Abstract
Objective
A functional repeat polymorphism in the SNCA promoter (REP1) conveys susceptibility for Parkinson’s disease (PD). There is also increasing evidence that SNPs elsewhere in the gene associate with risk. We sought to further explore the disease association, determine whether evidence of allelic heterogeneity exists, and examine the correlation between PD-associated variants and plasma α-synuclein levels.
Methods
We performed a two-tiered analysis of 1,956 PD patients and 2,112 controls from the NeuroGenetics Research Consortium using a comprehensive tagSNP approach. Previously published REP1 genotypes were also included. Plasma α-synuclein was assayed in 86 cases and 78 controls using a highly sensitive Luminex assay.
Results
Five of the 15 SNPs genotyped were associated with PD under an additive model in Tier 1 (α=0.05). Of these, four were successfully replicated in Tier 2. In the combined sample, the most significant marker was rs356219 (OR, 1.41; CI, 1.28–1.55; p = 1.6 × 10−12) located ~ 9 kb downstream from the gene. A regression model containing rs356219 alone best fit the data. The linkage disequilibrium correlation coefficient between this SNP and REP1 was low (r2=0.09). The risk-associated C allele of rs356219 was also correlated with higher transformed plasma α-synuclein levels in cases under an adjusted additive model (p = 0.005).
Conclusions
Our data suggest that one or more unidentified functional SNCA variants modify risk for PD, and that the effect is larger than, and independent of, REP1. This variant(s), tagged by rs356219, might act by upregulating SNCA expression in a dose-dependent manner.
Introduction
The SNCA gene encodes α-synuclein, a small (140 amino acid) protein localized, in part, to presynaptic terminals that modulates vesicle trafficking and neurotransmitter release.1–2 A link between α-synuclein and Parkinson's disease (PD) was first demonstrated in 1997 when a missense mutation (A53T) in SNCA was reported to cause autosomal dominant parkinsonism in a large Italian family (the Contursi Kindred).3 Shortly thereafter, α-synuclein was shown to be a major component of Lewy bodies, the pathologic hallmark of both familial and sporadic PD.4 Triplication of the gene was later discovered to cause dominant early-onset PD, indicating that overexpression of wild-type synuclein was sufficient to cause disease.5–6 However, missense mutations and multiplications of SNCA proved to be quite rare, which raised the question of whether common variants with more subtle functional effects might modify susceptibility for PD. Our group and others have reported an association between PD and "REP1" (D4S3481), a complex repeat polymorphism located approximately 10 kb upstream from the SNCA translation start site.7–9 REP1 is essentially triallelic and in comparison to the intermediate-length allele (“261”), the longest allele ("263") is associated with increased risk and the shortest ("259") with decreased risk for PD. In vitro data suggest that rather than simply serving as a genetic marker, REP1 alleles might differentially regulate SNCA transcription, possibly through interactions with the DNA-binding protein PARP-1.10–11 Association analyses of SNCA single nucleotide polymorphisms (SNPs), including two small genomewide association studies (GWAS), yielded mixed results over the past decade.9, 12–14 However, three larger GWAS published in the past year all reported strong association signals from SNPs within SNCA.15–17
Although α-synuclein was initially thought to be neuron-specific,18 SNCA mRNA is highly expressed in erythroid cells and the protein is detectable in all blood components including packed red blood cells (RBCs), peripheral blood mononuclear cells (PBMCs), platelets, and plasma.19–21 Whole blood α-synuclein levels in individuals with SNCA triplications are approximately double those of both their mutation-negative relatives and unrelated controls.22 Whether a correlation exists between common risk-associated SNCA polymorphisms and peripheral α-synuclein levels is not yet known. Examining this relationship could provide complementary evidence for use in refining the association signal and identifying the true risk variant(s) at the SNCA locus.
The goals of the present study were to (1) further explore the association between common SNCA SNPs and PD susceptibility, (2) assess whether the signal from any risk-associated SNPs is independent of REP1, and (3) test for correlation between PD-associated variants and plasma α-synuclein levels.
Materials and Methods
Subjects
For the SNCA association analysis we studied 1,956 PD patients (mean age at onset [AAO], 58.7 ± 11.9 years; mean age at enrollment, 67.9 ± 10.6 years; male, 67.8%) and 2,112 controls (mean age at enrollment, 67.0 ± 18.3 years; male, 37.4%) enrolled through the NeuroGenetics Research Consortium (NGRC) which includes movement disorder clinics in Albany, NY, Atlanta, GA, Portland, OR, and Seattle, WA. All patients met UK PD Society Brain Bank (UKPDSBB) clinical diagnostic criteria for PD as determined by a movement disorder specialist23 and were consecutively recruited except that patients who had an AAO <20 years, whose race was not solely classified as "white" (by self-report), or who carried pathogenic mutations in LRRK2 or PARK2 (homozygotes/compound heterozygotes) were excluded from the sample. Among patients, 22.6% reported a family history of PD in at least one first or second degree relative and were classified as "familial" PD for the purpose of this analysis. Controls had no history of parkinsonism and were either spouses of PD patients or community volunteers.
