Abstract
Objectives
The RAR-related orphan receptor alpha (RORalpha) gene is implicated as a candidate for age-related macular degeneration (AMD) through a previous microarray expression study, linkage data, biological plausibility, and two clinic-based cross sectional studies. We aimed to determine if common variants in RORalpha predict future risk of neovascular AMD.
Methods
We measured genotypes for 18 variants in intron 1 of the RORalpha gene among 164 cases who developed neovascular AMD and 485 age- and sex-matched controls in a prospective nested case-control study within the Nurses’ Health Study and the Health Professionals Follow-up Study. We determined the incidence rate ratios (IRR) and 95% confidence intervals (CI) for neovascular AMD for each variant, and examined interactions with other AMD-associated variants and modifiable risk factors.
Results
We identified a single SNP (rs12900948) that was significantly associated with increased incidence of neovascular AMD. Participants with one and two copies of the “G” allele were 1.73 (CI= 1.32–2.27) and 2.99 (CI=1.74–5.14) times more likely to develop neovascular AMD. Individuals homozygous for both the “G” allele of rs12900948 and ARMS2 A69S had a 40.8-fold increased risk of neovascular AMD (CI=10.1–164; P for interaction=0.017). Cigarette smokers who carried two copies of the “G” allele had a 9.89-fold risk of neovascular AMD, but the interaction was not significant (P=0.08). We identified a significant AMD-associated haplotype block containing SNPs rs730754, rs8034864, and rs12900948, with P-values for ACA=1.16 × 10−9, ACG=5.85 × 10−12, and GAA=0.0001 when compared to all other haplotypes.
Conclusion
Common variants and haplotypes within the RORalpha gene appear to act synergistically with the ARMS2 A69S polymorphism to increase risk of neovascular AMD. These data add further evidence of a high level of complexity linking genetic and modifiable risk factors to AMD development and should help efforts at risk prediction.
Introduction
The leading cause of blindness among whites in the US and other industrialized countries,1 age-related macular degeneration (AMD) has emerged as a paradigmatic example of a common complex disease caused by the interplay of genetic predisposition and exposure to modifiable risk factors.2 A large number of studies have established relationships between lifestyle factors such as diet, cigarette smoking, and obesity as well as common variants within a handful of genes and risk of AMD in multiple populations. Among the genetic risk factors, common variants in two genes, complement factor H (CFH) (1q32) and ARMS2/HTRA serine peptidase 1 (HTRA1) (10q26), have strong and consistent associations with risk of AMD and are estimated to contribute to a strikingly large proportion of AMD cases in the US population.2 Nonetheless, not every individual with a given risk factor or set of risk factors will develop AMD, and efforts at developing risk prediction models based on currently understood risk factors are insufficient to reliably predict the development and progression of AMD among individuals.3 Refinements in exposure assessment for epidemiological risk factors such as cigarette smoking, diet, and obesity, as well as the identification of other disease associated variants, may improve the predictive ability and lead to clinically relevant interventions to identify individuals that require closer follow-up and earlier or more targeted forms of intervention.
With such a goal in mind, the authors previously performed linkage analysis and gene expression microarray analysis on a family based cohort comprised of extremely discordant sibling pairs (EDSP) (that is pairs where the unaffected siblings had normal maculae at an age older ( ≥ 65 years) than that at which the index patient was first diagnosed with neovascular AMD.4 The EDSPs were used as a discovery cohort to identify novel candidate genes and pathways with biological relevance.5 Based on the results of these studies, the candidate gene RAR-related orphan receptor alpha (RORalpha) was chosen for further analysis. RORalpha is a retinoid-related orphan receptor and member of a distinct subfamily of nuclear receptors.6 RORalpha is known to be involved in a number of biological processes with potential relevance to AMD, including immunity/inflammation, angiogenesis, and lipid and cholesterol metabolism.7–17 It is also located within a linkage peak identified previously by two independent studies.18, 19 In an initial study, we identified common variants (rs4335725, rs12900948, and 2 haplotypes) within intron 1 of RORalpha that were associated with AMD in two independent cross-sectional clinic-based study populations.5
In the present study, we investigated 18 common intron 1 variants of RORalpha in a prospective nested case-control study of neovascular AMD within the Nurses’ Health Study (NHS) and Health Professionals Follow-up Study (HPFS) cohorts. We aimed to replicate the association between the RORalpha gene and neovascular AMD, and to further clarify the magnitude and inter-relationships of this gene and other risk factors in a prospective study population.
Methods
The NHS is an ongoing prospective study of 121,700 primarily white US female registered nurses aged 30–55 years in 1976. The HPFS includes 51,529 largely white US male health professionals who have been followed prospectively since 1986. At that time, HPFS participants ranged in age from 40 to 75 years. From 1989 to 1990, we obtained blood samples from 32,826 NHS participants; and between 1993 and 1995 from 18,162 HPFS participants, who form the study base. The Institutional Review Board of Brigham and Women’s Hospital and the Harvard School of Public Health’s Human Subjects Committee approved the study protocol.
From the time of enrollment, NHS and HPFS participants completed a mailed questionnaire every 2 years, on which we obtained information on lifestyle factors including height and weight from which we calculated the body mass index, cigarette smoking history 20, and diet assessed via validated semi-quantitative food frequency questionnaires 21, 22. For the present study, we used exposure information at the time of blood collection.
Ascertainment of AMD Cases & Control Selection
We used a validated two-stage procedure to document incident cases of AMD.20, 22 Briefly, we asked participants on each biennial study questionnaire about the diagnosis of AMD. When AMD was reported, we requested permission to review medical records. If permission was granted, we sent a letter to the participant’s eye doctor to obtain information on the date of AMD diagnosis, best-corrected visual acuity at the most recent exam, and the chorio-retinal lesions present (drusen; RPE changes including atrophy, hypertrophy and RPE detachment; geographic atrophy; subretinal neovascular membrane; disciform scar), and other information. We classified cases as neovascular AMD if there was an RPE detachment, subretinal neovascular membrane, or disciform scar not due to other causes (e.g. histoplasmosis, choroidal rupture). Only those participants in whom we confirmed the presence neovascular AMD with a visual acuity of 20/30 or worse attributable to AMD, who were first diagnosed after the date of receipt of the baseline blood specimen and were aged 50 years or older, were selected as cases for the present study. We classified participants based on the most severely affected eye.
We selected three controls for each case of neovascular AMD at random from study participants in the same cohort as the case who were still at risk of AMD at the time the case was diagnosed, who were of the same age within 1 year, and who reported having an eye exam in the past two years. Multiple controls were used to increase study power. We previously demonstrated significant associations between the Y402H polymorphism in CFH as well as the A69S polymorphism in ARMS2 in these cases and controls.
