11-321G3 is known.
4. Discussion
In the current report, we sought to determine whether serum antibody responses to a panel of prostate tissue- and prostate cancer-associated antigens might be developed as a diagnostic tool to evaluate immune responses elicited following immune-active therapies, and further to determine whether this might be developed in the future as a biomarker of clinical response. Using sera obtained from patients treated with three different therapies, we found that antigen-specific IgG responses could be detected, likely elicited as a result of therapy. The patterns of response differed with respect to the individual therapy, and recognition of specific antigens was most evident at a later (12 months following treatment) than at an earlier time point (3 months following treatment). Using a ML-BBN model to evaluate groups of IgG responses detected three months after treatment, we prioritized a cohort of antigens, immune responses to which were most associated with PSA decline. These findings suggest that, with data from larger populations of subjects, models could be developed to assist in the detection of potentially therapeutic immune responses resulting from immune-based therapies.
Our results demonstrate that immune-active therapies, including androgen deprivation, elicit IgG responses to individual prostate-associated antigens. This has already been demonstrated in previous studies [18, 20]; however, the IgG responses from androgen deprivation therapy were most obvious many months after treatment, where responses to individual antigens were common and predominantly induced rather than lost. It is conceivable that some immune responses wax and wane over time, and in fact the detection of frequent gains and losses of immune responses to individual antigens, common across treatment groups detected earlier at three months, suggests that this can happen with some antigens. Ideally, to control for this, we would have preferred to have sera samples from men without prostate cancer and/or not undergoing active treatment over the same periods of time. In the absence of this, however, we did have cohorts of subjects treated with different therapies. Given that different individual and sets of antigens were specifically recognized following these different therapies suggests that the responses observed were not purely by chance or due to the waxing and waning of responses to individual antigens. The antigens recognized following androgen deprivation, in particular, were ones previously demonstrated to be commonly recognized by IgG in patients with prostate cancer or inflammatory conditions of the prostate [8, 15]. We did not observe IgG responses to PAP in patients receiving the PAP-targeted vaccine, nor IgG responses to PSA in patients receiving the PSA-targeted vaccine. This was actually not unexpected, as we have previously reported that these vaccines, while able to elicit antigen-specific T-cell responses, do not elicit robust antigen-specific IgG in patients as detected by more sensitive ELISA methods [6, 22]. The observation of IgG responses elicited with these treatments to other antigens suggests that they may be presented by cross-presentation following immune-mediated tumor cell targeting.
Of interest was the observation that the antigens recognized following androgen deprivation were different from those recognized following vaccine treatment. Theoretically, the recognition of other nonvaccine target antigens represents antigen spread induced by immune targeting and presentation of other tissue-associated antigens. The recognition of different antigens suggests different mechanisms of antigen spread, or potentially recognition of other tissue-derived antigens, since most of the antigens in this panel are not prostate specific in expression. At present it is unclear whether the generation of such responses is favorable or not; at least one report has suggested that the generation of IgG responses to non-target antigens might be associated with a worse outcome [23]. Future studies will explore whether the antigens recognized are shared among different vaccine approaches, suggesting common mechanisms of antigen spread, or whether different vaccine therapies elicit specific responses to different “off-target” antigens. With larger group sizes, we also hope to address whether responses to these antigens are associated with measures of T-cell immune responses to the target antigen, further implicating antigen spread as the mechanism of their recognition. In addition, with larger group sizes we hope to answer whether these are clearly associated with improved clinical benefit or not, or whether this is dependent on treatment context and the specific antigen(s) recognized, as we expect.