Subjects for the plasma α-synuclein analysis were derived from a separate study on PD biomarkers. The PD patients (n = 86; mean AAO, 56.9 ± 11.2 years; mean age at enrollment, 66.3 ± 9.4 years; male, 76.7%) all met UKPDSBB clinical diagnostic criteria and were enrolled at NGRC clinics in Portland, OR and Seattle, WA. The controls (n = 78; mean age at enrollment, 65.1 ± 10.3 years; male, 43.6%) had no evidence of parkinsonism by examination and were enrolled through the Alzheimer’s Disease Research Centers of Oregon Health and Science University (Portland, OR), the University of California-San Diego (La Jolla, CA), and the University of Washington (Seattle, WA). All blood samples from these subjects were drawn in the morning after an overnight fast and the plasma was processed and frozen at −70° C within 90 minutes.
The institutional review boards at each participating site approved the study and all subjects gave informed consent.
Marker Selection and Genotyping
We divided the sample for the SNCA association analysis into two tiers. Tier 1 was composed of 685 cases and 673 controls closely matched for age and sex, while the remainder of the sample (cases, n = 1,271; controls, n = 1,439) was placed in Tier 2. We used the LD-select algorithm, as implemented on the SeattleSNPs Genome Variation Server (http://gvs.gs.washington.edu/GVS/), to choose tagSNPs at the SNCA locus based on data from the International HapMap Project CEU population (http://www.hapmap.org). The r2 threshold for bins was 0.80, the minor-allele frequency cutoff was 5%, and the region covered included 10 kb of upstream and downstream sequence (136 kb in total). We selected one tagSNP from each of 13 bins, and two additional SNPs reported to associate with PD that were not included in the HapMap-CEU population (rs2301135 and rs2619363), for genotyping in Tier 1. SNPs associated with PD in Tier 1 (α=0.05) were then replicated in Tier 2.
SNP genotyping in Tier 1 was performed using matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF MS; Sequenom, Inc., San Diego, CA). Genotyping for Tier 2 (and for SNPs that failed in Tier 1) was done using TaqMan assays on an ABI 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA). REP1 was genotyped by PCR using fluorescently labeled primers; the amplification products were separated on an ABI PRISM 3130 Genetic Analyzer and analyzed using GeneMapper 4.0 software (Applied Biosystems).
Plasma α-synuclein, Hemoglobin, and Soluble P-Selectin Measurements
Plasma α-synuclein levels were measured using a highly sensitive Luminex assay recently developed by our group,24 with minor modifications. Briefly, plasma samples were treated with equal volume of 2× RIPA buffer and then diluted with 0.1% BSA/PBS (pH 7.4) for a final dilution of 1:100 before incubating with capturing antibody-coupled beads. A series of recombinant, full-length human α-synuclein (rPeptide, Athens, GA) standards were diluted in a serum matrix-mimicking buffer (2% FBS/0.1% BSA/PBS, pH 7.4) and run in parallel. The incubation time with the detection antibody was 4 hours. All samples were analyzed using a LiquiChip Luminex 200™ Workstation (Qiagen, Valencia, CA).
Because the concentration of α-synuclein in RBCs and platelets is higher than plasma,20–21 residual amounts of these two blood components could result in spuriously elevated plasma α-synuclein levels. To account for this, we measured markers for RBCs (hemoglobin [Hb]) and platelets (soluble P-selectin [sP-selectin]) in plasma. Hb was assayed using a Human Hb ELISA Quantitation Kit (Bethyl Laboratorires, Inc., Montgomery, TX) and sP-selectin was measured with a Human sP-Selectin/CD62P ELISA Quantitation Kit (R&D Systems, Inc., Minneapolis, MN) according to manufacturer instructions.
Data Analysis
Each SNP was assessed for Hardy-Weinberg equilibrium (HWE) in cases and controls (separately) using an exact test. We used logistic regression to test for association between SNCA genotype and PD under an additive model, adjusting for sex and age at enrollment (divided into quartiles). Homozygotes for the more common allele were used as the reference. P-values were generated through a Wald test. The Breslow-Day test was used to test the homogeneity of the odds ratios (ORs) for selected SNPs across enrollment sites. For age-stratified analyses, we grouped cases into early onset (AAO ≤ 50 years) or late onset (AAO > 50 years). Controls were assigned to either a “younger” or “older” group. This was done by first parsing the whole sample into 5-year age at enrollment intervals, then determining the proportion of cases in each interval with early onset disease, and finally assigning a proportional random sample of controls from each age at enrollment interval to the “younger control” group. The remaining controls were assigned to the “older control” group. Early and late onset cases were compared with “younger” and “older” controls, respectively.
