Summary
Bacterial chemotaxis is mediated by signaling complexes of chemoreceptors, histidine kinase CheA and coupling protein CheW. Interactions in complexes profoundly affect the kinase. We investigated effects of these interactions on chemoreceptors by comparing receptors alone and in complexes. Assays of initial rates of methylation indicated that signaling complexes shifted receptor conformation toward the methylation-on, higher ligand affinity, kinase-off state, tuning receptors for greater sensitivity. In contrast, transmembrane and conformational signaling within chemoreceptors was essentially unaltered, consistent with other evidence identifying receptor dimers as the fundamental units of such signaling. In signaling complexes, coupling of ligand binding to kinase activity is cooperative and the dynamic range of kinase control expanded >100-fold by receptor adaptational modification. We observed no cooperativity in influence of ligand on receptor conformation, only on kinase activity. However, receptor modification generated increased dynamic range in a stepwise fashion, partly in coupling ligand to receptor conformation and partly in coupling receptor conformation to kinase activity. Thus, receptors and kinase were not equivalently affected by interactions in signaling complexes or by ligand binding and adaptational modification, indicating asymmetrical coupling between them. This has implications for mechanisms of precise adaptation. Coupling might vary, providing a previously unappreciated locus for sensory control.
Keywords: Bacterial Chemotaxis, Transmembrane Signaling, Sensory Adaptation, Conformational Change, Protein Methylation
Introduction
Bacterial chemotaxis is mediated by signaling complexes containing chemoreceptors, histidine kinase CheA and the coupling protein CheW (Hazelbauer & Lai, 2010, Hazelbauer et al., 2008). Interactions in these complexes profoundly affect the kinase, substantially enhancing its activity and placing that enhanced activity under the control of chemoreceptors. Do interactions in signaling complexes affect the functional activities of chemoreceptors? Are receptors and kinase in signaling complexes sufficiently coupled that they are comparably affected by ligand occupancy and adaptational modification? We addressed these questions by measuring receptor activities in vitro in conditions optimized for kinase activation and thus formation of functional signaling complexes, and comparing those values to receptor activities in the same conditions but without the CheA and CheW binding partners. To provide context for our studies, we provide a brief summary of salient features of chemoreceptors and signaling complexes.
The fundamental structural unit of a chemoreceptor is a homodimer (Milburn et al., 1991, Kim et al., 1999, Park et al., 2006, Pollard et al., 2009). Dimers can interact to form trimers of dimers (Kim et al., 1999, Studdert & Parkinson, 2004) and this organization is common and perhaps universal across bacterial diversity (Briegel et al., 2009). Chemoreceptors form noncovalent signaling complexes with the chemotaxis histidine kinase CheA and the coupling protein CheW (Fig. 1A). In signaling complexes the inherently low activity of CheA is enhanced ~100-fold and placed under the control of chemoreceptor ligand occupancy and adaptational modification. An increase in occupancy of an attractant molecule at a specific binding site at one end of the receptor dramatically reduces the activity of the kinase associated with the other end of the elongated receptors, a process that is both transmembrane signaling and a shift in the conformational equilibrium of the receptor and the signaling complex. Ligand-induced reduction in kinase activity is transient because the feedback of sensory adaptation restores the kinase to its initial level of activity. Adaptation is mediated by covalent modification of the receptor cytoplasmic domain, formation of methyl esters at specific glutamyl residues, catalyzed by the chemotaxis-specific methyltransferase CheR, and demethylation of those methyl esters, catalyzed by a chemotaxis-specific methylesterase CheB. The conformational shift induced in receptors by ligand occupancy results in an increased propensity for methylation, conveniently detected in vitro as an increase in initial rate of the modification. Methylation counteracts the effects of attractant binding and thus restores the null signaling state of the receptor and the signaling complex by generating a transmembrane signal that opposes the signal generated by attractant occupancy. This shifts the conformational equilibrium of the receptor and the signaling complex in the opposite direction induced by attractant binding (Lai et al., 2006). In E. coli, chemoreceptors contain 4–6 methyl-accepting glutamates. Two of these residues are initially glutamines, subsequently deamidated by CheB. Glutamines at methyl-accepting positions are in essence functionally equivalent to methylesters (Lai et al., 2006). Fig. 1A shows the sensory components and interactions relevant to this study.
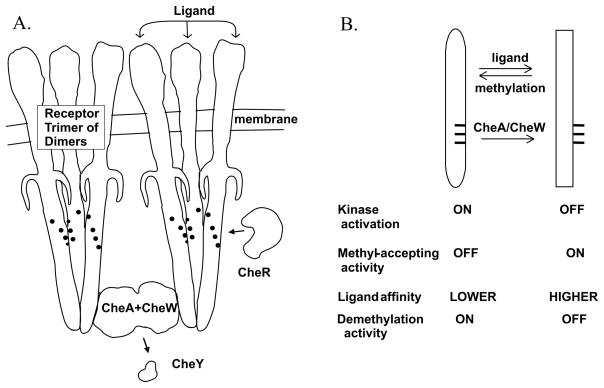
Fig. 1. Chemoreceptors and signaling complexes.
A. Core unit of chemotaxis signaling complexes. The cartoon shows two trimers of chemoreceptor dimers in complex with CheA and CheW in an organization consistent with recent models (Khursigara et al., 2008, Bhatnagar et al., 2010, Erbse & Falke, 2009). Also shown are two soluble chemotaxis proteins relevant to this study, methyltransferase CheR and response regulator CheY, and the positions of methyl-accepting sites (black dots on receptors). B. Diagram of the chemoreceptor conformational equilibrium. The diagram lists relevant features of the two conformations and the parameters that influence the equilibrium between them. The indicated influence of signaling complexes (CheA/CheW) on receptor conformation is documented in the current study.
Many observations about the chemotactic sensory system of E. coli can be explained by a simple model in which attractant occupancy and adaptational modification alter activities of chemoreceptors and signaling complexes by shifting in opposite directions the equilibrium between two conformations (Fig. 1B). The “kinase-on”, “methylation-off” conformation, favored by methylation or amidation and disfavored by attractant occupancy, is thought to activate the histidine kinase, be a substrate for demethylation but not for methylation, and have lowered affinity for attractant ligands. The “kinase-off”, “methylation-on” conformation, favored by attractant occupancy and disfavored by methylation or amidation, has the inverse pattern: it does not activate kinase or serve as a substrate for demethylation but serves as a substrate for methylation and has higher affinity for attractant.
Investigation of the influence on chemoreceptors of interactions in signaling complexes required an assay that did not depend on interactions with CheA and CheW. We recently showed that measurements of initial rates of receptor methylation were sufficiently sensitive and reproducible to determine operational affinities for ligand as well as the relative coupling of ligand occupancy and adaptational modification to receptor conformation and signaling within the receptor (Amin & Hazelbauer, 2010). These initial rates can be determined in the absence of CheA and CheW, and reflect the proportion of the receptor population in the “methylation-competent, kinase-off” state versus the “methylation-incompetent, kinase-on” state (Amin & Hazelbauer, 2010). In addition, the assays provide direct measures of the effectiveness of receptors as substrates for modification. Thus we used measurements of initial rates of methylation to investigate the effects on chemoreceptors of interactions with the components of signaling complexes.