Given the small sample size and the multitude of IgG response data points, we sought to identify if the use of ML-BBN modeling was feasible to identify biomarker cohorts in our study data. We were able to use a stepwise process and BBN model structures to identify those biomarkers which had high information content for use in a selected subset for ML-BBN modeling. We were subsequently able to use this subset to train an ML-BBN including clinical response, however on cross-validation, our AUC for clinical response was poor. This is likely due to the fact that of the 52 vaccine subjects we only had 5 “responders” as defined. This resulted in a very small set of training outcomes, making models very sensitive to record deletion, as in the case of cross-validation. PSA response has itself not been validated as a surrogate clinical endpoint, and ADT itself elicits initial PSA responses in the vast majority of patients. Consequently, future studies will explore other better markers of clinical response. In addition, as further data are collected from additional subjects treated by vaccines, we expect this will produce a more robust predictive model.
In any case, the use of ML-BBN modeling appears to provide a promising method for identifying biomarkers in complex data sets that can then be selected for further analysis, as the same subset of biomarkers appeared to produce high information content in models across different populations. Further, once we have sufficient subjects to produce a robust model, tables of posterior estimates for clinical response given combinations of IgG response biomarkers can be developed. An example inference table is provided in Table 2, where those biomarkers that are predictive of clinical response can provide a posterior estimate of response. This type of inference could support the translation of this research into a clinical application for determining whether an individual patient has “responded” from a particular vaccine therapy or potentially whether ongoing immunization should be performed. Future modeling might further permit the selection of patients who would be appropriate to receive vaccine therapy based on pre-existing immunological response parameters.
Table 2.
Estimate of PSA decline given biomarker evidence. Inference table describing estimates of the posterior distribution of PSA decline (target) representing estimates of likelihood of increase (0), decrease (1), or unknown (MISSING). For example, the most common case involves no change in any of the independent features (biomarkers), occurs 66.7% and results in a 64.1% posterior probability of PSA increase. The case representing an increase in IgG responses to BAC RP11-321G3, a decrease in IgG response to chromosome 1 gene contig and no change in IgG response to RP11-738B7 DNA on chromosome 7 occurs 1.1% of the time and results in a 6.9% posterior estimate of PSA increase.
| Probability of case | Drivers | Target | ||||
|---|---|---|---|---|---|---|
| BAC RP11-321G3 CHANGE | Chromosome 1 gene contig CHANGE1 | RP11-738B7 DNA on chromosome 7 CHANGE | PSA Decline | |||
| 0.0 | 1.0 | MISSING | ||||
| 0.235% | 0.0 | −1.0 | −1.0 | 28.6 | 14.3 | 57.1 |
| 0.168% | 1.0 | −1.0 | −1.0 | 40.0 | 20.0 | 40.0 |
| 0.638% | 0.0 | 0.0 | −1.0 | 55.6 | 6.5 | 37.9 |
| 0.517% | 1.0 | 0.0 | −1.0 | 68.6 | 8.0 | 23.4 |
| 0.423% | 0.0 | 1.0 | −1.0 | 18.7 | 6.3 | 75.0 |
| 0.264% | 1.0 | 1.0 | −1.0 | 30.0 | 10.0 | 60.0 |
| 10.727% | 0.0 | −1.0 | 0.0 | 36.2 | 13.2 | 50.6 |
| 1.172% | 1.0 | −1.0 | 0.0 | 6.9 | 48.3 | 44.8 |
| 66.744% | 0.0 | 0.0 | 0.0 | 64.1 | 5.4 | 30.5 |
| 4.311% | 1.0 | 0.0 | 0.0 | 20.7 | 33.6 | 45.7 |
| 11.448% | 0.0 | 1.0 | 0.0 | 24.8 | 6.0 | 69.2 |
| 1.101% | 1.0 | 1.0 | 0.0 | 5.4 | 25.0 | 69.6 |
| 0.235% | 0.0 | −1.0 | 1.0 | 28.6 | 14.3 | 57.1 |
| 0.168% | 1.0 | −1.0 | 1.0 | 40.0 | 20.0 | 40.0 |
| 0.638% | 0.0 | 0.0 | 1.0 | 55.6 | 6.5 | 37.9 |
| 0.517% | 1.0 | 0.0 | 1.0 | 68.6 | 8.0 | 23.4 |
| 0.423% | 0.0 | 1.0 | 1.0 | 18.7 | 6.3 | 75.0 |
| 0.264% | 1.0 | 1.0 | 1.0 | 30.0 | 10.0 | 60.0 |
Supplementary Material
Supplementary Table: Prostate-associated antigen panel. Shown are the lambda phage-encoded antigens, and GenBank accession numbers, used for the current studies and obtained from previous studies and used for the immunoblot studies. These antigens derived from studies of patients with chronic prostatitis, patients with prostate cancer treated with androgen deprivation therapy, patients with prostate cancer treated with various other therapies, or specific cancer-testis antigens (CTA).