We performed stepwise logistic regression using Akaike's Information Criterion (AIC) to assess the relative contributions of REP1 and all SNPs replicated in Tier 2 to PD risk. REP 1 alleles shorter than 259 or longer than 263, which together occur at a frequency of < 1%, were excluded from the dataset.
We used HPlus 3.1 to reconstruct haplotypes from unphased genotype data and to test for haplotype-disease associations before and after adjustment for age and sex. Haplotypes with an estimated frequency <0.01 in cases and controls were excluded from the analysis. We used the coefficient estimates and standard errors calculated by the program to construct ORs and 95% confidence intervals (CI). Pairwise LD (measured as D' and the correlation coefficient r2) between markers was calculated using Haploview.25
The association of plasma α-synuclein with SNCA genotype in cases and controls was examined using linear regression, adjusting for age, sex, and plasma levels of Hb and sP-selectin. To ensure that the assumptions of linear regression analysis were met, we used the Shapiro-Wilk test to assess for departures from normality and the Breusch-Pagan/Cook-Weisberg test to test for heteroskedasticity of the residuals. The difference in mean α-synuclein level between cases and controls was evaluated using a t-test.
Results
Two of the 15 SNPs selected for Tier 1 failed initial genotyping by MALDI-TOF MS, but both were then successfully genotyped by TaqMan. The mean and maximum genotyping failure rates for Tier 1 were 0.9% and 2.3%. One of the 13 tagSNPs (rs10002435) and one of the two unbinned SNPs (rs2619363) were out of HWE in controls (p<0.001) and were excluded from further analysis. Of the remaining SNPs, five were significantly associated with PD (p < 0.05) in Tier 1 (Supplementary Table S1) and were then genotyped in Tier 2. After adjusting for age and sex, the association with PD was replicated for four of the five SNPs in Tier 2 (Table 1). There was no significant evidence of heterogeneity for any of these SNPs across recruitment sites (p ≥ 0.28). The most robust association observed in the combined sample (Tier 1 + Tier 2) was for rs356219 (OR, 1.41; CI, 1.28–1.55; p = 1.6 × 10−12), which is located ~ 9 kb downstream from the SNCA gene.
Table 1.
SNCA Variants Associated with Parkinson’s Disease
| Tier 1a | Tier 2b | Combined | |||||
|---|---|---|---|---|---|---|---|
| Positionc | Variant | OR (95% CI) | p-value | OR (95% CI) | p-value | OR (95% CI) | p-value |
| 90986194 | REP1d | NA | 1.28 (1.07–1.53) | 0.008 | |||
| 90978910 | rs2619364 | 1.31 (1.10–1.55) | 0.002 | 1.26 (1.10–1.44) | 0.001 | 1.26 (1.14–1.40) | 9.4 × 10−6 |
| 90953751 | rs2119787 | 0.82 (0.71–0.96) | 0.013 | 0.89 (0.79–1.02) | 0.085 | 0.88 (0.80–0.96) | 0.006 |
| 90930793 | rs2737029 | 1.36 (1.17–1.58) | 7.9 × 10−5 | 1.32 (1.16–1.50) | 1.2 × 10−5 | 1.31 (1.19–1.44) | 1.6 × 10−8 |
| 90897821 | rs2572324 | 1.42 (1.20–1.68) | 3.9 × 10−5 | 1.30 (1.14–1.49) | 7.9 × 10−5 | 1.32 (1.19–1.46) | 7.2 × 10−8 |
| 90856624 | rs356219 | 1.52 (1.30–1.78) | 2.0 × 10−7 | 1.41 (1.24–1.60) | 6.9 × 10−8 | 1.41 (1.28–1.55) | 1.6 × 10−12 |
Data presented are for an additive model adjusted for age and sex
N = 685 cases and 673 controls
N = 1,271 cases and 1,439 controls
Position from NCBI genome build 36
Genotypes for 1,751 cases and 1,990 controls were derived from a previous publication7
OR = odds ratio; CI = confidence interval; NA = not applicable.
We then examined the relative contribution of these four SNPs, and REP1, to PD risk in our combined sample with stepwise logistic regression. Using a forward selection procedure, the best model (lowest AIC) was one containing rs356219 alone. There was no significant improvement in the fit of the model by sequential addition of rs2572324, rs2737029, rs2619364, and REP1 (in that order). A backward elimination procedure yielded similar results.
We constructed haplotypes comprised of REP1 and the four SNPs replicated in Tier 2 (Table 2). To facilitate comparison with previous studies,9, 17 we recoded REP1 as a binary variable, grouping the 259 and 261 alleles together. The most common haplotype (#1) was used as the reference. Two haplotypes were strongly associated with PD risk (#2, OR, 1.43, p = 4.1 × 10−11; #3, OR, 1.49, p = 3.7 × 10−4), and another (#5, OR, 1.24, p = 0.055) was marginally associated. The only allele shared by all three haplotypes was the C allele of rs356219.
Table 2.
