Abstract
BMP signaling plays many important roles during organ development, including palatogenesis. Loss of BMP signaling leads to cleft palate formation. During development, BMP activities are finely tuned by a number of modulators at the extracellular and intracellular levels. Among the extracellular BMP antagonists is Noggin, which preferentialy binds to BMP2, BMP4 and BMP7, all of which are expressed in the developing palatal shelves. Here we use targeted Noggin mutant mice as a model for gain of BMP signaling function to investigate the role of BMP signaling in palate development. We find prominent Noggin expression in the palatal epithelium along the anterior-posterior axis during early palate development. Loss of Noggin function leads to overactive BMP signaling, particularly in the palatal epithelium. This results in disregulation of cell proliferation, excessive cell death, and changes in gene expression, leading to formation of complete palatal cleft. The excessive cell death in the epithelium disrupts the palatal epithelium integrity, which in turn leads to an abnormal palate-mandible fusion and prevents palatal shelf elevation. This phenotype is recapitulated by ectopic expression of a constitutively active form of BMPR-IA but not BMPR-IB in the epithelium of the developing palate; this suggests a role for BMPR-IA in mediating overactive BMP signaling in the absence of Noggin. Together with the evidence that overexpression of Noggin in the palatal epithelium does not cause a cleft palate defect, we conclude from our results that Noggin mediated modulation of BMP signaling is essential for palatal epithelium integrity and for normal palate development.
Keywords: Bmp signaling, Noggin, cleft palate, palatal epithelium, palate development
Introduction
Cleft palate, one of the most frequent congenital birth defects in the human, results from genetic or environmental perturbations in palate development. Development of the mammalian secondary palate is a multiple staged process, which begins in mice at embryonic day 11.5 (E11.5), when the palatal shelves grow out of the bilateral maxillary processes. The palatal shelves continue to grow vertically along the developing tongue until E14.0, and then bend abruptly to a horizontal position above the tongue. At E14.5, the growing palatal shelves meet each other and fuse in the midline, separating the oral cavity from the nasal cavity. Each step of palate development, like the formation of many other mammalian organs, is directed by reciprocal and sequential epithelial-mesenchymal interactions (Ferguson and Honig, 1984; Ferguson, 1988; Hall, 1992).
The palatal shelves are composed of the epithelial covering and the mesenchymal tissue that is derived largely from cranial neural crest cells, and also from cranial paraxial mesoderm (Ferguson, 1988; Ito et al., 2003). The palatal epithelium, consisting of a basal columnar cell layer and covering periderm cells, is a heterogeneous structure. Based on the cell morphology, position, and genetic markers, the palatal epithelium can be divided into nasal, oral, and medial edge epithelium (MEE) (Ferguson, 1988). Prior to palatal elevation, the nasal portion of the palatal epithelium differentiates into pseudostratified nasal epithelial cells, and the oral portion differentiates into squamous oral epithelial cells. The MEE, which is positioned between the oral and nasal regions, develops into a single layered epithelial seam upon the contact and fusion of two palatal shelves, and ultimately diminishes to form an intact palatal shelf. During palate development, the integrity of palatal epithelium is essential for palate elevation. Disruption of this integrity would usually lead to abnormal adhesion or fusion between the elevating palatal shelves with adjacent structures, such as tongue and mandible, resulting in delayed or failed palate elevation, and consequently, generating a cleft palate defect (Rice et al., 2004; Alappat et al., 2005; Richardson et al., 2006; Xiong et al., 2009).
The Bone Morphogenetic Proteins (BMPs) have been implicated in mammalian palate development (Nie et al., 2006). Several Bmp genes, including Bmp2, Bmp4 and Bmp7, are expressed in the developing mouse palatal shelves (Lu et al., 2000; Zhang et al., 2002; Nie, 2005; Levi et al., 2006). The requirement for BMP signaling in palate development was initially demonstrated in Msx1 mutant mice, which exhibit the cleft palate phenotype (Zhang et al., 2002). In the Msx1 mutant palate, Bmp4 expression is abrogated and ectopic expression of its human ortholog rescues the cleft palate phenotype in Msx1 mutant. Furthermore, tissue-specific inactivation of the genes encoding type I BMP I receptors, such as Bmpr-IA (Alk3) and ActRIa (Alk2), or overexpression of the BMP antagonist Noggin in the palatal mesenchyme leads to cleft palate formation in corresponding mouse models (Dudas et al., 2004; Liu et al., 2005; Xiong et al., 2009). These lines of evidence indicate an essential role for BMP signaling in palate development. On the other hand, inactivation of the promiscuous TGFβ antagonist Follistatin causes a cleft palate phenotype, raising the possibility that an elevated level of BMP signaling also impairs palate development (Matzuk et al., 1995).