Results
Assessing effects of interactions in signaling complexes on chemoreceptors
Initial rates of methylation were determined for chemoreceptor Tar in the absence and presence of CheA plus CheW (CheA/CheW), as well as CheY, in conditions that produced maximum kinase activation and thus presumably maximal formation of functional signaling complexes. We measured these rates as a function of the attractant ligand aspartate over a range from 0.01 to 1000 μM and as a function of adaptational modification, testing representative forms of Tar with 0 to 3 of its four methyl-accepting sites modified by introduction of a glutamine in place of glutamates, a substitution that mimics the functional effects of a glutamyl methylester (Lai et al., 2006). Because our aim was to investigate effects on Tar of interactions with the components of signaling complexes by comparing receptors in the presence and absence of CheA/CheW, it was not necessary to assay all possible combinations of one, two or three modifications among the four sites. Instead we tested representative combinations: for the two-modification receptor we used Tar(QEQE), the product of the wild-type tar gene and for the one-and three-modification receptors we used Tar(QEEE) and Tar(QEQQ), respectively, each related to the wild-type gene product by a single amino acid substitution. Of course Tar with four modifications could not be tested using rates of methylation since it does not have an unmodified methyl-accepting site.
Effects of signaling complexes on receptor conformation and signaling
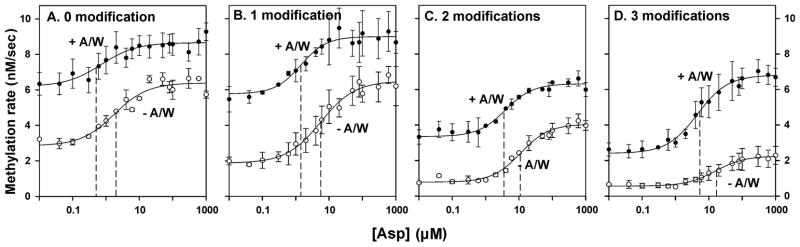
In Fig. 2, mean values for initial rates of Tar methylation are plotted as a function of ligand concentration for receptors in the absence and presence of CheA/CheW. Separate panels show the results for Tar carrying 0, 1, 2 or 3 adaptational modifications. We fit each data set for the effect of ligand concentration on methylation rate to a stimulus-response relationship (see Experimental procedures) from which were derived values for vu, the unstimulated initial rate of methylation; vs, the rate at maximal stimulation by aspartate; Asp½, the ligand concentration at which the initial rate is half-way between vu and vs; vs−vu, the increase in rate generated by saturation with aspartate; and n, the Hill coefficient (Table 1). Ligand occupancy in the periplasmic domain of Tar should shift receptor conformation in a concentration-dependent manner toward the methylation-on, kinase-off conformation. We detected that conformational shift as an increase in initial rate of methylation of the cytoplasmic domain as a function of ligand concentration (Fig. 2). This coupling of ligand binding to conformational change on the opposite side of the membrane, i.e. transmembrane signaling, was observed for Tar at each extent of adaptational modification and in the absence and presence of CheA/CheW. Hill coefficients derived from the fits of the data indicated that there was no significant cooperativity for receptors in any condition (Table 1). Increasing adaptational modification of Tar should shift its conformational equilibrium toward the methylation-off, kinase-on conformation. We detected that conformational shift as a progressive displacement of the dose-response curves to lower rates of methylation as the number of receptor modifications increased, both in the absence and presence of CheA/CheW (Figs. 2A–D). Plotting vu, the unstimulated rates of methylation, as a function of receptor modification revealed that the effect of modification on receptor conformation was only slightly, if at all, enhanced by signaling complexes, as illustrated by the just greater slope for the +A/W plots (Fig. 3A). We conclude transmembrane signaling and conformational coupling within Tar is not crucially dependent on or fundamentally altered by interactions with CheA and/or CheW.
Fig. 2. Initial rates of methylation as a function of adaptational modification and ligand concentration.
Rates were determined for Tar embedded in native membrane vesicles, carrying 0 (EEEE, 0 modification), 1 (QEEE, 1 modification), 2 (QEQE, 2 modifications) or 3 (QEQQ, 3 modifications) glutamines at the indicated aspartate concentrations and in the absence of CheA, CheW and CheY (−A/W, open circles) or in their presence in conditions that produced maximum kinase activation and thus the maximal level of functional signaling complexes (+A/W, solid circles). The plots show means (circles) and standard deviations (error bars) for ≥3 independent experiments. The curves are fits of the data to a simple dose-response relationship (Table 1, legend). The dashed lines show the respective values for [Asp]½.
TABLE 1.
Parameters from fits of initial rates of methylation as a function of aspartate concentration a
| + CheA/CheW | Modifications | [Asp]½ μM | vu nM/s | vs nM/s | vs−vu nM/s | Hill Coefficient |
|---|---|---|---|---|---|---|
| No | 0 (EEEE) | 2.0 ± 0.3 | 3.0 ± 0.2 | 6.4 ± 0.1 | 3.4 ± 0.2 | 0.9 ± 0.1 |
| 1 (QEEE) | 5.5 ± 1.5 | 1.9 ± 0.1 | 6.3 ± 0.8 | 4.4 ± 0.8 | 0.9 ± 0.2 | |
| 2 (QEQE) | 11.2 ± 1.1 | 0.8 ± 0.1 | 4.0 ± 0.4 | 3.2 ± 0.4 | 1.0 ± 0.1 | |
| 3 (QEQQ) | 17.0 ± 1.2 | 0.6 ± 0.1 | 2.3 ± 0.6 | 1.7 ± 0.6 | 1.0 ± 0.1 | |
| Yes | 0 (EEEE) | 0.5 ± 0.2 | 6.2 ± 0.6 | 8.8 ± 0.7 | 2.6 ± 0.9 | 1.0 ± 0.2 |
| 1 (QEEE) | 1.4 ± 0.1 | 5.8 ± 0.3 | 9.0 ± 1.0 | 5.2 ± 1.0 | 1.3 ± 0.2 | |
| 2 (QEQE) | 3.6 ± 0.9 | 3.6 ± 0.5 | 6.2 ± 0.1 | 3.6 ± 0.5 | 1.1 ± 0.1 | |
| 3 (QEQQ) | 5.3 ± 2.0 | 2.5 ± 0.5 | 6.9 ± 0.5 | 4.4 ± 0.7 | 1.0 ± 0.4 |
Data shown in Fig 2 were fit to the relationship v = vu + (vs [Asp] n)/([Asp]½ n + [Asp]n) where v is the initial rate of methylation at a given aspartate concentration, vu is the rate in the absence of aspartate, vs is the rate at saturating aspartate, [Asp]½ is the aspartate concentration at which stimulation of the initial rate is half maximum and n is the Hill coefficient. The magnitude of the rate increase generated by saturation with aspartate is vs− vu. The table shows mean values and standard deviations for these parameters derived from fits for at least three independent experiments determining kinase activity for Tar with 0 to 4 adaptational modifications in the presence of CheA, CheW and CheY at concentrations that produced maximum kinase activation and thus the highest level of functional signaling complexes.