SNCA Haplotype Analysis
| #a | Haplotypeb | Frequency (cases) |
Frequency (controls) |
OR (95% CI) | p-value | ||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 259 or 261 | T | A | T | T | 0.483 | 0.554 | Reference | |
| 2 | 259 or 261 | C | G | C | C | 0.273 | 0.223 | 1.43 (1.29–1.59) | 4.1 × 10−11 |
| 3 | 263 | T | G | T | C | 0.070 | 0.053 | 1.49 (1.20–1.86) | 3.7 × 10−4 |
| 4 | 259 or 261 | T | G | T | T | 0.042 | 0.050 | 0.93 (0.73–1.20) | 0.600 |
| 5 | 259 or 261 | T | A | C | C | 0.048 | 0.046 | 1.24 (0.99–1.55) | 0.055 |
| 6 | 259 or 261 | C | G | T | T | 0.028 | 0.029 | 1.16 (0.88–1.53) | 0.292 |
| 7 | 259 or 261 | T | G | T | C | 0.023 | 0.020 | 1.30 (0.96–1.76) | 0.094 |
Data presented are for the combined sample in an additive model adjusted for age and sex
Haplotypes are numbered in order of descending frequency in controls
Haplotypes comprised of REP1, rs2619364, rs2737029, rs2572324, and rs356219 (in that order). REP1 alleles 259 and 261 were grouped together. Haplotypes with an estimated frequency < 0.01 in cases and controls were excluded.
OR = odds ratio; CI = confidence interval
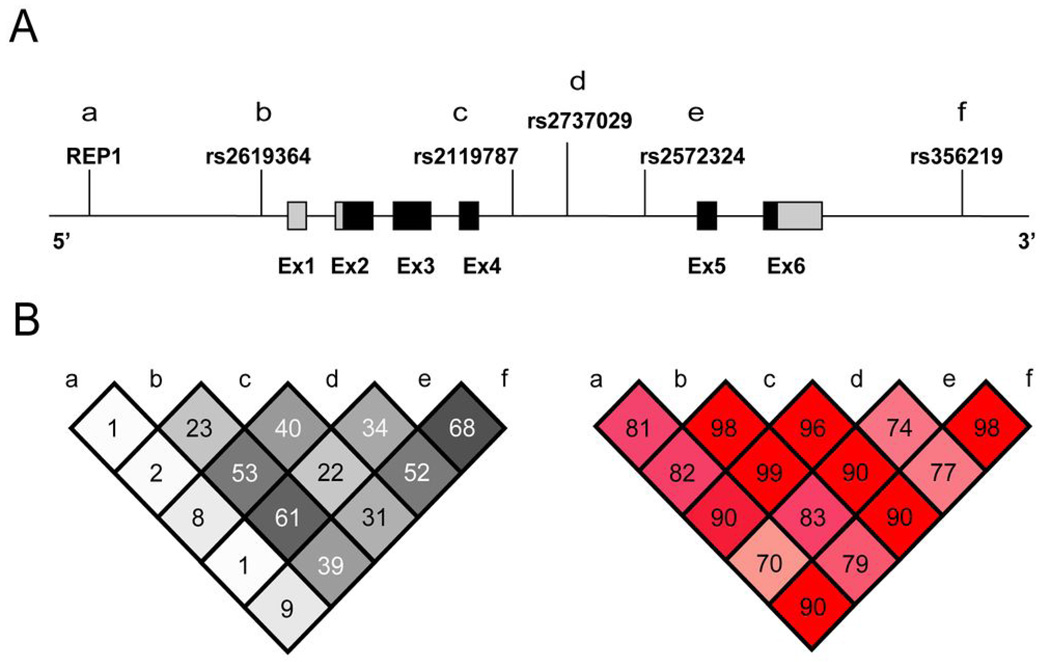
Figure 1 depicts LD between REP1 and the five PD-associated SNPs from Tier 1 in the combined control sample (grouping the REP1 259 and 261 alleles together). There were modest to high levels of correlation among all SNPs (r2 = 0.22 – 0.68; Figure 1B, left panel), but only low levels between REP1 and each SNP, including rs356219 (r2 = 0.09). Low correlation between REP1 and rs356219 was also observed when alternate combinations of REP1 alleles were grouped together (259 and 263, r2 < 0.01; 261 and 263, r2 = 0.03). The pattern of LD among these six markers in cases was very similar to that seen in controls (data not shown).
Figure 1.
Schematic of the SNCA gene and patterns of LD. (A) The positions of REP1 and the five SNPs significantly associated with PD in Tier 1 are shown. Coding and non-coding sequence is indicated by black and light gray shading, respectively. (B) Pairwise LD between REP1 and these five SNPs as measured by r2 (left) and D’ (right) in the combined control sample. REP1 alleles 259 and 261 were grouped together.
The association between PD and rs356219 was further explored by performing regression analyses stratified by family history and AAO (Table 3). A significant effect was seen in both familial and sporadic subgroups, and in early and late-onset PD.
Table 3.