During embryonic development, BMP signaling is finely tuned by a number of modulators at different levels (Gazzerro and Canalis, 2006). The intracellular modulators, such as Smad6, Smurf and Tob, can prevent R-Smads activation by interrupting their binding to receptors and Smad4, or mediate R-Smad degradation. At the extracellular level, BMP antagonists modulate BMP signaling activity by blocking selective ligands from binding to their receptors. A number of such extracellular antagonists have been documented, including Noggin, Chordin, Follistatin, and other molecules (Balemans and Van Hul, 2002; Gazzerro and Canalis, 2006). Among them, Noggin is a secretory polypeptide that binds preferentially to BMP2, BMP4, and BMP7 to prevent their signaling (Zimmerman et al., 1996; Groppe et al., 2002; Chen et al., 2004). Noggin (Nog) deficient mice exhibit a series of defects in organogenesis (Brunet et al., 1998; McMahon et al., 1998; Bachiller et al., 2000), including a spectrum of craniofacial defects, accompanied with upregulation of BMP activities (Bachiller et al., 2000; Stottmann et al., 2001; Anderson et al., 2006). However, a cleft palate defect in Nog-/- mice has not been reported.
We have previously investigated the role of BMP signaling in palate development using loss-of-function models (Zhang et al., 2002; Xiong et al., 2009). In this study, we used a conventionally gene-targeted Noggin mutant line (Nog-/-) (McMahon et al., 1998) as a gain-of-BMP function model to further evaluate the role of BMP signaling in palate development. We report here that Nog-/- mice exhibit a cleft palate defect. BMP/Smad signaling is ectopically activated in the Nog-/- palatal epithelium, consistent with the restricted Noggin expression pattern in the developing palate. Our results show that palatal epithelium integrity is disrupted in the Nog-/- palate and this disruption leads to an abnormal palate-mandible fusion, preventing the normal palate elevation. This phenotype is recapitulated in a transgenic model in which BMP receptor-IA mediated signaling is ectopically activated in the developing palate. In contrast, overexpression of Noggin in the palatal epithelium does not cause a cleft palate defect. We therefore conclude that Noggin-mediated repression of BMP signaling in the palatal epithelium is required for normal palate development.
Materials and Methods
Animals
The generation and genotyping of Nog+/-, K14Cre and pMesNog mice have been described previously (McMahon et al., 1998; Andl et al., 2004; Xiong et al., 2009). In Nog mutant mice, a null Nog allele was created by fusing the first 10 amino acids of the Noggin coding sequence to the lacZ gene so that the LacZ expression is under the control of the Nog regulatory elements (McMahon et al., 1998). The pMescaBmpr-IA and pMescaBmpr-IB transgenic mice were generated in a strategy similar to pMesNog mice (Xiong et al., 2009). Briefly, a constitutively active form of the chick Bmpr-IA (caBmpr-IA) with Gln-233 to Asp replacement and a constitutively active form of Bmpr-IB (caBmpr-IB) with Gln-203 to Asp change (Zou et al., 1997) were cloned into pMes-IRES-Egfp vector at the 5’ end of the IRES-Egfp sequence, and the 3’ end of the LoxP flanked STOP cassette, which is under the control of the chick β-actin promoter. Pronuclear injection was performed to generate transgenic founders. Transgene expression in each potential transgenic line was identified by Egfp expression and further confirmed by in situ hybridization. Embryos were collected from time-mated pregnant mice and dissected in ice-cold PBS treated with diethyl pyrocarbonate (DEPC). Embryonic head samples were then separated from the trunk, fixed in 4% paraformaldehyde (PFA) overnight at 4°C, and processed for paraffin section or frozen section for immunostaining. A tail sample from each embryo was used for PCR-based genotyping (primer information available upon request). Nog+/- mice were maintained in a C57/B6 background. The pMesNog, pMescaIA, and pMesBmpr-IB transgenic mice were maintained in an outbred CD1 background. All animals and procedures used in this study were approved by the Tulane University Institutional Animal Care and Use Committee.
In vitro organ culture
Paired secondary palatal shelves from individual E13.5 embryos were isolated; the anterior halves of the palatal shelves were dissected and collected. Paired anterior palatal shelves were placed in Trowell type organ culture, and were orientated so that the MEE of each palatal shelf was in contact, as described previously (Taya et al., 1999; Zhang et al., 2002). Samples were cultured in DMEM media supplemented with 20 % FCS at 37°C in an incubator filled with 5% CO2 for 3 days, and were then harvested for histological examination.