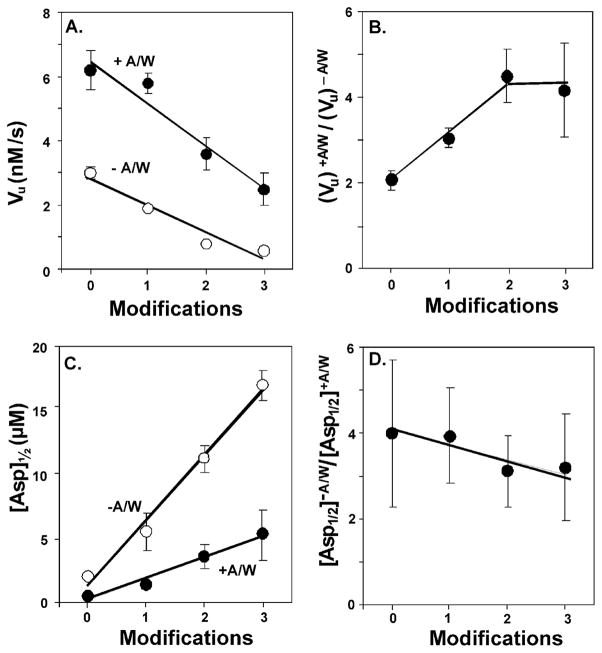
Fig. 3. Effects of adaptational modification on parameters derived from dose-response curves.
A. Means (circles) and standard deviations (error bars) for vu, initial velocity of methylation, derived from the fits of the data in Fig. 2 (see Table 1) plotted as a function of adaptational modification for Tar in the absence (−A/W, open circles) or presence (+A/W, solid circles) of signaling complexes. Lines are least square fits of the data. B. Ratios of vu in the presence (vu)+A/W versus the absence (vu)−A/W of the other components of signaling complexes. Error bars represent uncertainties estimated by error propagation. The line is presented to aid the eye. C. Means (circles) and standard deviations (error bars) for [Asp]½, derived from the fits of the data in Fig. 2 (see Table 1) plotted as a function of adaptational modification for Tar in the absence (−A/W, open circles) or presence (+A/W, solid circles) of signaling complexes. Lines are least square fits of the data. D. Ratios of [Asp]½ in the presence ([Asp]½ +A/W) versus the absence ([ASP]½−A/W) of the other components of signaling complexes. Error bars represent uncertainties estimated by error propagation. The line is a least square fit of the data.
However, Tar was clearly affected by interactions in signaling complexes. The presence of CheA/CheW shifted stimulus-response curves for Tar to higher initial rates, a effect observed at each level of adaptational modification (Fig. 2 and 3A; Table 1 The magnitudes of these shifts (Fig. 3B) were comparable to those generated by saturating Tar with ligand (Fig. 2; Table 1).
Effects of signaling complexes on ligand binding
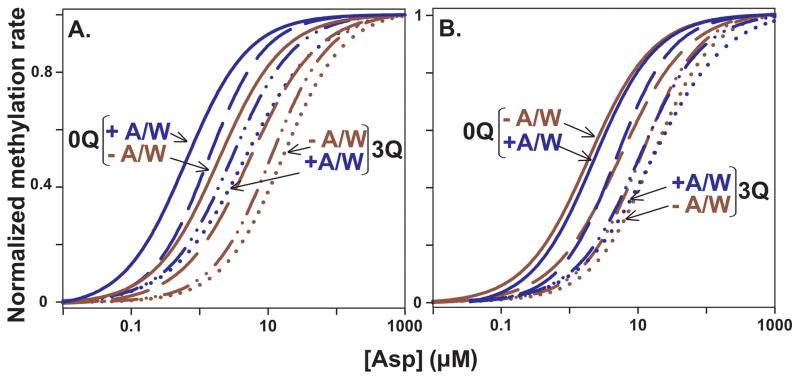
The two alternative receptor conformations are thought to differ not only in propensity for methylation but also in ligand affinity, with the kinase-on, methylation-off conformation having lower affinity. As the receptor population is shifted toward that lower affinity conformation by increased adaptational modification, the position of the respective stimulus-response curves should shift to higher aspartate concentrations and the concentration for half-maximal stimulation, [Asp]½ would increase. Our data showed that each increase in the extent of Tar adaptational modification shifted the stimulus-response curve to a higher ligand concentration (see Fig. 4A in which the curves shown in Fig 2 are normalized to the respective unstimulated and maximally stimulated rates). These effects of adaptational modification occurred whether or not CheA/CheW was present. However, interactions with signaling complex components shifted each stimulus-response curve and thus each [Asp]½ value to lower ligand concentrations (Figs. 4A and 3C). Comparing the set of curves in the absence and presence of CheA/CheW by putting the two sets in register for the respective [Asp]½ values for Tar with two modifications (Fig. 4B) or by plotting the ratio of [Asp]½ values in the absence versus the presence of CheA plus CheW (Fig. 3D) revealed that receptor interactions with CheA/CheW had little effect on the extent to which adaptational modification shifted functional ligand affinity.
Fig. 4. Normalized fits of initial rates of methylation data as a function of adaptational modification and ligand concentration.
A. The dose-response curves fitted to the data in Fig. 2 were normalized to their respective vu and vs values. Curves for 0 (solid), 1 (dashed), 2 (dot-dash) and 3 (dotted) receptor modifications (0Q, 1Q, 2Q and 3Q) are shown for Tar in the absence (−A/W, red) and presence (+A/W, blue) of CheA/CheW plus CheY to form signaling complexes. Only the 0Q and 3Q curves are labeled. B. The two sets of curves shown in Fig. 4A (−A/W, red and +A/W, blue) were aligned using the respective [Asp]½ values for Tar(2Q). Labels are as for Fig. 4A.