Stratified Analyses for rs356219
| Na | MAF | OR (95% CI) | p-value | |
|---|---|---|---|---|
| Total PD | 1942 | 0.44 | 1.41 (1.28–1.55) | 1.6 × 10−12 |
| Familial PDb | 431 | 0.44 | 1.40 (1.20–1.62) | 1.5 × 10−5 |
| Sporadic PDc | 1511 | 0.44 | 1.40 (1.27–1.54) | 1.2 × 10−11 |
| Total Controls | 2060 | 0.36 | - | - |
| Early-onset PDd | 489 | 0.44 | 1.37 (1.14–1.63) | 5.2 × 10−4 |
| Younger Controlse | 591 | 0.37 | - | - |
| Late-onset PDf | 1453 | 0.44 | 1.41 (1.27–1.56) | 2.2 × 10−10 |
| Older Controlsg | 1469 | 0.36 | - | - |
Data presented are for the combined sample in an additive, unadjusted model
Fourteen cases and 52 controls failed genotyping for rs356219
Patients with at least one first or second-degree relative with PD
Patients with no first or second-degree relatives with PD
Age at onset ≤ 50 years
Random sample of controls selected in proportion to the age at enrollment distribution of early-onset cases
Age at onset > 50 years
Total controls - younger controls
MAF = minor allele frequency; OR = odds ratio; CI = confidence interval; PD = Parkinson's disease
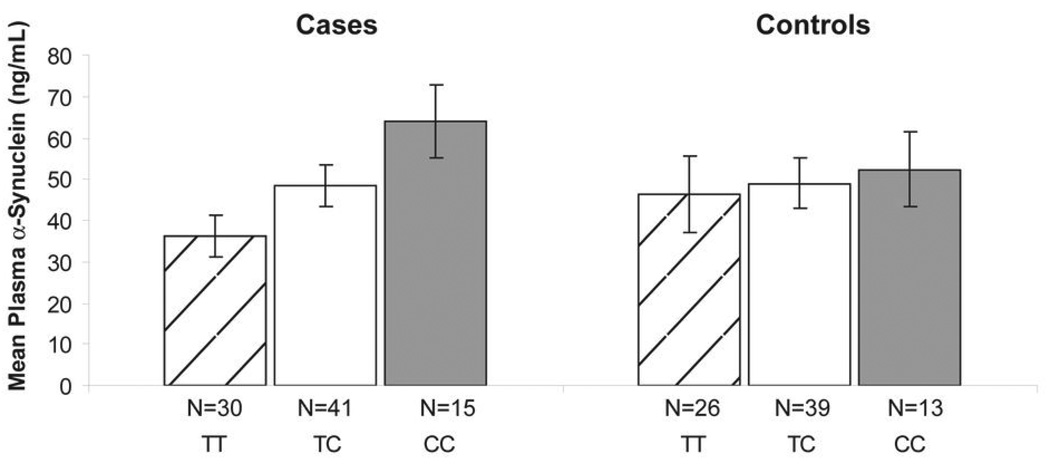
There was no significant difference in mean plasma α-synuclein levels between cases and controls (46.9 ± 32.6 vs. 48.6 ± 39.9 ng/ml; p = 0.76). We then examined the relationship between SNCA genotype and plasma rs356219, separately, in cases and controls. Because our data suggested that the association between SNCA and PD was large driven by rs356219, we focused only on this SNP. The distribution of plasma α-synuclein by rs356219 genotype deviated significantly from normality in both cases (p = 0.001) and controls (p < 0.001). We used a square root transformation in cases, and a natural log transformation in controls, to approximate a normal distribution. There was no significant evidence of heteroskedasticity in the transformed data for cases (p = 0.70) or controls (p = 0.48), and thus the transformed datasets were used for linear regression analysis. In cases, rs356219 was associated with square root plasma α-synuclein under an additive model after adjusting for age, sex, and RBC/platelet contamination (p = 0.005). The effect can be visualized by inspection of the raw data, which are plotted in Figure 2. Heterozygous cases had α-synuclein levels that were intermediate between the T/T and C/C homozygous groups. While a slight trend in the same direction can be seen in controls (Figure 2), rs356219 was not significantly associated with log plasma α-synuclein in controls in an adjusted additive model (p = 0.25).
Figure 2.
Mean plasma α-synuclein levels by rs356219 genotype in cases and controls. Error bars indicate one standard error of the mean. Genotype was significantly associated with transformed plasma α-synuclein levels in cases (p = 0.005) but not controls (p = 0.25) in an additive model adjusting for age, sex, and RBC/platelet contamination.
Discussion
Our findings strongly suggest that common variation within SNCA modifies susceptibility for PD. The association signal in our dataset emanated primarily from a single SNP (rs356219) whose effect was similar in early onset, late onset, familial, and sporadic disease. The signal from this SNP was largely independent of REP1, the only putative functional polymorphism identified to date within the SNCA locus. Finally, we have demonstrated for the first time that a risk-associated SNCA allele correlates with increased levels of α-synuclein protein in vivo, in a dose-dependent manner.