Genotyping
We examined 18 single nucleotide polymorphisms (SNPs) within intron 1 of the RORalpha gene. Seven of these SNPs (rs12916023, rs730754, rs8034864, rs12900948, rs12591914, rs17237514, and rs4335725) were shown to be significant, either individually or as part of a haplotype, in a previous study of 150 sib-pairs who were extremely discordant for AMD; that is the index patient with the neovascular form of AMD who had an unaffected sibling with normal maculae over the age of 65 years (detailed information on the EDSP cohort is described elsewhere 4, 23). These seven significant SNPs were derived from an initial group of 148 SNPs chosen approximately every 3000 to 5000 base pairs to represent variation within the 730 kilobases of the RORalpha gene.5 For this analysis, we chose 11 additional tagSNPs that surrounded the region encompassing the 7 significant SNPs using the HapMap (www.hapmap.org). In choosing the tagSNPs, each SNP must have had 1) a minor allele frequency of 10% or greater, and 2) an r2 value of at least 0.8. DNA was extracted from the buffy coat fraction of centrifuged blood specimens using the QIAmp Blood Kit (Qiagen). Sequenom SpectroDESIGNER software (version 3.0.0.3) (Sequenom, San Diego, CA) was used to design Multiplex PCR assays using sequence containing the SNP site and 100 bp of flanking sequence on either side of the SNP. Briefly, 10 ng genomic DNA was amplified in a 5 ul reaction containing 1X HotStar Taq PCR buffer (Qiagen, Valencia, CA), 1.625 mM MgCl2, 500 uM each dNTP, 100 nM each PCR primer, 0.5 U HotStar Taq (Qiagen). The reaction was incubated at 94°C for 15 minutes followed by 45 cycles of 94°C for 20 seconds, 56°C for 30 seconds, 72°C for 1 minute, followed by 3 minutes at 72°C. Excess dNTPs were then removed from the reaction by incubation with 0.3 U shrimp alkaline phosphatase (USB, Cleveland, OH) at 37°C for 40 minutes followed by 5 minutes at 85°C to deactivate the enzyme. Single primer extension over the SNP was carried out in a final concentration of between 0.625 uM and 1.5 uM for each extension primer (depending on the mass of the probe), iPLEX termination mix (Sequenom) and 1.35 U iPLEX enzyme (Sequenom) and cycled using a two-step 200 short cycles program; 94°C for 30 seconds followed by 40 cycles of 94°C for 5 seconds, 5 cycles of 52°C for 5 seconds, and 80°C for 5 seconds, then 72°C for 3 minutes. The reaction was then desalted by addition of 6 mg cation exchange resin followed by mixing and centrifugation to settle the contents of the tube. The extension product was then spotted onto a 384 well spectroCHIP before being flown in the MALDI-TOF mass spectrometer. Data were collected during real time, using SpectroTYPER Analyzer 3.3.0.15, SpectraAQUIRE 3.3.1.1 and SpectroCALLER 3.3.0.14 (Sequenom). Genotypes for each subject were also checked manually as an additional quality control measure. All laboratory personnel were blinded to case/control status.
Statistical Analysis
We initially examined allele distributions and used chi-square tests for Hardy-Weinberg equilibrium (HWE). We then compared genotype and allele frequencies between cases and controls using chi-square tests -- we used Armitage’s trend test to examine evidence for an additive allele effect on AMD susceptibility, and the genotype case-control test which tests for both additive and dominance (nonadditive) allelic effects (Nielsen and Weir 1999).
We then used logistic regression under additive, dominant, and recessive genetic models to estimate the incidence rate ratios (IRR) and 95% confidence intervals (CI) for each genotype adjusted for other risk factors. We first obtained separate estimates of the IRR in each cohort and tested for heterogeneity using Cochrane’s Q test. As there was no evidence for heterogeneity between cohorts (P for each SNP≥0.3), we present only pooled data from the two prospectively ascertained cohorts. Controlling for age and sex, we modeled the allelic effects using a multiplicative (i.e. log-additive) coding scheme using a single variable for each SNP coded 0 for subjects homozygous for the major allele (or, alternatively for the candidate SNPs, the allele previously found to be associated with the lowest risk of AMD), 1 for heterozygotes, and 2 for subjects homozygous for the minor allele (or, alternatively for the candidate SNPs, the allele previously found to be associated with increased risk of AMD). We next fit unconstrained (i.e. co-dominant) models using separate indicator variables for subjects who were heterozygous, and subjects who were homozygous for the risk allele. To arrive at the best-fitting model, we compared these alternative models using Akaike’s Information Criteria (AIC).24 As a rule of thumb, two models are statistically indistinguishable if the AIC difference is less than 2.
For SNPs that showed significant associations in the models controlling for age and sex, we extended the preferred models to control for potential confounding by other risk factors including cigarette smoking (current, yes versus no), obesity (BMI≥30 kg/m2 versus <30 kg/m2), regular aspirin use (yes versus no), alcohol intake (continuous), consumption of fruits (continuous), and the ratio of omega-6/omega-3 fatty acids in the diet (continuous). In the next step, we retained any significant risk factors and fit additional models controlling for the CFH Y402H and ARMS2/HTRA1 A69S variants, which were previously shown to be strongly associated with AMD in these cohorts.
For significant RORalpha SNPs, we additionally examined interactions between RORalpha and CFH Y402H, ARMS2 A69S, cigarette smoking, and obesity, and fit additional models to simultaneously estimate the stratum-specific IRR (CI) for the joint effects of the RORA variant and these other risk factors.
We used a model-based method based on the observed data to calculate the attributable fraction in the population as a measure of the proportion of AMD cases to which each polymorphism contributes.25–27
We estimated linkage disequilibrium (LD) (both r2 and D’) between each pair of SNPs and constructed haplotype blocks in Haploview (http://www.broad.mit.edu/mpg/haploview/) using the method proposed by Gabriel et al.28 We inferred individual haplotypes and tested these for association with AMD in Haploview.
Results
The study population included the 164 cases of incident neovascular AMD matched with 485 controls that were previously studied for association of CFH Y402H and ARMS2/HTRA1 A69S. The mean age at AMD diagnosis was 68.7(±6) years. Of the 19 RORalpha SNPS, genotype data were successfully obtained for each SNP in ≥98% of cases and controls, with the exception of rs7177611, which was successfully genotyped in 92% of cases and controls, and rs11071570, for which genotype data were available in 97% of cases and 94% of controls. We found no significant departures from Hardy-Weinberg equilibrium for any of the 18 SNPs among the control group (each P>0.05).
Chi square tests for single SNP analysis showed that one SNP, rs12900948, was significantly associated with increased risk of neovascular AMD (P for genotype=0.00018, P for allelic trend=0.00003), Table 1. This SNP was part of a haplotype block shown to be significant in a previous study of 150 extremely discordant sib-pairs (EDSP) and in an unrelated case control cohort from central Greece (Silvera et al, in press). In addition, these analyses identified participants with neovascular AMD were more likely to be heterozygous for two SNPs, rs730754 (P for genotype=0.038) and rs975501 (P for genotype=0.033).
Table 1.