Histology, in situ hybridization, immunostaining, and X-gal staining
After fixation, staged embryonic head samples were dehydrated through gradient ethanol series, cleared in xylene, and embedded in paraffin. Coronal sections at 10 µm were collected for either Hematoxylin/Eosin staining or non-radioactive in situ hybridization, as described previously (St. Amand et al., 2000). Three independent experiments were carried out for gene expression by in situ hybridization. Immunostaining was performed as described previously (Xiong et al., 2009). Phospho-Smad1 (Ser463/465)/Smad5 (Ser463/465)/Smad8 (Ser426/428) antibody from Cell Signaling (catalog # 9511) was used to detect Smad dependent BMP signaling activities. For X-gal staining, Nog+/- embryos were collected from mating of Nog+/- mice with wild type mice at designated time points, and the tail of each embryo was used for genotyping. Embryonic heads were removed, labeled, and individually fixed in 0.2% glutaraldehyde for overnight at 4°C. After genotyping, Nog+/- embryonic heads were washed in ice cold PBS, washed in 30% sucrose/PBS solution overnight, and embedded in O.C.T (Tissue-Tek). Cryosections at 10 μm were processed for X-gal staining, as described previously (Ito et al., 2003). For whole mount X-gal staining, samples were fixed for 20 min in 0.2% glutaraldehyde, and subjected to staining according to the standard protocol (Chai et al., 2000).
Cell proliferation and TUNEL assays
Bromodeoxyuridine (BrdU) Labeling and Detection Kit (Roche Diagnostics Corporation, Indianapolis) was used to measure cell proliferation rate. Briefly, BrdU solution was injected intraperitoneally into timed pregnant female mice (1.5ml/100g body weight) one hour before embryos were harvested. Embryonic heads were fixed in Carnoy's fixative and then processed into paraffin sections at 5-μm for immunostaining according to manufacturer's instruction. For BrdU-labeled cell counting, an arbitrary area was defined and both BrdU labeled cells and total cells were counted in that area. The outcome of BrdU labeling was presented as percentage of labeled cells among total nuclei in the fixed region. Collected from nine continuous sections of three individual samples of wild type controls and mutants, respectively, data were subjected to Student's t-test to determine the significance of differences. For TUNEL assays, samples were processed into 5 μm paraffin sections and apoptotic cells were detected as described previously (Alappat et al., 2005).
Results
The role of BMP signaling in palate development has been previously studied using various loss-of-function models (Zhang et al., 2002; Liu et al., 2005; Xiong et al., 2009). The cleft palate defects in mice deficient for Follistatin further suggest a requirement for a finely tuned level of TGFβ ligand activity in normal palate development (Matzuk et al., 1995), including that of BMPs. Noggin is a specific extracellular antagonist of BMP signaling that modulates BMP activities during embryonic development. Noggin-deficient mice therefore provide a gain-of-function model to study the role of BMP signaling in organ development. Besides defects found in many other developing organs, Nog-/- mice also exhibit a spectrum of craniofacial defects, suggesting a role for Noggin in palate development (McMahon et al., 1998; Bachiller et al., 2000; Stottmann et al., 2001; Anderson et al., 2006). However, neither expression nor function of Noggin has been documented in mammalian palate development.
Expression of Noggin in the developing palate
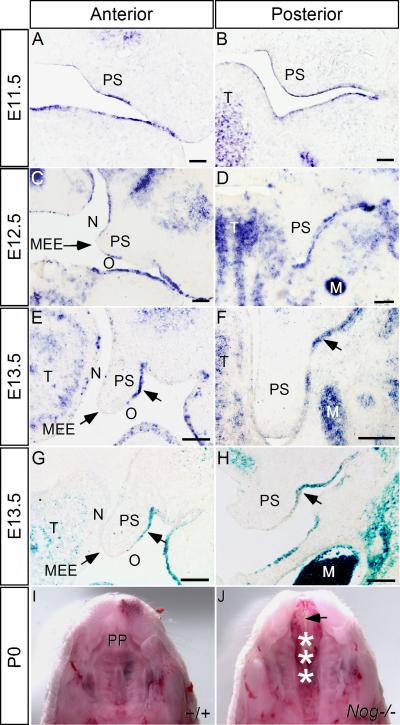
To investigate the potential role of Noggin and BMP signaling in palatogenesis, we began with an examination of Noggin expression by in situ hybridization. We found a dynamic Noggin expression pattern, primarily encompassing the palatal epithelium (Fig. 1). At E11.5, Noggin is expressed in the entire palatal epithelium along the anterior-posterior (A-P) axis (Fig. 1A, B). At E12.5, in the anterior portion of the palatal shelves, Noggin expression remains in the epithelium, but is relatively weak in the nasal side and the MEE (Fig. 1C), while in the posterior portion, Noggin expression was only detected in the oral side palatal epithelium (Fig. 1D). At E13.5, Noggin expression is restricted to the oral side epithelium in both the anterior and posterior palate (Fig. 1E, F). In the craniofacial region, Noggin mRNAs were also detected in the oral epithelium, maxillary mesenchyme, tongue and Meckel's cartilage (Fig. 1). We further assayed the Noggin expression pattern by X-gal staining in Nog+/- embryos, which carry a LacZ knock-in maker (Fig. 1G, H; and Supplemental Fig. 1A-D; McMahon et al., 1998); these results confirmed the expression pattern detected by in situ hybridization. The spatiotemporal profile of Noggin expression suggests an involvement of Noggin in palate development, prompting us to examine potential palate phenotypes in Nog-/- mice.