Few large studies on SNCA using a comprehensive tagSNP (or comparable) approach have been published in populations of European origin. The largest was a recent GWAS by Simón-Sánchez et al. of 1,713 cases and 3,978 controls in which strong association signals were observed from two genes, SNCA and MAPT.17 Several SNPs within SNCA were then successfully replicated in 3,361 cases and 4,573 controls. The most significant SNP in their combined sample was rs2736990 in intron 4 (OR = 1.23, p = 2.24 × 10−16). This finding is in agreement with our study, as the LD correlation coefficient between rs2736990 and rs356219 (our top SNP) was high (HapMap CEU sample, r2 = 0.82). In two smaller GWAS of 857 cases and 867 controls,15 and 604 cases and 619 controls,26 no markers met genomewide significance after correction for multiple testing. However, in the former study SNCA was ranked ninth by strength of association among all gene regions (under an additive model), and the most significant SNCA SNP was rs356229 (OR, 1.35; p = 5.5 × 10−5), located ~ 40 kb downstream from the gene.15 In the latter study, SNCA showed the highest association of all genes, and the strongest signal came from rs356220 (OR = 1.48; p = 2.7 × 10−6), located ~5 kb downstream.26 Again, both of these studies were consistent with our results, as the top SNCA SNP from each one was at least moderately well-correlated with rs356219 based on HapMap CEU data (rs356229, r2 = 0.56; rs356220, r2 = 0.96).
Results from our stepwise regression analysis, together with the low intermarker correlation observed (r2 = 0.09) indicate that the association signal from rs356219 is likely independent from that of REP1. This suggests that allelic heterogeneity exists and that one or more functional variants, in addition to REP1, act at the SNCA locus to convey risk for PD. The additional risk variant(s) is unlikely to be rs356219 itself, but rather a polymorphism in LD with it. Inspection of the HapMap CEU sample data that we used to select tagSNPs revealed seven other SNPs within the bin containing rs356219. However, none of these SNPs were obvious candidates to have direct biological effects. One (rs356165) is in the 3’ UTR, but not in a microRNA binding site predicted by the MicroCosm Targets web resource (http://www.ebi.ac.uk/enright-srv/microcosm/htdocs/targets/v5/), and the others are located downstream or deep within intron 4. Thus, the identity of the functional variant(s) in question remains to be determined. Our data are consistent with a recent analysis of 397 cases and 270 controls from Europe in which rs356165 was found to associate with PD independent of REP1, as there was essentially no correlation (r2 = 0) between the two polymorphisms.14 In contrast, Simón-Sánchez and colleagues examined REP1 in a subset of 1,774 samples from their GWAS and concluded that the association signal from REP1 and several SNCA SNPs was not independent and “may be the result of residual LD between these loci.” As evidence, the authors cited an r2 value of 0.37 between REP1 and rs3857059, but did not provide this information for the most significant SNCA SNP in their dataset, rs2736990, which was highly correlated with our top SNP (rs356219). Thus, it is difficult to directly compare our findings with theirs.
We found that the risk associated “C” allele of rs356219 was correlated with increased levels of plasma α-synuclein in PD patients in an additive manner (Figure 2). This suggests that the true functional risk variant tagged by this SNP might confer risk by upregulating neuronal SNCA expression, assuming that plasma α-synuclein (derived mainly from RBCs) and brain α-synuclein levels are correlated. Whether this assumption is valid in typical late onset PD remains to be determined, but carriers of SNCA triplications have a similar two-fold increase in both whole blood and brain α-synuclein.22 While a trend for higher plasma α-synuclein levels with increasing number of copies of the C allele was visible in our control group, this did not reach significance. One possible explanation is that at baseline, allele specific differences in SNCA expression are modest, but become accentuated beginning in the preclinical phase of the disease through gene×gene or gene × environment interactions.
Fuchs and colleagues examined the relationship between three SNCA polymorphisms (REP1, rs2583988, and rs356219) and SNCA mRNA and α-synuclein protein levels in substantia nigra (n = 8 cases and 14 controls), cerebellum (mRNA, n = 5 cases and 5 controls; protein, n= 17 cases and 24 controls), and PBMCs (n = 36 cases and 79 controls).27 They reported that the protective REP1 259/259 genotype was associated with lower PBMC protein levels, but did not see an association with the 263 risk allele. REP1 was not correlated with PBMC mRNA or brain mRNA/protein levels. Rs356219 was not associated with PBMC mRNA/protein, or with brain protein levels. However, the protective TT genotype was associated with higher mRNA levels in cerebellum. In substantia nigra, mRNA was highest in samples of the CT genotype, with lower and similar levels for the TT and CC genotypes. Rs2583988 was not associated with mRNA or protein levels in any of the tissues studied. We found these data difficult to interpret for a number of reasons, including the seemingly opposite effects of the protective genotypes across tissues, the small sample sizes, and the fact that cases and controls were pooled together in all analyses.