Chi-square tests and age- and sex-adjusted logistic regression estimates for association between 18 SNPs within intron 1 of the RORA gene and risk of neovascular AMD among 164 cases of neovascular AMD and 485 controls in the NHS and HPFS cohorts
| Locus | Controls | Cases | P-value | Additive Model | Dominant Model | Recessive Model | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| N (%) | N (%) | Genotype (2 df) |
Trend (1 df) Additive Genetic Model |
Odds Ratio (CI) | p value | Odds Ratio (CI) | p value | Odds Ratio (CI) | p value | |
| rs12916023 | ||||||||||
| C/C | 117 (24.7) | 36 (22.1) | ||||||||
| C/T | 248 (52.3) | 94 (57.7) | ||||||||
| T/T | 109 (23.0) | 33 (20.2) | 0.50 | 0.98 | 1.00 (0.77 – 1.30) | 0.98 | 1.18 (0.76 – 1.83) | 0.47 | 0.86 (0.56 – 1.32) | 0.50 |
| rs4583176 | ||||||||||
| C/C | 279 (58.4) | 88 (54.3) | ||||||||
| C/T | 169 (35.4) | 68 (42.0) | ||||||||
| T/T | 30 (6.3) | 6 (3.7) | 0.20 | 0.79 | 1.04 (0.78 – 1.40) | 0.78 | 1.18 (0.83 – 1.69) | 0.36 | 0.58 (0.24 – 1.41) | 0.23 |
| rs730754 | ||||||||||
| A/A | 173 (36.6) | 47 (28.7) | ||||||||
| A/G | 225 (47.6) | 97 (59.2) | ||||||||
| G/G | 75 (15.9) | 20 (12.2) | 0.038 | 0.49 | 1.10 (0.84 – 1.43) | 0.48 | 1.44 (0.98 – 2.13) | 0.07 | 0.74 (0.43 – 1.25) | 0.26 |
| rs8034864 | ||||||||||
| A/A | 20 (4.2) | 6 (3.7) | ||||||||
| A/C | 162 (34.3) | 61 (37.4) | ||||||||
| C/C | 291 (61.5) | 96 (58.9) | 0.75 | 0.69 | 1.07 (0.78 – 1.45)` | 0.69 | 1.12 (0.78 – 1.60) | 0.56 | 0.87 (0.34 – 2.19) | 0.76 |
| rs975501 | ||||||||||
| A/A | 171 (35.7) | 45 (27.8) | ||||||||
| A/G | 230 (48.0) | 97 (59.9) | ||||||||
| G/G | 78 (16.3) | 20 (12.4) | 0.033 | 0.52 | 1.09 (0.84 – 1.42) | 0.51 | 1.45 (0.98 – 2.14) | 0.06 | 0.73 (0.43 – 1.23) | 0.23 |
| rs12900948 | ||||||||||
| A/A | 128 (27.0) | 24 (14.7) | ||||||||
| A/G | 216 (45.6) | 69 (42.3) | ||||||||
| G/G | 130 (27.4) | 70 (42.9) | 0.00018 | 0.00003 | 1.76 (1.36 – 2.29) | 2.2 × 10−5 | 2.22 (1.36 – 3.62) | 0.0013 | 2.07 (1.42 – 3.04) | 0.0002 |
| rs782925 | ||||||||||
| A/A | 16 (3.4) | 6 (3.7) | ||||||||
| A/G | 172 (36.1) | 52 (32.1) | ||||||||
| G/G | 289 (60.6) | 104 (64.2) | 0.66 | 0.52 | 0.90 (0.65 – 1.24) | 0.51 | 0.85 (0.59 – 1.24) | 0.40 | 1.10 (0.42 – 2.87) | 0.84 |
| rs7177611 | ||||||||||
| C/C | 222 (49.8) | 69 (45.7) | ||||||||
| C/T | 192 (43.1) | 66 (43.7) | ||||||||
| T/T | 32 (7.2) | 16 (10.6) | 0.36 | 0.21 | 1.21 (0.91 – 1.61) | 0.20 | 1.18 (0.82 – 1.71) | 0.38 | 1.55 (0.82 – 2.92) | 0.17 |
| rs10403737 | ||||||||||
| C/C | 239 (49.5) | 86 (52.8) | ||||||||
| C/T | 206 (42.7) | 62 (38.0) | ||||||||
| T/T | 38 (7.9) | 15 (9.2) | 0.56 | 0.74 | 0.95 (0.72 – 1.26) | 0.74 | 0.88 (0.62 – 1.25) | 0.47 | 1.19 (0.64 – 2.23) | 0.59 |
| rs12591914 | ||||||||||
| G/G | 241 (50.4) | 72 (44.4) | ||||||||
| G/T | 207 (43.3) | 75 (46.3) | ||||||||
| T/T | 30 (6.3) | 15 (9.3) | 0.26 | 0.11 | 1.26 (0.94 – 1.67) | 0.12 | 1.27 (0.87 – 1.82) | 0.19 | 1.52 (0.79 – 2.90) | 0.21 |
| rs16943429 | ||||||||||
| A/A | 327 (68.7) | 102 (63.0) | ||||||||
| A/G | 134 (28.2) | 54 (33.3) | ||||||||
| G/G | 15 (3.2) | 6 (3.7) | 0.41 | 0.20 | 1.23 (0.89 – 1.69) | 0.21 | 1.29 (0.89 – 1.88) | 0.18 | 1.17 (0.45 – 3.08) | 0.74 |
| rs7495128 | ||||||||||
| A/A | 19 (4.0) | 2 (1.2) | ||||||||
| A/G | 183 (38.4) | 61 (37.7) | ||||||||
| G/G | 275 (57.7) | 99 (61.1) | 0.22 | 0.22 | 0.82 (0.59 – 1.14) | 0.23 | 0.87 (0.60 – 1.26) | 0.46 | 0.30 (0.07 – 1.31) | 0.11 |
| rs2414687 | ||||||||||
| G/G | 257 (53.8) | 96 (59.3) | ||||||||
| G/T | 185 (38.7) | 57 (35.2) | ||||||||
| T/T | 36 (7.5) | 9 (5.6) | 0.42 | 0.19 | 0.82 (0.61 – 1.10) | 0.19 | 0.80 (0.56 – 1.15) | 0.22 | 0.72 (0.34 – 1.53) | 0.40 |
| rs17237514 | ||||||||||
| A/A | 333 (70.0) | 115 (70.1) | ||||||||
| A/G | 129 (26.9) | 43 (26.2) | ||||||||
| G/G | 17 (3.6) | 6 (3.7) | 0.98 | 0.92 | 0.99 (0.71 – 1.37) | 0.93 | 0.97 (0.66 – 1.44) | 0.89 | 1.03 (0.40 – 2.67) | 0.95 |
| rs4335725 | ||||||||||
| A/A | 42 (8.9) | 18 (11.0) | ||||||||
| A/G | 218 (46.1) | 68 (41.7) | ||||||||
| G/G | 213 (45.0) | 77 (47.2) | 0.54 | 0.99 | 1.00 (0.76 – 1.32) | >0.99 | 0.92 (0.64 – 1.31) | 0.63 | 1.28 (0.71 – 2.29) | 0.41 |
| rs17270640 | ||||||||||
| C/C | 226 (47.5) | 68 (42.0) | ||||||||
| C/G | 201 (31.5) | 75 (46.3) | ||||||||
| G/G | 49 (10.3) | 19 (11.7) | 0.47 | 0.25 | 1.17 (0.89 – 1.52) | 0.26 | 1.25 (0.87 – 1.79) | 0.23 | 1.16 (0.66 – 2.04) | 0.61 |
| rs11071570 | ||||||||||
| C/C | 72 (15.7) | 26 (16.5) | ||||||||
| C/G | 208 (45.4) | 76 (47.8) | ||||||||
| G/G | 178 (38.7) | 57 (35.9) | 0.80 | 0.57 | 1.07 (0.83 – 1.39) | 0.61 | 1.13 (0.77 – 1.65) | 0.54 | 1.04 (0.64 – 1.71) | 0.87 |
| rs6494231 | ||||||||||
| A/A | 85 (17.9) | 27 (16.6) | ||||||||
| A/G | 220 (46.2) | 67 (41.1) | ||||||||
| G/G | 171 (35.9) | 69 (42.3) | 0.34 | 0.24 | 0.86 (0.67 – 1.11) | 0.24 | 0.76 (0.53 – 1.10) | 0.15 | 0.91 (0.57 – 1.47) | 0.71 |
In age- and sex-adjusted logistic regression models, we found that the “G” allele of SNP rs12900948 was significantly associated with increased risk of AMD under all three genetic models tested– additive (P=2.2 × 10−5), dominant (P=0.0013) and recessive (P=0.0002), Table 1. Odds ratio estimates from the additive model indicated a 1.76-fold increased risk of neovascular AMD for heterozygotes, and a 3.10-fold increased risk for participants who were homozygous for the “G” allele (Table 2). None of the other 17 RORalpha SNPs was significantly associated with risk of neovascular AMD under age- and sex-adjusted additive, dominant, or recessive models (Table 1).
Table 2.