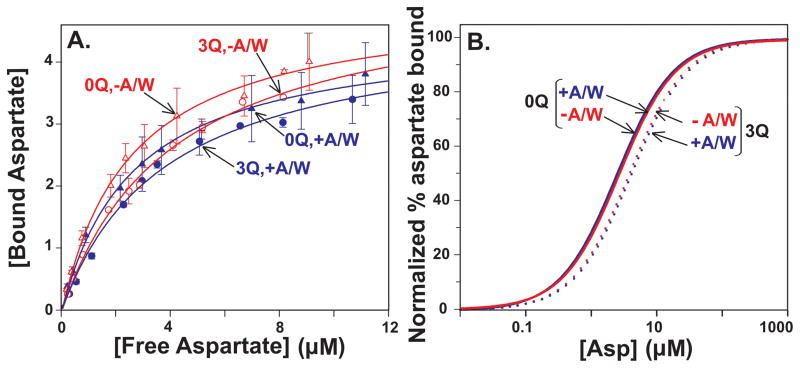
As a complement to determination of functional aspartate affinity using initial rates of methylation, we measured direct binding of this attractant ligand to Tar, in the absence and presence of CheA/CheW as well as CheY. Fig. 5A shows the primary data (mean values with standard deviations) for aspartate bound by Tar containing 0 or 3 adaptational modifications as a function of aspartate concentration, in the absence or presence of CheA/CheW plus fits of each data set to a simple binding isotherm (see Experimental procedures). Fig. 5B displays the fits in the double-normalized format used for Fig. 4. Increasing the extent of Tar adaptational modification resulted in a very slight decrease in the affinity of the Tar binding site for aspartate, seen as a shift of the binding curve to slightly higher aspartate concentrations. This modest effect is consistent with previous observations of modest changes in ligand affinity generated by receptor modification (Dunten & Koshland, 1991, Borkovich et al., 1992, Lin et al., 1994, Levit & Stock, 2002). The presence of CheA/CheW did not significantly affect aspartate binding to Tar nor did formation of signaling complexes affect the slight reduction in affinity generated by increased adaptational modification. The minimal effects of CheA/CheW on aspartate binding by Tar are consistent with the modest affect of signaling complex formation on serine binding by Tsr (Levit & Stock, 2002).
Fig. 5. Aspartate binding by Tar as a function of adaptational modification and ligand concentration.
A. Binding of radiolabeled aspartate to Tar embedded in native membrane vesicles and carrying 0 (EEEE, 0Q, triangles) or 3 (QEQQ, 3Q, circles) glutamines in the absence (−A/W, red, open symbols) or presence (+A/W, blue, solid symbols) of the other components of core signaling complexes plus CheY in conditions that produced maximum kinase activation and thus the highest level of functional signaling complexes. The plots show means (symbols) and standard deviations (error bars) for ≥3 independent experiments. The curves are fits of the data to a simple dose-response relationship (Experimental Procedures). Respective [Asp]½ values derived from these fits were −A/W: 0Q = 2.8 ± 0.1, 3 Q = 3.5 ± 0.9; +A/W: 0Q = 2.7 ± 0.5, 3 Q = 3.9 ± 0.3. B. Curves shown in Fig. 5A normalized to their respective maximal binding values.
Kinase activation and control
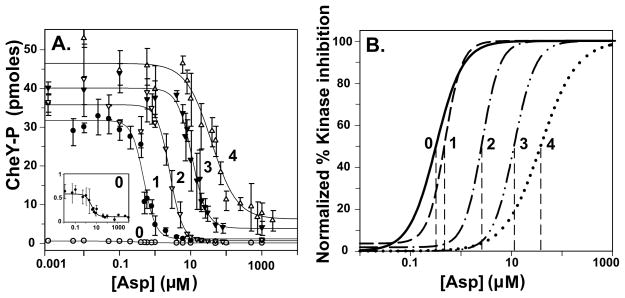
Ligand occupancy and adaptational modification of chemoreceptors in signaling complexes have been observed to generate large changes in kinase activity (Borkovich et al., 1989, Borkovich et al., 1992, Li & Weis, 2000, Bornhorst & Falke, 2000, Bornhorst & Falke, 2001, Levit & Stock, 2002), changes of much greater magnitude than those we observed for effects of ligand occupancy or modification on initial rates of Tar methylation in the presence of CheA/CheW (and also in their absence). We tested signaling complexes formed using our experimental conditions and materials for kinase activation and for control of that activation by aspartate and adaptational modification. Fig. 6A shows mean values and standard deviations for kinase activity as a function of aspartate concentration and adaptational modification with fits of those data to a relationship related to the one used to fit the data for methylation rates (Experimental procedures). Besides Tar with 0 to 3 modifications we also tested Tar(4Q), a form of the receptor that could not be tested for initial rate of methylation. To facilitate comparisons of effects on different receptor properties we plotted the fitted curves using the double-normalized format (Fig. 6B). Activation of kinase and control of kinase activity by occupancy or adaptational modification for our preparations of Tar from Escherichia coli were comparable to activation and control observed for Tar from Salmonella (Bornhorst & Falke, 2000, Bornhorst & Falke, 2001). Specifically, saturation of Tar with aspartate decreased kinase activity ~100-fold, the stimulus-response relationships exhibited cooperativity (Hill coefficients between ~1.5 and 2.5) and increasing receptor modification from none to maximum shifted functional affinity for aspartate over 100-fold (Fig. 5B and Table 2). As considered in the Discussion, ligand binding and adaptational modification of receptors in signaling complexes had substantially greater effects on kinase activity (Fig. 6) than on receptor conformation as assayed by initial rates of methylation (Figs 2–5).
Fig. 6. Initial rates of phosphorylation as a function of adaptational modification and ligand concentration.
Rates were determined for the forms of Tar used for the experiments shown in Fig. 2 plus Tar4Q (QQQQ) at the indicated aspartate concentrations in the presence of CheA/CheW in conditions that produced maximum kinase activation and thus the highest level of functional signaling complexes. The plots show means (circles) and standard deviations (error bars) for ≥3 independent experiments. The curves are fits of the data to a simple dose-response relationship (Table 2, legend). B. Curves in Fig. 6A normalized to their respective maximal and minimal values. The dashed lines show the respective values for [Asp]½.
Table 2.
Parameters from fits of kinase activity as a function of aspartate concentration a
| Modifications | [Asp]½ μM | Fold Kinase Activation | Hill Coefficient |
|---|---|---|---|
| 0 (EEEE) | 0.3 ± 0.2 | 0.7 ± 0.5 | 1.5 ± 0.4 |
| 1 (QEEE) | 0.5 ± 0.1 | 88 ± 8.0 | 2.5 ± 0.3 |
| 2 (QEQE) | 2.7 ± 0.5 | 102 ± 9.6 | 2.5 ± 0.1 |
| 3 (QEQQ) | 11.1 ± 1.7 | 113 ± 4.4 | 2.2 ± 0.1 |
| 4 (QQQQ) | 45.9 ± 2.5 | 133 ± 2.4 | 1.4 ± 0.4 |
Data shown in Fig 6A were fit to the relationship p = pu + (ps [Asp]n)/([Asp]½ n + [Asp]n) where p is the rate of phosphorylation at a given aspartate concentration, pu is the rate in the absence of aspartate, ps is the rate at saturating aspartate, [Asp]½ is the aspartate concentration at which stimulation of the initial rate is half maximum and n is the Hill coefficient. Fold Kinase Activation is (pu − pA)/pA where pA is the rate of phosphorylation of CheA in the absence of chemoreceptors and CheW. The table shows mean values and standard deviations for these parameters derived from fits for at least three independent experiments determining kinase activity for Tar with 0 to 4 adaptational modifications in the presence of CheA and CheW in conditions that form signaling complexes.