Acknowledgments
This work was supported by NIH (K23 RR16489), and by the US Army Medical Research and Materiel Command Prostate Cancer Research program (W81XWH-06-1-0184).
References
- 1.Chen H, Liakou CI, Kamat A, et al. Anti-CTLA-4 therapy results in higher CD4ICOS T cell frequency and IFN-γ levels in both nonmalignant and malignant prostate tissues. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(8):2729–2734. doi: 10.1073/pnas.0813175106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yuan J, Gnjatic S, Li H, et al. CTLA-4 blockade enhances polyfunctional NY-ESO-1 specific T cell responses in metastatic melanoma patients with clinical benefit. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(51):20410–20415. doi: 10.1073/pnas.0810114105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fong L, Kwek SS, O’Brien S, et al. Potentiating endogenous antitumor immunity to prostate cancer through combination immunotherapy with CTLA4 blockade and GM-CSF. Cancer Research. 2009;69(2):609–615. doi: 10.1158/0008-5472.CAN-08-3529. [DOI] [PubMed] [Google Scholar]
- 4.Dols A, Meijer SL, Hu HM, et al. Identification of tumor-specific antibodies in patients with breast cancer vaccinated with gene-modified allogeneic tumor cells. Journal of Immunotherapy. 2003;26(2):163–170. doi: 10.1097/00002371-200303000-00009. [DOI] [PubMed] [Google Scholar]
- 5.Gulley JL, Arlen PM, Madan RA, et al. Immunologic and prognostic factors associated with overall survival employing a poxviral-based PSA vaccine in metastatic castrate-resistant prostate cancer. Cancer Immunology, Immunotherapy. 2010;59(5):663–674. doi: 10.1007/s00262-009-0782-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McNeel DG, Dunphy EJ, Davies JG, et al. Safety and immunological efficacy of a DNA vaccine encoding prostatic acid phosphatase in patients with stage D0 prostate cancer. Journal of Clinical Oncology. 2009;27(25):4047–4054. doi: 10.1200/JCO.2008.19.9968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dunphy EJ, Johnson LE, Olson BM, Frye TP, McNeel DG. New approaches to identification of antigenic candidates for future prostate cancer immunotherapy. Update on Cancer Therapeutics. 2006;1(2):273–284. [Google Scholar]
- 8.Dunphy EJ, Eickhoff JC, Muller CH, Berger RE, McNeel DG. Identification of antigen-specific IgG in sera from patients with chronic prostatitis. Journal of Clinical Immunology. 2004;24(5):492–502. doi: 10.1023/B:JOCI.0000040920.96065.5a. [DOI] [PubMed] [Google Scholar]
- 9.Mooney CJ, Dunphy EJ, Stone B, McNeel DG. Identification of autoantibodies elicited in a patient with prostate cancer presenting as dermatomyositis. International Journal of Urology. 2006;13(3):211–217. doi: 10.1111/j.1442-2042.2006.01263.x. [DOI] [PubMed] [Google Scholar]
- 10.Dunphy EJ, McNeel DG. Antigen-specific IgG elicited in subjects with prostate cancer treated with Flt3 ligand. Journal of Immunotherapy. 2005;28(3):268–275. doi: 10.1097/01.cji.0000158853.15664.0c. [DOI] [PubMed] [Google Scholar]
- 11.Hoeppner LH, Dubovsky JA, Dunphy EJ, McNeel DG. Humoral immune responses to testis antigens in sera from patients with prostate cancer. Cancer Immunity. 2006;6:1–7. [PubMed] [Google Scholar]
- 12.Dubovsky JA, McNeel DG. Inducible expression of a prostate cancer-testis antigen, SSX-2, following treatment with a DNA methylation inhibitor. Prostate. 2007;67(16):1781–1790. doi: 10.1002/pros.20665. [DOI] [PubMed] [Google Scholar]