Figure 1.
Expression of Noggin in the developing mouse palatal shelves. (A, B) At E11.5, Noggin mRNA is detected in the epithelium of the oral-nasal cavity, including the anterior (A) and posterior (B) palatal shelves. (C, D) In the anterior palatal shelves at E12.5, Noggin expression is detected in the nasal side and oral side palatal epithelium, but not in the MEE (C), while in the posterior palate, Noggin mRNA is restricted to the oral side palatal epithelium (D). (E-H) At E13.5, Noggin expression (arrows) is only detected in the oral side palatal epithelium (arrows) in both the anterior (E) and posterior (F) palatal shelves. LacZ reporter expression in the Nog+/- palate recapitulates the expression pattern of Noggin mRNA (G, H). (I, J) At P0, the wild type mouse develops an intact palatal shelf (I), while Nog-/- mouse exhibits a complete cleft of the secondary palate (asterisks in J) and a lack of the primary palate (arrow in J). M, Meckel's cartilage; MEE, medial edge epithelium; PP, primary palate; PS, palatal shelf; T, tongue. N designates nasal side of the palatal shelf, and O designates oral side of the palatal shelf. Scale bars represent 100 μm.
Nog-/- mice exhibit complete cleft palate phenotype
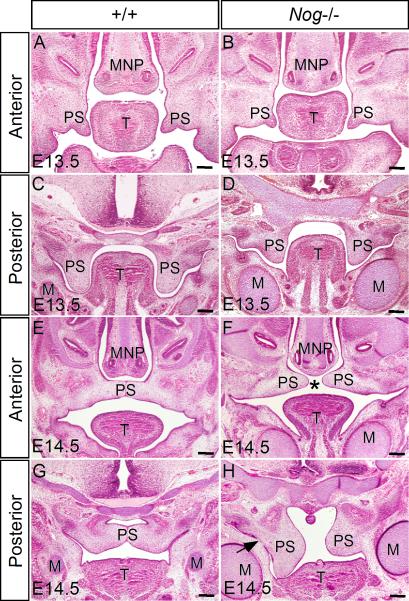
Gross examination of Nog-/- mice at postnatal day 0 (P0) identified a complete clefting of the secondary palate with 100% penetrance (Fig. 1J). Histological analyses revealed that the Nog-/- palatal shelves exhibit morphology comparable to the wild type control until E12.5, when the anterior palatal shelves looked normal but posterior palatal shelves appeared slightly smaller than their wild type counterparts (data not shown). At E13.5, the anterior portion of the palatal shelves in the mutants appeared smaller than in the wild type controls, and the posterior palate also appeared slightly shorter (Fig. 2A-D). During normal embryogenesis, the palatal shelves elevate and fuse at the midline at E14.5 (Fig. 2E, G). In Nog-/- embryos, we observed that the anterior palatal shelves elevated to horizontal position above the tongue, but did not meet medially. In contrast, the posterior portion of the palatal shelves fused with the mandible and did not elevate at all (Fig. 2F, H). In addition to the defect in palate development, significantly enlarged Meckel's cartilage was also observed in Nog-/- embryos (Fig. 2), consistent with Noggin expression in Meckel's cartilage and its general role in skeletal development (Brunet et al., 1998; McMahon et al., 1998).
Figure 2.