Discussion
Effects of signaling complexes on chemoreceptors
The presence of CheA/CheW in conditions that generated maximal kinase activation and thus by definition maximal formation of operational signaling complexes increased initial rates of Tar methylation and shifted dose-response curves to lower ligand concentration (Figs 2 and 4; Table 1). Thus interactions with the components of the signaling complex, presumably in such complexes, altered chemoreceptors. Our data confirm and substantially extend observations of enhanced methylation of chemoreceptor Tsr in signaling complexes (Levit & Stock, 2002, Chalah & Weis, 2005) or in the presence of CheW alone (Chalah & Weis, 2005). Interactions in signaling complexes could increase initial rates of methylation by 1) a conformational shift of receptor dimers toward the “methylation-on” state or 2) altered positioning between receptor dimers in timers, providing increased accessibility of modification sites to the methyltransferase and thus increased initial rates of methylation. Altered positioning was suggested as an explanation for the enhancement of receptor methylation by high concentrations of CheW (Chalah & Weis, 2005). We tested the two alternatives by assaying initial rates of demethylation, reasoning that a shift in receptor conformation to the methylation-on, higher ligand affinity, kinase-off conformation would reduce demethylation rates since that conformation is also demethylation-off, whereas increased accessibility of modification sites should increase rates of both methylation and demethylation. We found that formation of signaling complexes reduced initial rates of demethylation to approximately 50% of initial rates in the absence of CheA/CheW (see Experimental procedures for details). This supported the conformational shift hypothesis but not the altered positioning/increased accessibility explanation. In addition presence of CheA/CheW increased functional affinity for ligand (Fig. 4A), an effect not predicted by the altered positioning, but inherent in the conformational shift postulate. Taken together, these observations indicate that interactions in signaling complexes shift receptor conformation in the same direction as ligand binding, toward the methylation-on, higher ligand affinity, demethylation-off, kinase-off conformation (Fig. 1B). This shift reduces slightly the large magnitude of kinase activation created by formation of signaling complexes, but this modest reduction could be functionally appropriate because of the accompanying increase in ligand affinity which may tune receptors for greater sensitivity. The extent of this tuning could be influenced by other sensory inputs and components, providing a previously unappreciated locus for sensory control.
The clearly detectable effects of CheA/CheW on Tar indicated that a significant proportion of receptor dimers were functionally parts of signaling complexes. Saturation with aspartate, which should affect all receptors, increased initial rates of methylation 1.5 to 5 times, depending on the extent of receptor adaptational modification (Fig. 2; Table 1). CheA/CheW increased rates 2 to 4.5 times (Figs. 2 and 3B; Table 1). The comparable magnitudes imply that in our experimental conditions most receptors were involved in complexes, an implication consistent with stoichiometries of binding (see Experimental procedures). This makes it unlikely that the observed differences between coupling of ligand binding and modification to receptor conformation versus kinase activity (see below) reflected nothing more than such a small proportion of receptors actually in signaling complexes that their influence was undetected.
The presence of CheA/CheW did not affect all receptor activities. We did not observe significant effects on the magnitude of the conformational shifts generated by the sensory inputs of ligand occupancy (Table 1, Fig. 2) or on the shifts in operational affinity generated by adaptational modification (Table 1, Fig. 4B). Thus transmembrane and conformational signaling within chemoreceptors neither required nor were significantly altered by interactions with other components of the signaling complex, just as such signaling is essentially independent of interactions among receptor dimers (Amin & Hazelbauer, 2010). These observations reinforce the notion that the chemoreceptor homodimer is the fundamental unit of transmembrane and intra-receptor conformational signaling.
Coupling among receptors
For Tar alone or in signaling complexes, the relationship of ligand concentration to the conformation of the Tar dimer, as measured by initial rate of methylation, was not cooperative (Table 1) and the extent to which adaptational modification increased dynamic range was significantly less than observed for kinase activity (Fig. 7A). The dose-response data for initial rate of receptor methylation as a function of aspartate concentration could be fit, at every extent of adaptational modification, by modeling receptors as independent, two-state elements. A similar pattern, relatively narrow dynamic range as a function of modification, no cooperativity and action as independent, two-state elements, was observed for dose-response curves relating ligand concentration to the separation between the receptor homodimers within trimers (Vaknin & Berg, 2007). The authors of that study suggested that changes in dimer separation would likely be generated by conformational changes within the individual homodimers. By assessing dimer conformation using initial rates of methylation, we were able to detect these changes. The striking similarity of effects of sensory inputs on dimer conformation and dimer separation imply a direct link between conformational changes in the homodimer and relative movements of dimers within a trimer. The data for dimer movements within a trimer were collected in the absence of CheA/CheW, a situation that left open the possibility that interactions in signaling complexes might introduce cooperativity and the increased dynamic range created by adaptational modification (Vaknin & Berg, 2007). However, formation of signaling complexes did not induce cooperativity or increase dynamic range in the relationship between ligand concentration and dimer conformation, implying they would not do so for the movement of dimers within a trimer.
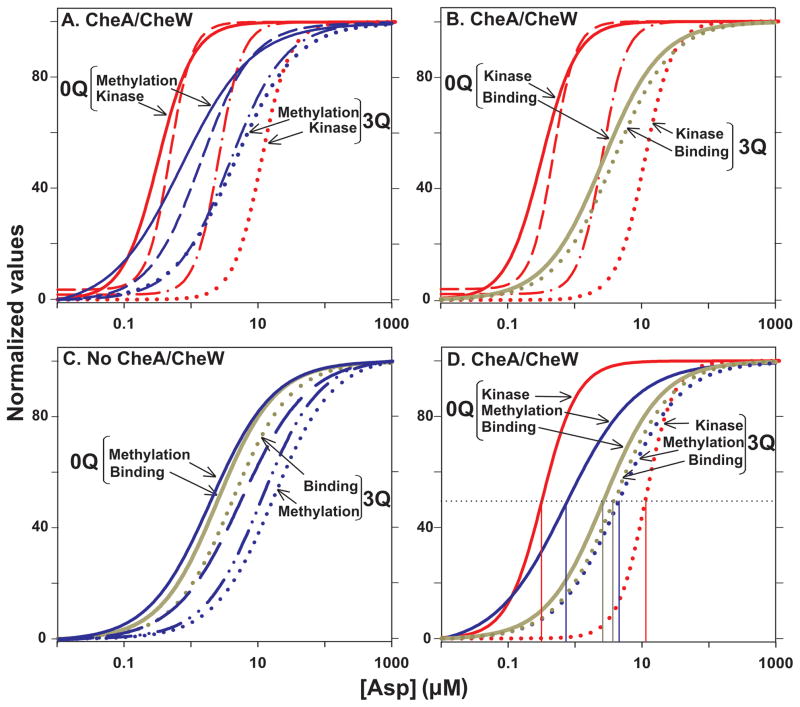
Fig. 7. Comparison of dose-response curves.