Odds ratio estimates for RORA rs12900948 in the Nurses’ Health Study and Health Professionals Follow-up Study Cohorts
| Factors Controlled For | Genetic Model for rs12900948 |
OR (CI) for rs12900948 | |
|---|---|---|---|
| GA | GG | ||
| Age, sex | Additive | 1.76 (1.36 – 2.29) | 3.10 (1.84 – 5.25) |
| Age, sex | Co-dominant | 1.78 (1.06 – 2.99) | 3.12 (1.81 – 5.37) |
| Age, sex, pack-years of cigarette smoking, obesity | Additive | 1.73 (1.32 – 2.27) | 2.99 (1.74 – 5.14) |
| Age, sex, pack-years of cigarette smoking, obesity, CFH Y402H, ARMS2 A69S | Additive | 1.47 (1.08 – 1.99) | 2.15 (1.17 – 3.97) |
We next fit an unconstrained model for SNP rs12900948 to distinguish between the log-additive model and a co-dominant model. Comparison of AIC from these models indicated the log-additive model was the best fitting model for these data (AIC difference = 2.0). Using log-additive models for the genetic effect, we extended the models for rs12900948 to control for additional risk factors for AMD. After controlling for cigarette smoking and obesity, the two strongest lifestyle risk factors for AMD in these cohorts, the odds ratios (95% CI) for rs12900948 were: 1.73 (1.32 – 2.27) for one “G” allele, and 2.99 (1.74 – 5.14) among those with two copies of the “G” allele. Estimates for rs12900948 remained significantly associated with incidence of neovascular AMD in models further controlling for other lifestyle risk factors (data not shown), or the genetic factors CFH Y402H and ARMS2/HTRA1 A69S (Table 2). After controlling for these additional genetic risk factors, we observed an approximate 30% reduction in the magnitude of the odds ratio estimates for rs12900948 suggesting the possibility of shared biological pathways.
We tested for statistical interaction on the multiplicative scale between the rs12900948 SNP and CFH Y402H and ARMS2/HTRA1 A69S, and found no evidence for interaction with CFH Y402H (P=0.37). In contrast, we identified a statistically significant departure from multiplicative effects between rs12900948 of RORalpha and ARMS2 A69S (P=0.017). Estimates from this model indicated a 20-fold increased risk of neovascular AMD for participants who were homozygous for both risk-associated variants. In an alternative model in which we simultaneously estimated the stratum-specific IRR of neovascular AMD for each possible combination of rs12900948 and ARMS2 A69S genotypes (Table 3), the estimated relative risk of neovascular AMD was >40-fold higher among subjects who were homozygous for the risk-associated variant at both loci compared to subjects with no risk-associated alleles at either locus, although the confidence interval was wide (CI=10.1 to 164).
Table 3.
Joint effects of RORA rs12900948 and ARMS2 A69S genotypes on the incidence of neovascular AMD.
| LOC387715 A69S Genotype | RORA rs12900948 Genotype |
|||
|---|---|---|---|---|
| A/A | A/G | G/G | ||
| A/A | IRR (CI)* | 1.00 | 1.67 (0.78 – 3.58) | 1.47 (0.64 – 3.38) |
| N cases/ N controls | 11 / 79 | 27 / 120 | 17 / 88 | |
| A/S | IRR (CI) | 1.97 (0.75 – 5.19) | 2.43 (1.11 – 5.30) | 6.75 (3.02 – 15.1) |
| N cases/ N controls | 9 / 33 | 25 / 76 | 32 / 36 | |
| S/S | IRR (CI) | 5.34 (1.05 – 27.2) | 7.53 (2.62 – 21.6) | 40.8 (10.1 – 164) |
| N cases/ N controls | 3 / 4 | 11 / 11 | 16 / 3 | |
| P for interaction = 0.017 | ||||
Estimates of the incidence rate ratios (IRR) and 95% confidence intervals (CI) were estimated simultaneously from a logistic regression model with an indicator for each possible combination of genotypes at the two loci, using a common referent group of those homozygous for the major allele at both loci (or, alternatively for the candidate SNPs, the allele previously found to be associated with the lowest risk of AMD), and controlling for age and sex.
Testing of multiplicative interaction terms demonstrated no statistically significant departures from multiplicative joint effects between rs12900948 and either cigarette smoking (P for interaction=0.08) or obesity (P for interaction=0.56), Table 4. We calculated the odds ratios for each genotype-risk factor combination to identify whether any of subgroups of subjects at particularly high risk of AMD could be identified (Table4). Though statistically consistent with multiplicative effects, these estimates indicate that compared to participants who never smoked and had no risk-associated alleles at rs12900948, there is a 4.6-fold increased incidence of neovascular AMD among non-smoking participants with two “G” alleles, whereas the risk is nearly 10-fold higher among participants who were current smokers and carried two “G” alleles.
Table 4.
Incidence rate ratios (IRR) and 95% confidence intervals (CI) of developing neovascular AMD according to the genotype at RORA rs12900948 and modifiable risk factors for neovascular AMD
| RORA rs12900948 Genotype |
|||||
|---|---|---|---|---|---|
| Risk Factor | A/A | A/G | G/G | P for interaction |
|
| Body Mass Index |
|||||
| <30 kg/m2 | IRR (CI)* | 1.00 | 1.79 (1.04–3.07) | 3.00 (1.70–5.30) | |
|
N cases/N controls |
22/118 | 62/193 | 60/116 | ||
| ≥30 kg/m2 | IRR (CI) | 1.05 (0.21–5.11) | 1.81 (0.68–4.80 | 4.24 (1.65–11.0) | 0.49 |
|
N cases/N controls |
2/10 | 7/23 | 10/14 | ||
| Cigarette Smoking |
|||||
| Non- smoker |
IRR (CI) | 1.00 | 1.92 (1.00–3.71) | 4.58 (2.28–9.19) | |
|
N cases/N controls |
15/95 | 38/129 | 39/58 | ||
| Current smoker |
IRR (CI) | 3.45 (0.91–13.1) | 4.28 (1.69–10.8) | 9.89 (3.71–26.3) | 0.08 |
|
N cases/N controls |
4/8 | 13/22 | 17/13 | ||
Estimates of the incidence rate ratios (IRR) and 95% confidence intervals (CI) are from separate logistic regression models for the interaction of cigarette smoking or obesity with rs12900948, controlling for age and sex. Each unique combination of risk factor and genotype was coded with a separate indicator variable, using the jointly unexposed group as a common referent, to allow for the simultaneous estimation of stratum specific IRR. Departures from multiplicative interaction were tested using the alternative coding scheme comprised of an indicator variable for the risk factor and for genotype (for which we used an ordinal variable, representative of the multiplicative genetic model), and a product term for their interaction. Significance of the product terms was evaluated by 2-sided Wald tests.
Calculation of the population attributable fractions using a model-based method showed an attributable fraction of 45% (CI= 32% to 59%) for rs12900948, after controlling for CFH Y402H and ARMS2/HTRA1 A69S. The attributable fraction for all three SNPS (rs12900948 together with CFH Y402H and ARMS2/HTRA1 A69S) was 84% (CI= 80% to 88%) for rs12900948.