Nog-/- embryos show defective palate development. (A, C) At E13.5, the wild type palatal shelves grow vertically along the tongue in the oral-nasal cavity. (B, D) In E13.5 mutant, the anterior palatal shelves appear smaller than the wild type controls (B); in the posterior portion, the mutant palatal shelves grow shorter but slightly wider than the wild type counterparts, accompanied by ectopic cartilage formation and hyperplastic Meckel's cartilage (D). (E, G) At E14.5, the palatal shelves in wild type embryos have elevated and fused to form an intact structure above the tongue. (F, H) In the mutant at E14.5, however, the palatal shelves fail to contact each other, forming a palatal cleft (asterisk). Note that in the mutant, the anterior palatal shelves have elevated but did not make contact (F), while the posterior palatal shelves remain in a vertical position, and show abnormal fusion (arrow) with the mandible (H). M, Meckel's cartilage; MNP, medial nasal process; PS, palatal shelf; T, tongue. Scale bars represent 200 μm.
The failed palatal shelf contact at the midline in the anterior region of Nog-/- palate could be caused by an impaired palatal fusion. To test this possibility, we used in vitro organ culture as described previously (Taya et al., 1999; Zhang et al., 2002). The anterior halves of the palatal shelves from E13.5 wild type controls and Nog-/- embryos were dissected and placed in contact as pairs with the MEE of each facing the other. After 3 days in culture, paired palatal shelves from both genotypes underwent fusion (5/5 for wild type control, and 6/6 for Noggin mutant), as revealed by the disappearance of the midline seam (Supplemental Fig. 2), indicating that failure of fusion is not the cause of cleft palate in Nog-/- embryo. Many factors are known to influence palatal shelf contact, including malformed craniofacial structures. Since micrognathia was rarely found in Nog-/- mutants (Stottmann et al., 2001), the cleft palate is not secondary to micrognathia where the tongue fails to sink down from between the palatal shelves. However, the lack of the primary palate (see below) and the formation of enlarged Meckel's cartilage could also attribute to the failure in palatal shelf contact. Nevertheless, these observations revealed a requirement for Noggin in palate development.
In addition to the cleft secondary palate formation, the primary palate was also absent completely in Noggin mutants (Fig. 1J), consistent with Noggin expression in the forming site of primary palate and surrounding tissue (Stottmann et al., 2001; Supplemental Fig. 1E, F). However, we did not observe any type of cleft lip formation, although we indeed found defective maxillary incisors that were fused at the midline to form a single tooth bud which was arrested at the late bud stage (unpublished results).
Alteration of BMP signaling in the Nog mutant palate
Noggin functions to antagonize the activities of selective specific BMP ligands, such as BMP2, BMP4, and BMP7, which signal primarily by phosphorylating Smad1/5/8 and activating the Smad4 dependent pathway. Although Smad-independent TGFβ/BMP signaling through p38 MAPK has also been implicated in palate development, this pathway mainly acts in the MEE region to regulate palatal fusion (Xu et al., 2008). We therefore used Smad1/5/8 phosphorylation as an indicator of BMP activated Smad signaling to determine changes in the level of BMP signaling in the Nog-/- palatal shelves. In the wild type palatal shelves, phosphorylated Smad1/5/8 (pSmad1/5/8) was detected in both anterior and posterior portion (Fig. 3A, E). In the anterior palate, we found abundant pSmad1/5/8 staining in the maxillary and palatal mesenchyme, with a few positive signals in the epithelium (Fig. 3A, C). In the posterior palate, pSmad1/5/8 was restricted to the nasal side palatal mesenchyme, with sporadic staining in the oral side palatal mesenchyme and epithelium, and completely negative staining in the Meckel's cartilage (Fig. 3E, G). In Nog-/- embryos, Smad1/5/8 phosphorylation was dramatically elevated in the maxillary mesenchyme and the Meckel's cartilage (Fig. 3B, F). However, in the mutant palate, we detected differential alterations in pSmad1/5/8 staining. In the anterior palatal mesenchyme, the staining was generally reduced (Fig. 3B, D). In contrast, in the posterior palatal mesenchyme, ectopic staining was found in the oral side region, while the nasal half remained comparable staining to the wild type controls (Fig. 3F, H). In the mutant palatal epithelium, we found consistently ectopic pSmad1/5/8 staining in the oral side epithelium along the A-P axis (Fig. 3), in accord with the Noggin expression pattern. The ectopic pSmad1/5/8 staining in the mutant epithelium indicates that Noggin is required to inhibit Smad dependent signaling in normal palate development. The different alterations in pSmad1/5/8 staining in the mutant palatal mesenchyme, on the other hand, suggest that palatal mesenchyme along the A-P axis responds differentially to the absence of Noggin, consistent with the tissue heterogeneity along the A-P axis of the developing palatal shelves (Hilliard et al., 2005; Okano et al., 2006).
Figure 3.