The normalized curves for initial rate of methylation from Fig 4A (Methylation), aspartate binding from Fig. 5B (Binding) and kinase activity from Fig. 6B (Kinase) as a function of aspartate concentration are compared in selected groupings. A. Comparison of initial rates of Tar methylation and kinase activity in conditions generating maximal formation of signaling complexes. Curves are shown for Tar with 0 to 3 modifications but only the extremes are labeled. B. Comparison of aspartate binding and kinase activity in conditions generating maximal formation of signaling complexes. Only the extremes of adaptational modification, no glutamines (0Q) and three glutamines (3Q) are labeled but curves are also shown for kinase activity with Tar carrying 1 and 2 modifications. C. Comparison of aspartate binding and initial rates of methylation for Tar in the absence of signaling complexes. As for Fig. 7A, only the extremes of adaptational modification, no glutamines (0Q) and three glutamines (3Q) are labeled but curves are also shown for initial rates of methylation activity with Tar carrying 1 and 2 modifications. D. Comparison of aspartate binding, initial rates of Tar methylation and kinase activity as a function of aspartate concentration in conditions generating maximal formation of signaling complexes. Curves are shown for Tar with 0 and 3 modifications, labeled as for the other panels. The thin vertical lines show the respective values for [Asp]½.
Coupling in signaling complexes
In Tar-containing signaling complexes we observed distinctly non-linear relationships between ligand binding and kinase inhibition, relationships that varied as a function of receptor adaptational modification (Fig. 7B), paralleling previous observations for chemoreceptor Tsr (Levit & Stock, 2002). For no or one adaptational modifications per Tar subunit, at low ligand concentrations relative to [Asp]½ small increases in ligand binding corresponded to large increases in kinase inhibition, i.e. the gain in the system was high and maximal kinase inhibition was achieved at less than 50% occupancy of the ligand-binding site. As the extent of modification was increased to two modifications, gain was negative below ~50% occupancy and positive until maximal inhibition was reached at ~80% occupancy (Fig. 7B). At three modifications per Tar, gain was negative for even greater extents of ligand occupancy and maximal kinase inhibition was reached at approximately maximal ligand occupancy. These relationships were the result of patterns of effects in which increased receptor adaptational modification reduced only modestly the affinity of Tar for aspartate but shifted the effect of aspartate on kinase activity as much as ~100-fold (Fig. 7B). In addition, the direct binding curve for aspartate was hyperbolic whereas kinase activity as a function of aspartate concentration was cooperative (compare the slopes of the curves for ligand binding versus kinase inhibition in Fig. 7B and Hill coefficients in Tables 1 and 2).
The non-linear relationship between changes in methylation rate and kinase activity has implications for the process of “precise adaptation” (Alon et al., 1999). The simplest form of precise adaptation implies close coupling between the kinase activity and propensity for methylation, the reaction that will restore kinase activity to the null state after an increase in ligand occupancy. However, our data indicate that changes in initial rates of methylation and changes in kinase activity are not coupled in a one-to-one manner and are not constant as a function of ligand occupancy or receptor modification (Fig. 7A). These features are likely to be important for a detailed understanding of precise adaptation.
At what point in the coupling of ligand binding to kinase activity are the features of cooperativity and wide dynamic range introduced? There was no significant cooperativity in the coupling of ligand binding to receptor conformation (Table 1) indicating that the cooperativity observed in the coupling of ligand concentration to kinase activity must be introduced no earlier than the coupling of receptor conformation to kinase activity. In contrast, generation of a wide dynamic range by adaptational modification appeared to occur stepwise, partly in the coupling of ligand occupancy to receptor conformation (Figs. 7C and D) and additionally in the coupling of receptor conformation to kinase activity (Fig. 7A and D). Coupling of receptor conformation to ligand binding was non-linear both in the absence (Fig. 7C) and presence (Fig. 7D) of CheA/CheW. However, because signaling complexes shifted dose-response curves for methylation rate as a function of aspartate to lower concentrations without shifting dose-response curves for direct binding of the ligand (Fig. 5B), the specific relationships between binding and receptor conformation at different extents of receptor modification were different for Tar alone (Fig. 7C) and in signaling complexes (Fig. 7D).
In summary, we find that cooperativity is introduced in the coupling of ligand binding to kinase activity at the level of signaling complexes whereas the increased dynamic range provided by receptor modification is introduced stepwise. In signaling complexes, CheA and chemoreceptors are not symmetrically affected by their mutual interactions or the sensory inputs of ligand binding and adaptational modification, indicating that receptors and the kinase CheA are not tightly coupled. Incomplete coupling implies that coupling could vary and thus be influenced by sensory inputs and interactions among signaling components.
Experimental procedures
Strains and Plasmids
Escherichia coli K-12 strain RP3098 (Parkinson & Houts, 1982) carries a chromosomal deletion from flhA to flhD, eliminating expression of genes for chemoreceptors and Che proteins. pAL67 carries a form of tar coding for Tar with a 6-histidine, carboxyl-terminal extension (Tar-6H) and a QEQE arrangement at the four methyl-accepting sites (Lai & Hazelbauer, 2005). Versions of pAL67 coding for Tar with 0, 1, 3 or 4 glutamines were created by PCR mutagenesis and the constructs verified by DNA sequencing. These were pAL529 (0Q; EEEE), pAL531 (1Q; QEEE), pAL532 (3Q; QEQQ) and pAL533 (4Q, QQQQ).
Membrane vesicle-borne Tar
Cytoplasmic membrane vesicles were prepared from RP3098 harboring an appropriate plasmid by osmotic lysis and sucrose gradient centrifugation, and analyzed for total protein, Tar and available methyl-accepting sites as described (Boldog et al., 2007, Amin & Hazelbauer, 2010). Approximately 68 to 85% of receptor cytoplasmic domains in these preparations could be modified to a level of maximal methylation by CheR, indicating both accessibility and native structure. All concentrations of Tar noted in this work are adjusted for that functional, accessible fraction.
Kinase activity
Kinase activity of CheA was determined as described (Bornhorst & Falke, 2000, Bornhorst, 2001 #1469) with the following modifications. 2 μM Tar-6H in membrane vesicles was incubated with purified 0.5 μM CheA, 2 μM CheW and 10 μM CheY in 50 mM Tris HCl pH 7.5, 50 mM KCl, 5 mM MgCl2 and 2 mM DTT for 45 min at room temperature to allow formation of the receptor signaling complexes. Samples were further incubated with or without aspartate (0 to 1 mM) in the same buffer for 15 min at room temperature. [γ-32P] ATP (Perkin Elmer, ~ 3.6 mCi/mmol) was added to 0.4 mM final concentration, the reaction terminated at 10 s by addition 2X SDS polyacrylamide gel electrophoresis sample buffer containing 25mM EDTA and submitted to SDS polyacrylamide gel electrophoresis on 15% polyacrylamide gels (Nowlin et al., 1987). Immediately following electrophoresis, gels were briefly stained, destained and dried. [32P] Phospho-CheY was determined by phosphorimaging.