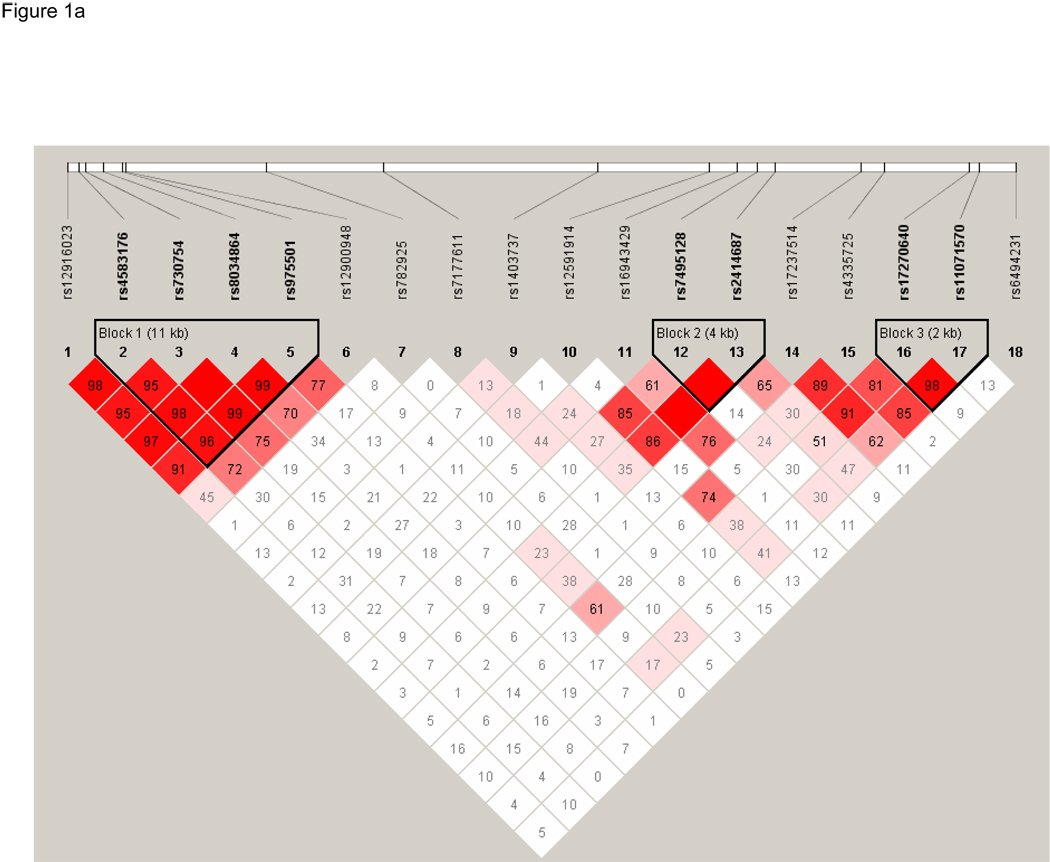
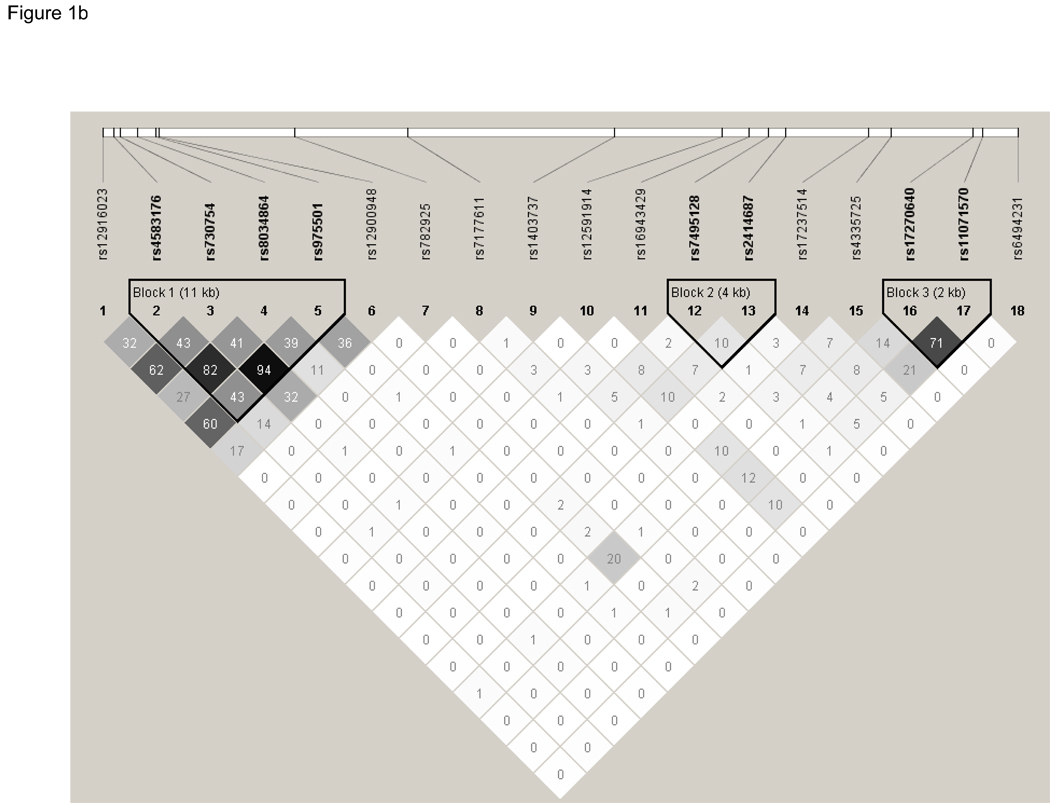
Finally, we constructed linkage disequilibrium plots of these 18 SNPs using Haploview (Figure 1). Three haplotype blocks were constructed based on confidence intervals using the method proposed by Gabriel et al.28 None of these three blocks contained the significant SNP, rs12900948. Association testing of these three haplotype blocks showed that one haplotype, h1 of Block 2, was significantly associated with AMD (P=0.03), Table 5. In an effort to replicate the haplotypes that were significant in the EDSP cohort, Block 1 and Block 4 as defined by Gabriel in the EDSP cohort were constructed on the NHS/HPFS cohort using Haploview. In this analysis, Block 1, which contained SNPs rs730754, rs8034864, and rs12900948, was shown to be significantly associated with AMD in the NHS/HPFS cohort. Specifically, h1 (P=1.16 × 10−9), h3 (P=5.85 × 10−12), and h5 (P=0.0001) were all significantly associated with AMD. Block 4, which contained SNPs rs17237514 and rs4335725, was not significantly associated with AMD in the NHS/HPFS cohorts (P>0.8 for all), Table 6.
Figure 1.
Linkage Disequilibrium (D’) between the 18 SNPs genotyped in the RORalpha gene in the Nurses' Health Study (NHS) and Health Professionals Follow-up Study (HPFS) cohorts. LD was determined using the program Haploview and LD blocks were defined using the Gabriel Rule. Boxes are shaded increasingly darker to represent higher percentage of LD and the numbers listed in each square represent the D’ (Part 1a) and r2 values (Part 1b). A completely shaded in box represents complete LD. Three haplotype blocks were generated for this cohort of NHS and HPFS participants.
Table 5.
Haplotype Analysis of the NHS and HPFS cohorts, blocks were defined by the method outlined by Gabriel et al. 2005
| Block 2 | rs7495128 | rs2414687 | Overall Frequency, Percent* |
Frequency Among Cases, Percent (Number of Chromosomes)† |
Frequency Among Controls, Percent (Number of Chromosomes) † |
Chi Square | P-Value‡ |
|---|---|---|---|---|---|---|---|
| h1 | G | G | 0.517 | 0.568 (184) | 0.499 (478) | 4.55 | 0.0329 |
| h2 | G | T | 0.259 | 0.231 ( 75) | 0.269 (258) | 1.77 | 0.1833 |
| h3 | A | G | 0.224 | 0.201 ( 65) | 0.232 (222) | 1.341 | 0.2469 |
Overall frequency of each haplotype in the study population (percentages may not total exactly 100% due to rounding)
Frequency of each haplotype and the estimated number of chromosomes containing each haplotype among cases and controls, respectively. Percentages may not total exactly 100% due to rounding.
P-value testing the significance of each haplotype versus all others.
Table 6.