Though some studies28–29 have proposed that SNCA is divided into two or more LD blocks, data from these same studies, as well as ours (Figure 1B, right panel), indicate that there is still substantial LD (measured by D’) between markers at the 5’ and 3’ ends of the gene. Therefore, future work aimed at discovering the true risk variant(s) tagged by rs356219, which could require deep resequencing, should include the entire gene and possibly regions some distance away. The yield from such efforts might be increased by incorporating measurements of plasma α-synuclein. For example, selecting a subset of patients for resequencing from the extremes of the distribution of plasma α-synuclein level might enrich the sample for chromosomes bearing susceptibility alleles. Defining the full spectrum of SNCA risk variants is an important step in better understanding the molecular mechanisms by which α-synuclein mediates neurodegeneration, and ultimately could prove useful for developing PD therapeutics aimed at modifying α-synuclein in vivo.
Supplementary Material
Acknowledgements
We thank Erica Martinez and Sydney Thomas for assistance with subject recruitment, and Carolyn Hutter and Jia Yin Wan for statistical support. This work was supported by grants from the American Parkinson Disease Association, the Department of Veterans Affairs (1I01BX000531), the Michael J. Fox Foundation, the National Institutes of Health (P30 AG008017, P42 ES004696, P50 NS062684, R01 AG033398, R01 NS065070, R01 NS036960, R01 NS057567), and the Parkinson’s Disease Foundation. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
Footnotes
Potential Conflict of Interest: Dr. Agarwal serves on advisory boards for Ipsen and Merz Pharmaceuticals, and has received compensation as a speaker for Boehringer Ingelheim, GlaxoSmithKline, Novartis, and Teva Neuroscience. Dr. Factor has received research grants from Ipsen, Schering Plough, and Teva Neurosciences, and is a consultant for Allergan, Boehringer Ingelheim, Lundbeck, and UCB. Dr. Samii has received compensation as a speaker for Boerhinger Ingelheim, Ipsen, and Teva Neuroscience.
References
- 1.Gitler AD, Bevis BJ, Shorter J, et al. The Parkinson's disease protein alpha-synuclein disrupts cellular Rab homeostasis. Proc Natl Acad Sci U S A. 2008;105(1):145–150. doi: 10.1073/pnas.0710685105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nemani VM, Lu W, Berge V, et al. Increased Expression of [alpha]-Synuclein Reduces Neurotransmitter Release by Inhibiting Synaptic Vesicle Reclustering after Endocytosis. Neuron. 2010;65(1):66–79. doi: 10.1016/j.neuron.2009.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Polymeropoulos MH, Lavedan C, Leroy E, et al. Mutation in the alpha-synuclein gene identified in families with Parkinson's disease. Science. 1997;276(5321):2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 4.Spillantini MG, Schmidt ML, Lee VM, Trojanowski JQ, Jakes R, Goedert M. Alpha-synuclein in Lewy bodies. Nature. 1997;388(6645):839–840. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- 5.Farrer M, Kachergus J, Forno L, et al. Comparison of kindreds with parkinsonism and alpha-synuclein genomic multiplications. Ann Neurol. 2004;55(2):174–179. doi: 10.1002/ana.10846. [DOI] [PubMed] [Google Scholar]
- 6.Singleton AB, Farrer M, Johnson J, et al. alpha-Synuclein locus triplication causes Parkinson's disease. Science. 2003;302(5646):841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- 7.Kay DM, Factor SA, Samii A, et al. Genetic association between alpha-synuclein and idiopathic Parkinson's disease. Am J Med Genet B Neuropsychiatr Genet. 2008;147B(7):1222–1230. doi: 10.1002/ajmg.b.30758. [DOI] [PubMed] [Google Scholar]
- 8.Kruger R, Vieira-Saecker AM, Kuhn W, et al. Increased susceptibility to sporadic Parkinson's disease by a certain combined alpha-synuclein/apolipoprotein E genotype. Ann Neurol. 1999;45(5):611–617. doi: 10.1002/1531-8249(199905)45:5<611::aid-ana9>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 9.Maraganore DM, de Andrade M, Elbaz A, et al. Collaborative analysis of alpha-synuclein gene promoter variability and Parkinson disease. JAMA. 2006;296(6):661–670. doi: 10.1001/jama.296.6.661. [DOI] [PubMed] [Google Scholar]