Initial rates of methylation
Assays of initial rates of Tar methylation were performed as described previously (Barnakov et al., 1998, Amin & Hazelbauer, 2010) with the following modifications. Tar-6H in membrane vesicles was incubated in the conditions described for formation of signaling complexes (see section on kinase activity) in absence or presence of CheA, CheW and CheY, and incubated for an additional 15 min at room temperature after addition of aspartate or the equivalent volume of buffer. These conditions differed from those used in a previous study (Amin & Hazelbauer, 2010) by the presence of 50 mM KCl, 5 mM MgCl2, which resulted in differences in absolute rates of methylation but not in the general patterns of effects of ligand or receptor modification. Methylation was initiated by the addition of a CheR-enriched cell extract to which had been added S-adenosyl-L-[methyl-3H] methionine (Amersham Pharmacia, ~1.1 Ci/mmol) to produce final concentration of 5 μM available methyl-accepting sites, 0.125 μM CheR, 50 μM AdoMet and 0 to 1 mM aspartate in 50 μL. At 10, 20, 30 and 40 s, 10 μL samples were removed, mixed with SDS gel electrophoresis sample buffer to stop the reaction and analyzed by SDS gel electrophoresis, excision of the region containing Tar, alkaline hydrolysis of radiolabeled glutamyl esters to yield radiolabeled methanol, vapor-phase diffusion and scintillation counting. The initial rates of methylation were determined by linear fits of time courses as described and illustrated in (Amin & Hazelbauer, 2010).
Measurement of binding of CheA and CheW to vesicle embedded Tar
We incubated Tar, CheA, CheW and CheY in the conditions we used to form signaling complexes and then centrifuged at 16,100 × g for 10 min in micro-centrifuge, a process that quantitatively sedimented receptor-containing membranes. Pellets were suspended in twice-concentrated SDS gel electrophoresis sample buffer, analyzed by SDS polyacrylamide gel electrophoresis and proteins quantified by immunoblotting (Li & Hazelbauer). Tests of selected examples of the Tar-containing membrane vesicles used in our studies showed a consistent ratio of CheR-accessible Tar, bound CheA and bound CheW of approximately 1 CheA2: 7 Tar2: 6 CheW. These ratios were within the range of values reported by other investigators for stoichiometries of in vitro formation of signaling complexes (see (Erbse & Falke, 2009) for a summary of those data). Notably, the ratio of ~7 Tar:1 CheA approximated a stoichiometry of two trimers of receptor dimers per CheA dimer suggested by recent electron microscopic (Khursigara et al., 2008), structural (Bhatnagar et al., 2010) and functional characterization (Erbse & Falke, 2009) of signaling complexes. Taken together we interpret the stoichiometry measurements and the comparable magnitudes of effects of signaling complex formation and ligand binding to mean that in our experimental conditions the proportion of receptors not affected by signaling complexes was small and thus it is unlikely we missed substantial effects of signaling complexes on receptors.
Aspartate binding to Tar
Ligand binding assays were performed by competition centrifugation as described previously (Clarke & Koshland, 1979) with the following modifications. 16 μM Tar-6H in membrane vesicles was incubated for 45 min with no further additions or in conditions for formation of signaling complexes. 3H-aspartate (GE healthcare, ~1.0 Ci/mmol) was added to duplicate samples at a concentration from 0 to 15 μM. After incubation for 15 min at room temperature, non-radioactive aspartate was added to one of each duplicate at a final concentration of 4 mM and samples were centrifuged 16 min at 30,000 rpm (109,368 × g) at 4°C in Ti42.2 rotor. Radioactivity in each supernatant was determined by scintillation counting and the calculated concentration of free aspartate used to determine bound and free ligand at each concentration of aspartate. Data were fit to the relationship b = bm ([Asp]n)/([Asp]½ n + [Asp]n) where b is the concentration of bound aspartate at a given aspartate concentration, bm is the maximal concentration of bound aspartate, [Asp]½ is the aspartate concentration at which aspartate binding is half maximum and n is the Hill coefficient.
Initial rates of demethylation
Demethylation was assayed essentially as described (Barnakov et al., 1999, Li & Hazelbauer, 2005) with the following modifications. Methylated receptor was prepared by incubation of 10 μM vesicle-borne receptor, 50 μM S-adenosyl-L-[methyl-3H]methionine (Amersham Pharmacia, ~1.1 Ci/mmol) and 10 μM CheR in a cell lysate for 2 hrs and stored at −30° C. Methylated receptor was incubated in the conditions described for formation of signaling complexes (see section on kinase activity) in absence or presence of CheA/CheW/CheY and incubated for an additional 15 min at room temperature after addition of aspartate or the equivalent volume of buffer. Demethylation was initiated by addition of a mixture to yield 0.25 μM CheB, 25mM phosphoramidate, 10 mM KCl and 10 mM MgCl2. At times after initiation, 10 μL samples were removed and mixed with 50 μL 5N acetic acid, and radioactive methanol released was quantified by the vapor-phase equilibrium method (Chelsky et al., 1984). To obtain the results cited in the first paragraph of the Discussion, we determined initial rates of demethylation by phospho-CheB in the absence or presence of CheA/CheW/CheY for Tar in five individual native membrane vesicle preparations carrying 0.7, 1.6, 2.6, 3.3 or 3.4 modifications (the sum of 0, 1, 2, 3 or 3 glutamines and 0.7, 0.6, 0.6, 0.3 or 0.4 radiolabeled methyl glutamates per polypeptide chain). Interactions in signaling complexes reduced the initial rate of demethylation, independent of the extent of modification. Rates for Tar in signaling complexes were 54% ± 9 % of the respective rates in the absence of signaling complexes.
Acknowledgments
We thank Angela Lilly for creating the strains and plasmids used, and Nicholas Bartelli for noticing an important issue of experimental design. This work was supported in part by grant GM29963 to GLH from the National Institute of General Medical Sciences.