Replication of Risk Haplotypes Defined in an Earlier Study of Extremely Discordant Sib Pairs5 in the NHS and HPFS Cohorts
| Block 1 | rs730754 | rs8034864 | rs12900948 | Overall Frequency* |
Frequency Among Cases, Percent (Number of Chromosomes) † |
Frequency Among Controls, Percent (Number of Chromosomes) † |
Chi Square |
P-Value‡ |
|---|---|---|---|---|---|---|---|---|
| h1 | A | C | A | 0.42 | 0.274 ( 90) | 0.467 (443) | 37.036 | 1.16E-09 |
| h2 | G | A | G | 0.19 | 0.166 ( 55) | 0.197 (187) | 1.448 | 0.2288 |
| h3 | A | C | G | 0.18 | 0.308 (101) | 0.138 (131) | 47.378 | 5.85E-12 |
| h4 | G | C | G | 0.17 | 0.165 ( 54) | 0.167 (159) | 0.013 | 0.9092 |
| h5 | G | A | A | 0.03 | 0.056 ( 18) | 0.016 ( 15) | 15.077 | 1.00E-04 |
| h6 | G | C | A | 0.02 | 0.030 ( 10) | 0.015 ( 15) | 3.009 | 0.0828 |
| Block 4 | rs17237514 | rs4335725 | Overall Frequency* | Frequency Among Cases, Percent (Number of Chromosomes) † |
Frequency Among Controls, Percent (Number of Chromosomes) † |
Chi Square |
P-Value‡ |
|---|---|---|---|---|---|---|---|
| h1 | A | G | 0.52 | 0.523 (172) | 0.516 (490) | 0.045 | 0.8314 |
| h2 | A | A | 0.31 | 0.309 (102) | 0.315 (300) | 0.038 | 0.8452 |
| h3 | G | G | 0.16 | 0.159 ( 52) | 0.164 (156) | 0.057 | 0.8112 |
Overall frequency of each haplotype in the study population (percentages may not total exactly 100% due to rounding)
Frequency of each haplotype and the estimated number of chromosomes containing each haplotype among cases and controls, respectively. Percentages may not total exactly 100% due to rounding.
P-value testing the significance of each haplotype versus all others.
Discussion
The present prospective study of incident cases of neovascular AMD confirms and extends our recent findings of a significant association between the RORalpha gene and this leading cause of blindness in US adults. We identified a single SNP, rs12900948 that was associated with 3-fold increased incidence of neovascular AMD among carriers of two “G” alleles at this locus. This SNP was part of a haplotype (GCG) in a haplotype block comprised of SNPs rs730754, rs8034864, and rs12900948 that we previously found to be significantly associated with neovascular AMD in a cohort of EDSP, as well as in two haplotypes (GAG and GCG) significantly associated with neovascular AMD in a separate unrelated group of 139 prevalent cases of neovascular AMD and 121 controls from central Greece.5 In the present study, haplotypes ACA, ACG, and GAA were significantly associated with neovascular AMD. We further observed a significant interaction between SNP rs12900948 and the ARMS2/HTRA1 A69S SNP. Based on the present study, individuals with two “G” alleles at rs12900948 as well as two copies of the ARMS2/HTRA1 A69S risk allele have an estimated 40-fold increase in incidence of neovascular AMD.
In exploring whether the effect of the rs12900948 variants is influenced by cigarette smoking or obesity, we found no statistical evidence for departures from multiplicative interaction between this SNP and these modifiable risk factors, but statistical power was low and the P-value for interaction with cigarette smoking was borderline at P=0.08. Estimates of the joint effects of SNP rs12900948 and cigarette smoking showed that individuals with two “G” alleles have a nearly 10-fold increased incidence of neovascular AMD if they also smoke cigarettes, compared to a 4.6-fold increased risk among never smokers.
Data from our controls indicates that this allele is very common in the US population, with 73% of our US-based controls having at least one copy of the AMD-associated “G” allele. The high prevalence of this allele contributes to a strong attributable risk estimate of 45%. The rs12900948 variant and haplotypes associated with neovascular AMD in this and two prior cross-sectional study populations lie within a well-conserved region of the first intron of the RORalpha-001 transcript (ENST00000335670). Based on the available evidence, we think it is most likely that the causal variation in this region (SNPs or insertion or deletion of copy number) has yet to be identified. In this regard, SNP rs12900948 was not individually associated with neovascular AMD in the initial discovery population of EDSP, though it was significant as part of a haplotype. Further sequencing of the exons and the acceptor/donor splice sites adjacent to this region could help resolve this issue. Although there is no apparent functional change, it is possible that the rs12900948 or other undiscovered variants within intron 1 of the RORalpha gene could cause sequence changes, insertions and/or deletions within modifying elements such as silencers and enhancers, or influence the splicing of the transcript, which could contribute to a functional effect.29–32
In the eye, RORalpha is involved in regulating the development of photoreceptors through the coordinated expression of several cone genes,33 and it is expressed in the ganglion cell layer and inner nuclear layer of the adult retina.34, 35 More generally, RORalpha participates in several biological pathways, including oxidative stress, inflammation, lipid metabolism, and angiogenesis, which have been implicated in the development of neovascular AMD. 10–17 A nuclear receptor, RORalpha has been shown to regulate production of cytokines including interleukin-6 (IL-6), interleukin-8 (IL-8), and tumor necrosis factor-alpha (TNF-α), as well as the cellular adhesion molecules vascular cell adhesion molecule (VCAM-1) and intracellular adhesion molecule, (ICAM-1).36 It is also involved in regulation and homeostasis of lipoproteins, such as high density lipoprotein (HDL), serum amyloid A, and apolipoprotein A1,37 and is thought to be a key modulator of fat accumulation.16 In this context, it is interesting to hypothesize based on the present findings that RORalpha influence the development of neovascular AMD through the genes it regulates, or indirectly through the genes that regulate RORalpha.
As a complex disease, it is thought that the risk of AMD is altered through a combination of environmental effects plus effects of variants within several genes that lead to alterations in their interactions with each other, as well as to alterations in their interactions with other genes and/or proteins.38 Because the genes involved in AMD have these pleiotropic effects, however, variants in these genes could also alter the clinical course or survival of individuals who carry certain alleles. Particularly when the alleles are common, such effects could introduce selection bias when prevalent rather than incident cases are studied.39 The present study, using a validated prospective nested case-control methodology therefore adds strength to our initial findings in two clinic-derived populations of prevalent AMD cases.
Although we cannot perform standardized clinical assessments of retinal status among participants of these large and geographically dispersed cohorts, we have demonstrated that our case ascertainment method has high specificity,22, 40 which ensures minimal bias in a prospective study.39 Furthermore, the consistency of our previous findings linking CFH Y402H and ARMS2/HTRA1 A69S with AMD, as well as our prior work on modifiable risk factors for AMD,21, 22, 40, 41 provides further reassurance of the validity of the present findings. The selected nature of these cohorts of health professionals may limit generalizability, but significant results have already been demonstrated in two other study populations, including a group of cases and controls from central Greece.5 Further study of the possibility that variants in RORalpha may influence the development of earlier stages of AMD also deserves to be studied, and these studies are currently underway in our prospective cohorts.
Whereas we observed a statistically significant interaction between rs12900948 and ARMS2/HTRA1 A69S, sparseness of data contributed to imprecision in stratum-specific effect estimates. This was a greater issue in terms of our ability to detect an interaction with cigarette smoking, which is less common in our cohorts of health professionals than in some other studies.42, 43 Cigarette smoking and obesity are known to promote inflammatory activity,44 and previous studies have suggested that in such situations in which underlying levels of inflammatory activity are likely to be elevated the impact of carrying AMD-associated risk alleles is magnified.2 Although not statistically significant, stratum specific estimates showed an elevated incidence of neovascular AMD among homozygous carriers of the “G” allele that were current versus never smokers. This coincides with evidence showing, for example, that homozygous staggerer mice that display decreased and dysfunctional RORA expression are more susceptible to (at least certain types of) inflammation.45
With regard to obesity, we hypothesized that obese individuals who carried the “G” allele might be more likely to develop AMD. In addition to possible shared pathways involving inflammation, our hypothesis was also based on evidence showing specific interactions of obesity and RORalpha mediated through alterations in lipoprotein pathways. For example, homozygous staggerer mice with decreased and dysfunctional RORA expression are resistant to diet-induced obesity.16 Moreover, RORalpha appears to participate in regulating plasma cholesterol levels, and positively regulates apolipoprotein (apo)A-I and apoC-III gene expression, whereas its activity is also regulated by cholesterol.46 A balanced translocation in RORalpha has also been associated with severe obesity in humans.47 In spite of this evidence for biological interplay between RORalpha and obesity, however, estimates for association with AMD were similar for homozygous carriers of the “G” allele whether the individuals were obese or not. Such observations provide support for the argument that it is important to separate the concept of joint biological effects from the issue of statistical testing of interaction terms.39, 48 Further study of interactions in prospective studies of larger sample size, as well as other types of studies to identify joint biological effects, will be necessary to address these complicated issues.