- 13.Wang X, Yu J, Sreekumar A, et al. Autoantibody signatures in prostate cancer. The New England Journal of Medicine. 2005;353(12):1224–1235. doi: 10.1056/NEJMoa051931. [DOI] [PubMed] [Google Scholar]
- 14.Wu L, Chang W, Zhao J, et al. Development of autoantibody signatures as novel diagnostic biomarkers of non-small cell lung cancer. Clinical Cancer Research. 2010;16(14):3760–3768. doi: 10.1158/1078-0432.CCR-10-0193. [DOI] [PubMed] [Google Scholar]
- 15.Maricque BB, Eickhoff JC, McNeel DG. Antibody responses to prostate-associated antigens in patients with prostatitis and prostate cancer. doi: 10.1002/pros.21229. The Prostate, http://www.ncbi.nlm.nih.gov/pubmed/20632317, In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nissan A, Protic M, Bilchik A, Eberhardt J, Peoples GE, Stojadinovic A. Predictive model of outcome of targeted nodal assessment in colorectal cancer. Annals of Surgery. 2010;251(2):265–274. doi: 10.1097/SLA.0b013e3181bd5187. [DOI] [PubMed] [Google Scholar]
- 17.Stojadinovic A, Eberhardt C, Henry L, et al. Development of a Bayesian classifier for breast cancer risk stratification: a feasibility study. Eplasty. 2010;10, article e25 [PMC free article] [PubMed] [Google Scholar]
- 18.Nesslinger NJ, Sahota RA, Stone B, et al. Standard treatments induce antigen-specific immune responses in prostate cancer. Clinical Cancer Research. 2007;13(5):1493–1502. doi: 10.1158/1078-0432.CCR-06-1772. [DOI] [PubMed] [Google Scholar]
- 19.Mercader M, Bodner BK, Moser MT, et al. T cell infiltration of the prostate induced by androgen withdrawal in patients with prostate cancer. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(25):14565–14570. doi: 10.1073/pnas.251140998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morse MD, McNeel DG. Prostate cancer patients on androgen deprivation therapy develop persistent changes in adaptive immune responses. Human Immunology. 2010;71(5):496–504. doi: 10.1016/j.humimm.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dubovsky JA, Albertini MR, McNeel DG. MAD-CT-2 identified as a novel melanoma cancer-testis antigen using phage immunoblot analysis. Journal of Immunotherapy. 2007;30(7):675–683. doi: 10.1097/CJI.0b013e3180de4d19. [DOI] [PubMed] [Google Scholar]
- 22.Gulley J, Chen AP, Dahut W, et al. Phase I study of a vaccine using recombinant vaccinia virus expressing PSA (rV-PSA) in patients with metastatic androgen-independent prostate cancer. Prostate. 2002;53(2):109–117. doi: 10.1002/pros.10130. [DOI] [PubMed] [Google Scholar]
- 23.Nesslinger NJ, Ng A, Tsang K-Y, et al. A viral vaccine encoding prostate-specific antigen induces antigen spreading to a common set of self-proteins in prostate cancer patients. Clinical Cancer Research. 2010;16(15):4046–4056. doi: 10.1158/1078-0432.CCR-10-0948. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table: Prostate-associated antigen panel. Shown are the lambda phage-encoded antigens, and GenBank accession numbers, used for the current studies and obtained from previous studies and used for the immunoblot studies. These antigens derived from studies of patients with chronic prostatitis, patients with prostate cancer treated with androgen deprivation therapy, patients with prostate cancer treated with various other therapies, or specific cancer-testis antigens (CTA).