References
- Alon U, Surette MG, Barkai N, Leibler S. Robustness in Bacterial Chemotaxis. Nature. 1999;397:168–171. doi: 10.1038/16483. [DOI] [PubMed] [Google Scholar]
- Amin DN, Hazelbauer GL. The Chemoreceptor Dimer Is the Unit of Conformational Coupling and Transmembrane Signaling. J Bacteriol. 2010;192:1193–1200. doi: 10.1128/JB.01391-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnakov A, Barnakova L, Hazelbauer G. Comparison in vitro of a high- and a low-abundance chemoreceptor of Escherichia coli: similar kinase activation but different methyl-accepting activities. J Bacteriol. 1998;180:6713–6718. doi: 10.1128/jb.180.24.6713-6718.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnakov AN, Barnakova LA, Hazelbauer GL. Efficient adaptational demethylation of chemoreceptors requires the same enzyme-docking site as efficient methylation. Proc Natl Acad Sci USA. 1999;96:10667–10672. doi: 10.1073/pnas.96.19.10667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatnagar J, Borbat PP, Pollard AM, Bilwes AM, Freed JH, Crane BR. Structure of the Ternary Complex Formed by a Chemotaxis Receptor Signaling Domain, the CheA Histidine Kinase, and the Coupling Protein CheW As Determined by Pulsed Dipolar ESR Spectroscopy. Biochemistry. 2010;49:3824–3841. doi: 10.1021/bi100055m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldog T, Li M, Hazelbauer GL. Using Nanodiscs to create water-soluble transmembrane chemoreceptors inserted in lipid bilayers. Methods Enzymol. 2007;423:317–335. doi: 10.1016/S0076-6879(07)23014-9. [DOI] [PubMed] [Google Scholar]
- Borkovich KA, Alex LA, Simon MI. Attenuation of sensory receptor signaling by covalent modification. Proc Natl Acad Sci USA. 1992;89:6756–6760. doi: 10.1073/pnas.89.15.6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkovich KA, Kaplan N, Hess JF, Simon MI. Transmembrane signal transduction in bacterial chemotaxis involves ligand-dependent activation of phosphate group transfer. Proc Natl Acad Sci USA. 1989;86:1208–1212. doi: 10.1073/pnas.86.4.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornhorst JA, Falke JJ. Attractant regulation of the aspartate receptor-kinase complex: limited cooperative interactions between receptors and effects of the receptor modification state. Biochemistry. 2000;39:9486–9493. doi: 10.1021/bi0002737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornhorst JA, Falke JJ. Evidence that both ligand binding and covalent adaptation drive a two-state equilibrium in the aspartate receptor signaling complex. J Gen Physiol. 2001;118:693–710. doi: 10.1085/jgp.118.6.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briegel A, Ortega DR, Tocheva EI, Wuichet K, Li Z, Chen S, Muller A, Iancu CV, Murphy GE, Dobro MJ, Zhulin IB, Jensen GJ. Universal architecture of bacterial chemoreceptor arrays. Proc Natl Acad Sci U S A. 2009;106:17181–17186. doi: 10.1073/pnas.0905181106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalah A, Weis R. Site-specific and synergistic stimulation of methylation on the bacterial chemotaxis receptor Tsr by serine and CheW. BMC Microbiology. 2005;5:12. doi: 10.1186/1471-2180-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelsky D, Gutterson NI, Koshland DE., Jr A diffusion assay for detection and quantitation of methyl-esterified proteins on polyacrylamide gels. Anal Biochem. 1984;141:143–148. doi: 10.1016/0003-2697(84)90437-8. [DOI] [PubMed] [Google Scholar]
- Clarke S, Koshland DE., Jr The effect of protein modification reagents on the chemotactic response in Salmonella typhimurium. Can J Biochem. 1979;57:1331–1336. [PubMed] [Google Scholar]
- Dunten P, Koshland DE., Jr Tuning the responsiveness of a sensory receptor via covalent modification. J Biol Chem. 1991;266:1491–1496. [PubMed] [Google Scholar]
- Erbse AH, Falke JJ. The Core Signaling Proteins of Bacterial Chemotaxis Assemble To Form an Ultrastable Complex. Biochemistry. 2009;48:6975–6987. doi: 10.1021/bi900641c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelbauer GL, Falke JJ, Parkinson JS. Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends Biochem Sci. 2008;33:9–19. doi: 10.1016/j.tibs.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelbauer GL, Lai W-C. Bacterial chemoreceptors: providing enhanced features to two-component signaling. Curr Opin Microbiol. 2010;13:124–132. doi: 10.1016/j.mib.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khursigara CM, Wu X, Subramaniam S. Chemoreceptors in Caulobacter crescentus: Trimers of Receptor Dimers in a Partially Ordered Hexagonally Packed Array. J Bacteriol. 2008;190:6805–6810. doi: 10.1128/JB.00640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KK, Yokota H, Kim SH. Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature. 1999;400:787–792. doi: 10.1038/23512. [DOI] [PubMed] [Google Scholar]
- Lai WC, Beel BD, Hazelbauer GL. Adaptational modification and ligand occupancy have opposite effects on positioning of the transmembrane signalling helix of a chemoreceptor. Mol Microbiol. 2006;61:1081–1090. doi: 10.1111/j.1365-2958.2006.05296.x. [DOI] [PubMed] [Google Scholar]
- Lai W-C, Hazelbauer GL. Carboxyl-terminal extensions beyond the conserved pentapeptide reduce rates of chemoreceptor adaptational modification. J Bacteriol. 2005;187:5115–5121. doi: 10.1128/JB.187.15.5115-5121.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levit MN, Stock JB. Receptor methylation controls the magnitude of stimulus-response coupling in bacterial chemotaxis. J Biol Chem. 2002;277:36760–36765. doi: 10.1074/jbc.M204325200. [DOI] [PubMed] [Google Scholar]
- Li G, Weis RM. Covalent modification regulates ligand binding to receptor complexes in the chemosensory system of Escherichia coli. Cell. 2000;100:357–365. doi: 10.1016/s0092-8674(00)80671-6. [DOI] [PubMed] [Google Scholar]
- Li M, Hazelbauer GL. Cellular stoichiometry of the components of the chemotaxis signaling complex. J Bacteriol. 2004;186:3687–3694. doi: 10.1128/JB.186.12.3687-3694.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Hazelbauer GL. Adaptational assistance in clusters of bacterial chemoreceptors. Mol Microbiol. 2005;56:1617–1626. doi: 10.1111/j.1365-2958.2005.04641.x. [DOI] [PubMed] [Google Scholar]
- Lin LN, Li J, Brandts JF, Weis RM. The serine receptor of bacterial chemotaxis exhibits half-site saturation for serine binding. Biochemistry. 1994;33:6564–6570. doi: 10.1021/bi00187a025. [DOI] [PubMed] [Google Scholar]
- Milburn MV, Prive GG, Milligan DL, Scott WG, Yeh J, Jancarik J, Koshland DE, Jr, Kim SH. Three-dimensional structures of the ligand-binding domain of the bacterial aspartate receptor with and without a ligand. Science. 1991;254:1342–1347. doi: 10.1126/science.1660187. [DOI] [PubMed] [Google Scholar]
- Nowlin DM, Bollinger J, Hazelbauer GL. Sites of covalent modification in Trg, a sensory transducer of Escherichia coli. J Biol Chem. 1987;262:6039–6045. [PubMed] [Google Scholar]
- Park SY, Borbat PP, Gonzalez-Bonet G, Bhatnagar J, Pollard AM, Freed JH, Bilwes AM, Crane BR. Reconstruction of the chemotaxis receptor-kinase assembly. Nat Struct Mol Biol. 2006;13:400–407. doi: 10.1038/nsmb1085. [DOI] [PubMed] [Google Scholar]
- Parkinson JS, Houts SE. Isolation and behavior of Escherichia coli deletion mutants lacking chemotaxis functions. J Bacteriol. 1982;151:106–113. doi: 10.1128/jb.151.1.106-113.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard AM, Bilwes AM, Crane BR. The Structure of a Soluble Chemoreceptor Suggests a Mechanism for Propagating Conformational Signals. Biochemistry. 2009;48:1936–1944. doi: 10.1021/bi801727m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studdert CA, Parkinson JS. Crosslinking snapshots of bacterial chemoreceptor squads. Proc Natl Acad Sci USA. 2004;101:2117–2122. doi: 10.1073/pnas.0308622100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaknin A, Berg HC. Physical responses of bacterial chemoreceptors. J Mol Biol. 2007;366:1416–1423. doi: 10.1016/j.jmb.2006.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]