The identification of a number of prevalent AMD-associated polymorphisms has raised the question of the utility of population-based or targeted genetic testing. However, as others have pointed out, predictive models for AMD based on currently known risk factors are still inaccurate.3 This is an expected problem for a complex disease such as AMD in which multiple common genetic and non-genetic factors influence risk. Improvement of predictive models may be accomplished through enhanced measurement of exposures, as well as the identification of additional genetic and non-genetic risk factors. The utility of this approach will of course ultimately depend on the development of effective strategies for preservation of vision among individuals identified as having high risk of AMD.
In summary, common variants and haplotypes within the RORalpha gene appear to increase incidence of neovascular AMD. There is significant evidence of a multiplicative interaction between the RORalpha SNP rs12900948 with the ARMS2/HTRA1 A69S polymorphism in AMD. Cigarette smoking may also confer excess risk among individuals who carry two copies of the “G” allele at rs12900948, though further study of larger groups is needed to refine this estimate. These data identify another major gene associated with risk of neovascular AMD, and add further evidence of the complex interplay among genetic and modifiable risk factors for AMD. Such information could lead to enhanced accuracy of risk prediction for neovascular AMD.
Acknowledgements
This work was supported by NIH grants EY017362, EY013834, EY009611, EY014458, CA87969, CA49449, HL35464, and the Lincy Fund.
References
- 1.Congdon N, O'Colmain B, Klaver CC, et al. Causes and prevalence of visual impairment among adults in the United States. Arch Ophthalmol. 2004 Apr;122(4):477–485. doi: 10.1001/archopht.122.4.477. [DOI] [PubMed] [Google Scholar]
- 2.Schaumberg DA, Hankinson SE, Guo Q, Rimm E, Hunter DJ. A prospective study of 2 major age-related macular degeneration susceptibility alleles and interactions with modifiable risk factors. Arch Ophthalmol. 2007 Jan;125(1):55–62. doi: 10.1001/archopht.125.1.55. [DOI] [PubMed] [Google Scholar]
- 3.Jakobsdottir J, Gorin MB, Conley YP, Ferrell RE, Weeks DE. Interpretation of genetic association studies: markers with replicated highly significant odds ratios may be poor classifiers. PLoS Genet. 2009 Feb;5(2):e1000337. doi: 10.1371/journal.pgen.1000337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeAngelis MM, Lane AM, Shah CP, Ott J, Dryja TP, Miller JW. Extremely discordant sib-pair study design to determine risk factors for neovascular age-related macular degeneration. Arch Ophthalmol. 2004 Apr;122(4):575–580. doi: 10.1001/archopht.122.4.575. [DOI] [PubMed] [Google Scholar]
- 5.Silveira AC, Morrison MA, Ji F, et al. Convergence of linkage, gene expression and association data demonstrates the influence of the RAR-related orphan receptor alpha (RORA) gene on neovascular AMD: A systems biology based approach. Vision Res. 2009 Sep 26; doi: 10.1016/j.visres.2009.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hubbard TJ, Aken BL, Ayling S, et al. Ensembl 2009. Nucleic Acids Res. 2009 Jan;37:D690–D697. doi: 10.1093/nar/gkn828. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anderson DH, Mullins RF, Hageman GS, Johnson LV. A role for local inflammation in the formation of drusen in the aging eye. Am J Ophthalmol. 2002 Sep;134(3):411–431. doi: 10.1016/s0002-9394(02)01624-0. [DOI] [PubMed] [Google Scholar]
- 8.Klein R, Klein BE, Tomany SC, Cruickshanks KJ. The association of cardiovascular disease with the long-term incidence of age-related maculopathy: the Beaver Dam eye study. Ophthalmology. 2003 Apr;110(4):636–643. doi: 10.1016/S0161-6420(02)01448-3. [DOI] [PubMed] [Google Scholar]
- 9.Schaumberg DA, Christen WG, Buring JE, Glynn RJ, Rifai N, Ridker PM. High-sensitivity C-reactive protein, other markers of inflammation, and the incidence of macular degeneration in women. Arch Ophthalmol. 2007 Mar;125(3):300–305. doi: 10.1001/archopht.125.3.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Besnard S, Bakouche J, Lemaigre-Dubreuil Y, Mariani J, Tedgui A, Henrion D. Smooth muscle dysfunction in resistance arteries of the staggerer mouse, a mutant of the nuclear receptor RORalpha. Circ Res. 2002 Apr 19;90(7):820–825. doi: 10.1161/01.res.0000014489.24705.71. [DOI] [PubMed] [Google Scholar]
- 11.Conley YP, Thalamuthu A, Jakobsdottir J, et al. Candidate gene analysis suggests a role for fatty acid biosynthesis and regulation of the complement system in the etiology of age-related maculopathy. Hum Mol Genet. 2005 Jul 15;14(14):1991–2002. doi: 10.1093/hmg/ddi204. [DOI] [PubMed] [Google Scholar]
- 12.Hageman GS, Anderson DH, Johnson LV, et al. A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration. Proc Natl Acad Sci U S A. 2005 May 17;102(20):7227–7232. doi: 10.1073/pnas.0501536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boukhtouche F, Vodjdani G, Jarvis CI, et al. Human retinoic acid receptor-related orphan receptor alpha1 overexpression protects neurones against oxidative stress-induced apoptosis. J Neurochem. 2006 Mar;96(6):1778–1789. doi: 10.1111/j.1471-4159.2006.03708.x. [DOI] [PubMed] [Google Scholar]
- 14.Boukhtouche F, Mariani J, Tedgui A. The "CholesteROR" protective pathway in the vascular system. Arterioscler Thromb Vasc Biol. 2004 Apr;24(4):637–643. doi: 10.1161/01.ATV.0000119355.56036.de. [DOI] [PubMed] [Google Scholar]
- 15.Zhu Y, McAvoy S, Kuhn R, Smith DI. RORA, a large common fragile site gene, is involved in cellular stress response. Oncogene. 2006 May 11;25(20):2901–2908. doi: 10.1038/sj.onc.1209314. [DOI] [PubMed] [Google Scholar]
- 16.Lau P, Fitzsimmons RL, Raichur S, Wang SC, Lechtken A, Muscat GE. The orphan nuclear receptor, RORalpha, regulates gene expression that controls lipid metabolism: staggerer (SG/SG) mice are resistant to diet-induced obesity. J Biol Chem. 2008 Jun 27;283(26):18411–18421. doi: 10.1074/jbc.M710526200. [DOI] [PubMed] [Google Scholar]
- 17.Besnard S, Silvestre JS, Duriez M, et al. Increased ischemia-induced angiogenesis in the staggerer mouse, a mutant of the nuclear receptor Roralpha. Circ Res. 2001 Dec 7;89(12):1209–1215. doi: 10.1161/hh2401.101755. [DOI] [PubMed] [Google Scholar]
- 18.Iyengar SK, Song D, Klein BE, et al. Dissection of genomewide-scan data in extended families reveals a major locus and oligogenic susceptibility for age-related macular degeneration. Am J Hum Genet. 2004 Jan;74(1):20–39. doi: 10.1086/380912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schick JH, Iyengar SK, Klein BE, et al. A whole-genome screen of a quantitative trait of age-related maculopathy in sibships from the Beaver Dam Eye Study. Am J Hum Genet. 2003 Jun;72(6):1412–1424. doi: 10.1086/375500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seddon JM, Willett WC, Speizer FE, Hankinson SE. A prospective study of cigarette smoking and age-related macular degeneration in women. JAMA. 1996;276(14):1141–1146. [PubMed] [Google Scholar]