- 10.Chiba-Falek O, Kowalak JA, Smulson ME, Nussbaum RL. Regulation of alpha-synuclein expression by poly (ADP ribose) polymerase-1 (PARP-1) binding to the NACP-Rep1 polymorphic site upstream of the SNCA gene. Am J Hum Genet. 2005;76(3):478–492. doi: 10.1086/428655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiba-Falek O, Nussbaum RL. Effect of allelic variation at the NACP-Rep1 repeat upstream of the alpha-synuclein gene (SNCA) on transcription in a cell culture luciferase reporter system. Hum Mol Genet. 2001;10(26):3101–3109. doi: 10.1093/hmg/10.26.3101. [DOI] [PubMed] [Google Scholar]
- 12.Fung HC, Scholz S, Matarin M, et al. Genome-wide genotyping in Parkinson's disease and neurologically normal controls: first stage analysis and public release of data. Lancet Neurol. 2006;5(11):911–916. doi: 10.1016/S1474-4422(06)70578-6. [DOI] [PubMed] [Google Scholar]
- 13.Maraganore DM, de Andrade M, Lesnick TG, et al. High-resolution whole-genome association study of Parkinson disease. Am J Hum Genet. 2005;77(5):685–693. doi: 10.1086/496902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Winkler S, Hagenah J, Lincoln S, et al. alpha-Synuclein and Parkinson disease susceptibility. Neurology. 2007;69(18):1745–1750. doi: 10.1212/01.wnl.0000275524.15125.f4. [DOI] [PubMed] [Google Scholar]
- 15.Pankratz N, Wilk JB, Latourelle JC, et al. Genomewide association study for susceptibility genes contributing to familial Parkinson disease. Hum Genet. 2009;124(6):593–605. doi: 10.1007/s00439-008-0582-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Satake W, Nakabayashi Y, Mizuta I, et al. Genome-wide association study identifies common variants at four loci as genetic risk factors for Parkinson's disease. Nat Genet. 2009;41(12):1303–1307. doi: 10.1038/ng.485. [DOI] [PubMed] [Google Scholar]
- 17.Simon-Sanchez J, Schulte C, Bras JM, et al. Genome-wide association study reveals genetic risk underlying Parkinson's disease. Nat Genet. 2009;41(12):1308–1312. doi: 10.1038/ng.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maroteaux L, Campanelli JT, Scheller RH. Synuclein: a neuron-specific protein localized to the nucleus and presynaptic nerve terminal. J Neurosci. 1988;8(8):2804–2815. doi: 10.1523/JNEUROSCI.08-08-02804.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Michell AW, Luheshi LM, Barker RA. Skin and platelet alpha-synuclein as peripheral biomarkers of Parkinson's disease. Neurosci Lett. 2005;381(3):294–298. doi: 10.1016/j.neulet.2005.02.030. [DOI] [PubMed] [Google Scholar]
- 20.Scherzer CR, Grass JA, Liao Z, et al. GATA transcription factors directly regulate the Parkinson's disease-linked gene alpha-synuclein. Proc Natl Acad Sci U S A. 2008;105(31):10907–10912. doi: 10.1073/pnas.0802437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barbour R, Kling K, Anderson JP, et al. Red blood cells are the major source of alpha-synuclein in blood. Neurodegener Dis. 2008;5(2):55–59. doi: 10.1159/000112832. [DOI] [PubMed] [Google Scholar]
- 22.Miller DW, Hague SM, Clarimon J, et al. Alpha-synuclein in blood and brain from familial Parkinson disease with SNCA locus triplication. Neurology. 2004;62(10):1835–1838. doi: 10.1212/01.wnl.0000127517.33208.f4. [DOI] [PubMed] [Google Scholar]
- 23.Gibb WR, Lees AJ. The relevance of the Lewy body to the pathogenesis of idiopathic Parkinson's disease. J Neurol Neurosurg Psychiatry. 1988;51(6):745–752. doi: 10.1136/jnnp.51.6.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hong Z, Shi M, Chung KA, et al. DJ-1 and {alpha}-synuclein in human cerebrospinal fluid as biomarkers of Parkinson's disease. Brain. 2010 doi: 10.1093/brain/awq008. doi:10.1093/brain/awq1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 26.Edwards TL, Scott WK, Almonte C, et al. Genome-Wide Association Study Confirms SNPs in SNCA and the MAPT Region as Common Risk Factors for Parkinson Disease. Ann Hum Genet. 2010 doi: 10.1111/j.1469-1809.2009.00560.x. DOI 10.1111/j.1469-1809.2009.00560.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fuchs J, Tichopad A, Golub Y, et al. Genetic variability in the SNCA gene influences alpha-synuclein levels in the blood and brain. FASEB J. 2008;22(5):1327–1334. doi: 10.1096/fj.07-9348com. [DOI] [PubMed] [Google Scholar]
- 28.Mueller JC, Fuchs J, Hofer A, et al. Multiple regions of alpha-synuclein are associated with Parkinson's disease. Ann Neurol. 2005;57(4):535–541. doi: 10.1002/ana.20438. [DOI] [PubMed] [Google Scholar]
- 29.Myhre R, Toft M, Kachergus J, et al. Multiple alpha-synuclein gene polymorphisms are associated with Parkinson's disease in a Norwegian population. Acta Neurol Scand. 2008;118(5):320–327. doi: 10.1111/j.1600-0404.2008.01019.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.