Noggin deficiency alters BMP/Smad signaling activity in the developing palate. (A, C, E, G) In the wild type control at E13.5, pSmad1/5/8 is detected in both the anterior and posterior palate. In the anterior palate, pSmad1/5/8 signal is found at high levels in the mesenchyme but is sparse in the epithelium (A, C); in the posterior palate, pSmad1/5/8/ activity is mainly restricted in the nasal side palatal mesenchyme, with a few positive signals in the oral side palatal epithelium (E, G). (B, D, F, H) In the Nog-/- palate at the comparable stage, Smad1/5/8 phosphorylation is enhanced in the oral side palatal epithelium, in both the anterior and posterior palate. As compared to the wild type control, the mutant anterior palatal mesenchyme exhibits downregulated pSmad1/5/8 signal, while the posterior palatal mesenchyme shows ectopic pSmad1/5/8 activity in the oral side. Note that the pSmad1/5/8 signal is significantly enhanced in the Nog-/- Meckel's cartilage (F). The straight white line in (E) and (F) divides the palatal mesenchyme into nasal and oral halves. The dash white lines in (C, D, G, H) demarcate the epithelial boundary. M, Meckel's cartilage; T, tongue. N designates nasal side of the palatal shelf, and O designates oral side of the palatal shelf. Scale bars represent 100 μm.
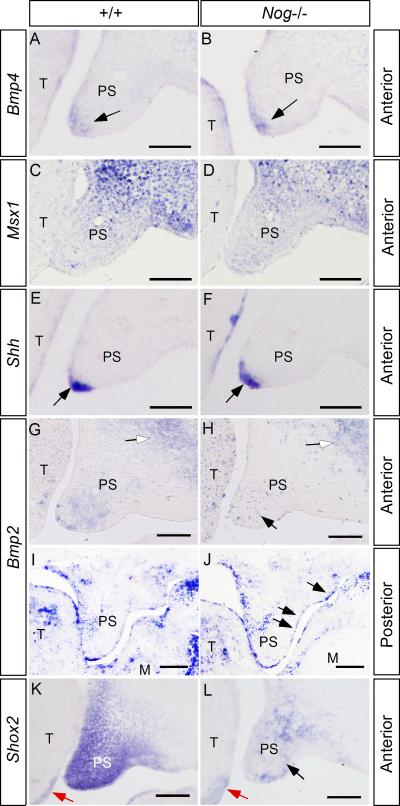
Activation of BMP/Smad signaling requires ligand-receptor binding. It has been well documented that Bmp2 and Bmp4 are expressed in the developing palate, and it has been shown that exogenous BMP can induce its antagonists expression, and vice versa (Lu et al., 2000; Stottmann et al., 2001; Ashique et al., 2002; Zhang et al., 2002; Nie, 2005). These lines of evidence promoted us to examine if inactivation of Noggin could also affect Bmp2 and Bmp4 expression in the developing palate. We found that the expression of Bmp4 is comparable to that in the wild type control (Fig. 4A, B). This observation is consistent with the unaltered expression patterns of Msx1 and Shh, downstream targets of Bmp4 in the anterior palate (Fig. 4C-F). In the wild type controls at E13.5, Bmp2 is expressed in the palatal mesenchyme of the anterior portion, and in the MEE as well as the nasal side palatal mesenchyme of the posterior portion (Fig. 4G, I). In the Nog-/- palate, we found that Bmp2 expression is reduced in the anterior palatal mesenchyme, but is ectopically activated in the oral side of posterior palatal epithelium (Fig. 4H, 4J), suggesting that Noggin regulates Bmp2 expression through different mechanisms along the A-P axis in the developing palate. In contrast, Bmp2 expression in the anterior maxillary mesenchyme and the posterior palate mesenchyme of the mutant was not affected (Fig. 4G-J). The down-regulation of Bmp2 expression in the anterior palatal mesenchyme is accompanied by a reduced expression of Shox2 (Fig. 4K, L), whose expression is restricted in the anterior palatal mesenchyme and is dependent on BMP activities (Yu et al., 2005). These results indicate that, in addition to its antagonizing property, Noggin is also able to regulate the expression of Bmp2, but not Bmp4, in the developing palate. The alteration in Bmp2 expression could at least partially account for the changes in pSmad1/5/8 activity in the mutant palate along the A-P axis, and a down-regulation of Shox2 expression in the anterior palatal mesenchyme.
Figure 4.