- 21.Cho E, Seddon JM, Rosner B, Willett WC, Hankinson SE. Prospective study of intake of fruits, vegetables, vitamins, and carotenoids and risk of age-related maculopathy. Arch Ophthalmol. 2004 Jun;122(6):883–892. doi: 10.1001/archopht.122.6.883. [DOI] [PubMed] [Google Scholar]
- 22.Cho E, Hung S, Willett WC, et al. Prospective study of dietary fat and the risk of age-related macular degeneration. Am J Clin Nutr. 2001 Feb;73(2):209–218. doi: 10.1093/ajcn/73.2.209. [DOI] [PubMed] [Google Scholar]
- 23.DeAngelis MM, Ji F, Kim IK, et al. Cigarette smoking, CFH, APOE, ELOVL4, and risk of neovascular age-related macular degeneration. Arch Ophthalmol. 2007 Jan;125(1):49–54. doi: 10.1001/archopht.125.1.49. [DOI] [PubMed] [Google Scholar]
- 24.Akaike H. Information theory and an extension of the maximum likelihood principle; Paper presented at: Second International Symposium on Information Theory; Budapest. 1973. [Google Scholar]
- 25.Walter SD. The estimation and interpretation of attributable risk in health research. Biometrics. 1976 Dec;32(4):829–849. [PubMed] [Google Scholar]
- 26.Bruzzi P, Green SB, Byar DP, Brinton LA, Schairer C. Estimating the population attributable risk for multiple risk factors using case-control data. Am J Epidemiol. 1985 Nov;122(5):904–914. doi: 10.1093/oxfordjournals.aje.a114174. [DOI] [PubMed] [Google Scholar]
- 27.Spiegelman D, Hertzmark E, Wand HC. Point and interval estimates of partial population attributable risks in cohort studies: examples and software. Cancer Causes Control. 2007 Jun;18(5):571–579. doi: 10.1007/s10552-006-0090-y. [DOI] [PubMed] [Google Scholar]
- 28.Gabriel SB, Schaffner SF, Nguyen H, et al. The structure of haplotype blocks in the human genome. Science. 2002 Jun 21;296(5576):2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 29.Margulies EH, Birney E. Approaches to comparative sequence analysis: towards a functional view of vertebrate genomes. Nat Rev Genet. 2008 Apr;9(4):303–313. doi: 10.1038/nrg2185. [DOI] [PubMed] [Google Scholar]
- 30.Maddox DM, Vessey KA, Yarbrough GL, et al. Allelic variance between GRM6 mutants, Grm6nob3 and Grm6nob4 results in differences in retinal ganglion cell visual responses. J Physiol. 2008 Sep 15;586(Pt 18):4409–4424. doi: 10.1113/jphysiol.2008.157289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Armengol L, Rabionet R, Estivill X. The emerging role of structural variations in common disorders: initial findings and discovery challenges. Cytogenet Genome Res. 2008;123(1–4):108–117. doi: 10.1159/000184698. [DOI] [PubMed] [Google Scholar]
- 32.Hastings PJ, Lupski JR, Rosenberg SM, Ira G. Mechanisms of change in gene copy number. Nat Rev Genet. 2009 Aug;10(8):551–564. doi: 10.1038/nrg2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fujieda H, Bremner R, Mears AJ, Sasaki H. Retinoic acid receptor-related orphan receptor alpha regulates a subset of cone genes during mouse retinal development. J Neurochem. 2009 Jan;108(1):91–101. doi: 10.1111/j.1471-4159.2008.05739.x. [DOI] [PubMed] [Google Scholar]
- 34.Steinmayr M, Andre E, Conquet F, et al. staggerer phenotype in retinoid-related orphan receptor alpha-deficient mice. Proc Natl Acad Sci U S A. 1998 Mar 31;95(7):3960–3965. doi: 10.1073/pnas.95.7.3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller DM, Espinosa-Heidmann DG, Legra J, et al. The association of prior cytomegalovirus infection with neovascular age-related macular degeneration. Am J Ophthalmol. 2004 Sep;138(3):323–328. doi: 10.1016/j.ajo.2004.03.018. [DOI] [PubMed] [Google Scholar]
- 36.Migita H, Satozawa N, Lin JH, Morser J, Kawai K. RORalpha1 and RORalpha4 suppress TNF-alpha-induced VCAM-1 and ICAM-1 expression in human endothelial cells. FEBS Lett. 2004 Jan 16;557(1–3):269–274. doi: 10.1016/s0014-5793(03)01502-3. [DOI] [PubMed] [Google Scholar]
- 37.Voyiaziakis E, Goldberg IJ, Plump AS, Rubin EM, Breslow JL, Huang LS. ApoA-I deficiency causes both hypertriglyceridemia and increased atherosclerosis in human apoB transgenic mice. J Lipid Res. 1998 Feb;39(2):313–321. [PubMed] [Google Scholar]
- 38.Chen Y, Zhu J, Lum PY, et al. Variations in DNA elucidate molecular networks that cause disease. Nature. 2008 Mar 27;452(7186):429–435. doi: 10.1038/nature06757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rothman KJ, Greenland S. Modern epidemiology. Second ed. Philadelphia: Lipincott-Raven; 1998. pp. 133–134. [Google Scholar]
- 40.Seddon JM, Rosner B, Sperduto RD, et al. Dietary fat and risk for advanced age-related macular degeneration. Arch Ophthalmol. 2001 Aug;119(8):1191–1199. doi: 10.1001/archopht.119.8.1191. [DOI] [PubMed] [Google Scholar]
- 41.Cho E, Hankinson SE, Willett WC, et al. Prospective study of alcohol consumption and the risk of age-related macular degeneration. Arch Ophthalmol. 2000 May;118(5):681–688. doi: 10.1001/archopht.118.5.681. [DOI] [PubMed] [Google Scholar]
- 42.Rivera A, Fisher SA, Fritsche LG, et al. Hypothetical LOC387715 is a second major susceptibility gene for age-related macular degeneration, contributing independently of complement factor H to disease risk. Hum Mol Genet. 2005 Nov 1;14(21):3227–3236. doi: 10.1093/hmg/ddi353. [DOI] [PubMed] [Google Scholar]
- 43.Schmidt S, Hauser MA, Scott WK, et al. Cigarette smoking strongly modifies the association of LOC387715 and age-related macular degeneration. Am J Hum Genet. 2006 May;78(5):852–864. doi: 10.1086/503822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garcia-Lorda P, Bullo M, Balanza R, Salas-Salvado J. C-reactive protein, adiposity and cardiovascular risk factors in a Mediterranean population. Int J Obes (Lond) 2005 Nov 29; doi: 10.1038/sj.ijo.0803182. [DOI] [PubMed] [Google Scholar]
- 45.Stapleton CM, Jaradat M, Dixon D, et al. Enhanced susceptibility of staggerer (RORalphasg/sg) mice to lipopolysaccharide-induced lung inflammation. Am J Physiol Lung Cell Mol Physiol. 2005 Jul;289(1):L144–L152. doi: 10.1152/ajplung.00348.2004. [DOI] [PubMed] [Google Scholar]
- 46.Jetten AM. Recent advances in the mechanisms of action and physiological functions of the retinoid-related orphan receptors (RORs) Curr Drug Targets Inflamm Allergy. 2004 Dec;3(4):395–412. doi: 10.2174/1568010042634497. [DOI] [PubMed] [Google Scholar]
- 47.Klar J, Asling B, Carlsson B, et al. RAR-related orphan receptor A isoform 1 (RORa1) is disrupted by a balanced translocation t(4;15)(q22.3;q21.3) associated with severe obesity. Eur J Hum Genet. 2005 Aug;13(8):928–934. doi: 10.1038/sj.ejhg.5201433. [DOI] [PubMed] [Google Scholar]
- 48.Andersson T, Alfredsson L, Kallberg H, Zdravkovic S, Ahlbom A. Calculating measures of biological interaction. Eur J Epidemiol. 2005;20(7):575–579. doi: 10.1007/s10654-005-7835-x. [DOI] [PubMed] [Google Scholar]