Gene expression in the wild type and Nog-/- palate. (A, B) At E13.5, Bmp4 is expressed in the anterior palatal mesenchyme (arrow) underlying MEE in the wild type palate (A); in the Nog-/- palate, comparable Bmp4 expression (arrow) is observed (B). (C, D) Comparable Msx1 expression is observed in the anterior palatal mesenchyme of E13.5 wild type (C) and Nog-/- embryo (D). (E, F) Shh expression (arrows) in the MEE of the anterior palatal shelves is not affected in Nog-/- embryo. (G, I) In E13.5 wild type controls, Bmp2 expression is detected in the anterior palatal mesenchyme and maxillary region (open arrow) (G); in the posterior palate, it is expressed in the nasal side palatal mesenchyme and the MEE region (I). (H) In the Nog-/- anterior palate, Bmp2 expression is downregulated in the palatal mesenchyme (black arrow), but is not affected in the maxillary region (open arrow). (J) In the posterior palate of the mutant, Bmp2 expression is not altered in the palatal mesenchyme and the MEE, but is ectopically activated in the oral side palatal epithelium (arrows). (K, L) Shox2 expression is significantly down-regulated in the anterior palatal mesenchyme of the mutant (L), as compared to the wild type control (K). Note comparable Shox2 expression at the ventral-lateral side of the developing tongue (red arrows). PS, palatal shelf; T, tongue. Scale bars represent 100 μm.
Cellular defects in the Nog-/- palatal shelves
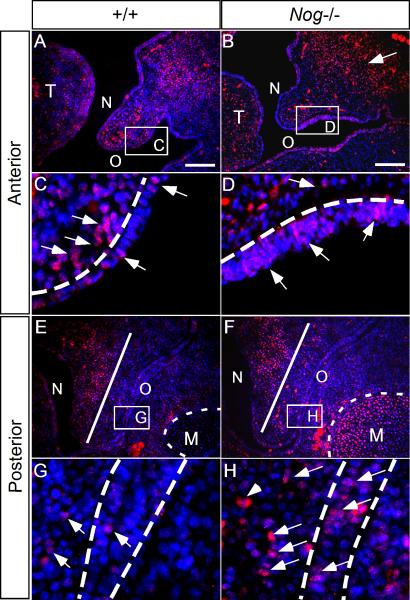
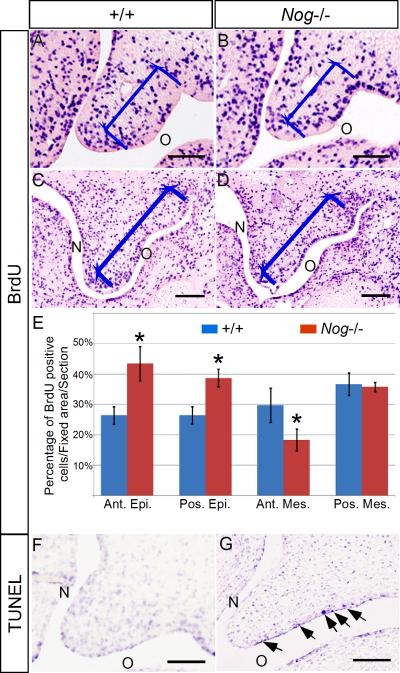
To reveal the underlying cellular mechanisms that lead to a cleft palate formation in the Nog-/- palate, we carried out BrdU labeling and TUNEL assays to examine cell proliferation rates and apoptosis levels, respectively. In this study, cell proliferation rate was measured by the ratio of proliferating cells against total nucleus number in a defined area. At E12.5, while slight deviation in cell proliferation rates was found in the both palatal epithelium and mesenchyme along the A-P axis in the wild type controls and mutants (data not shown), the difference was not significant, consistent with morphological analysis results. However, at E13.5 in the mutants, we found that the cell proliferation rate was significantly upregulated in the oral side palatal epithelium, in both the anterior and posterior portion, as compared to the wild type controls (Fig. 5A-E). This elevated cell proliferation rate appears to correlate with an enhanced pSmad1/5/8 staining in the Nog-/- palate, suggesting that BMP/Smad signaling positively modulated cell proliferation levels in the palatal epithelium. However, in the mutant mesenchyme, the cell proliferation rate was downregulated in the anterior region, but remained unchanged in the posterior portion despite ectopic pSmad1/5/8 staining in the oral half, as compared to the wild type controls (Fig. 5A-E). This observation is consistent with our previous findings that exogenous BMP induces cell proliferation in the anterior palatal mesenchyme but not in the posterior palatal mesenchyme (Hilliard et al., 2005), and that a reduced level of cell proliferation is observed in the anterior palatal mesenchyme of Shox2 mutant (Yu et al., 2005). We next carried out TUNEL assay to examine cell apoptosis in the wild type and Nog-/- palate at E13.5. We detected no apoptotic cell in the wild type control (Fig. 5F), but in the mutant, on the other hand, we found numerous apoptotic cells in the anterior palatal epithelium on the oral side, where Noggin is normally expressed (Fig. 5G). Ectopic apoptotic cells were not found in the nasal side epithelium. However, in the posterior palate of the mutant, we did not find apoptotic cells at this particular stage (data not shown). Thus alterations in cell proliferation rate and enhanced cell apoptosis contribute to abnormal growth and shaping in the mutant palatal shelves.
Figure 5.
The Nog-/- palatal shelves exhibit defective cell proliferation and excessive apoptosis. (A-D) Coronal sections of BrdU labeled palatal shelf in the wild type and Nog-/- palate at E13.5. The blue bracket defines the regions used for cell proliferation ratio statistics. (E) Statistical data analysis shows cell proliferation rate is upregulated in the mutant palatal epithelium (both anterior and posterior). Cell proliferation rate is downregulated in the mutant anterior palatal mesenchyme, but remains unaltered in the posterior palatal mesenchyme. (F, G) At E13.5, the anterior palate of mutant exhibits excessive apoptotic cells in the oral side epithelium, but not in the nasal side (G), as compared to the wild type control (F). Ant. Epi., epithelium of anterior palate; Ant. Mes., mesenchyme of anterior palate; Pos. Epi., epithelium of posterior palate; Pos. Mes., mesenchyme of posterior palate; *, P<0.01. Scale bars represent 100 μm.
Nog-/- mice exhibit abnormal fusion between the posterior palate and the mandible
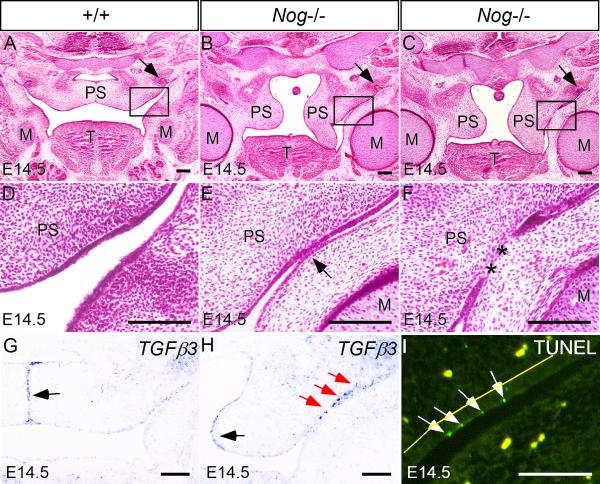
Palate elevation defects could arise from multiple etiologies, such as intrinsic elevation force deficiency, extrinsic blockage by the tongue, or abnormal fusion between palate shelf and adjacent structures (Gritli-Linde, 2007). As shown in Fig. 2, while the anterior portion of the mutant palate elevated normally, the posterior palate failed to elevate, apparently due to an aberrant palate-mandible fusion. To reveal the process of the abnormal fusion, we examined serial histological sections of the Nog-/- palatal shelves at E14.5 when they would have elevated to the horizontal position above the tongue (Fig. 6A). We observed an initial adhesion of the palatal epithelium with the mandibular epithelium, followed by an elimination of the adhered epithelia, forming a mesenchymal continuity between the palatal shelf and the mandible. Similar abnormal palate-mandible fusion phenotype has been reported in Fgf10-/- mice (Rice et al., 2004; Alappat et al., 2005). In Fgf10 mutants, cell apoptosis induced by ectopic Tgfβ3 expression was thought to attribute to the disruption of palatal epithelial integrity and abnormal fusion (Alappat et al., 2005). The phenotypic similarity between Nog-/- and Fgf10-/- mice promoted us to examine if the expression of Fgf10 and its downstream effectors was altered in the Nog-/- palate. We did not see a down-regulation of Fgf10 expression in the Nog-/- palate (data not shown). However, we indeed observed an ectopic Tgfβ3 expression in the oral side epithelium of the mutant posterior palatal shelves prior to adhesion (Fig. 6H), although Tgfβ3 expression in the mutant MEE remained comparable to that in wild type controls where palatal shelves just met at the midline (Fig. 6G, H). Associated with this ectopic activation of Tgfβ3 expression is aberrant induction of apoptosis in the periderm cells prior to adhesion (Fig. 6I). Together with our pSmad1/5/8 immunostaining results, these results suggest that Noggin functions downstream of or in parallel to Fgf10 in inhibiting Tgfβ3 expression in the oral side palatal epithelium, and an elevated BMP/Smad signaling in the posterior palate can activate Tgfβ3 expression and induce epithelial cell apoptosis, which consequently disrupts palatal epithelium integrity and leads to abnormal palate-mandible fusion.
Figure 6.
Nog-/- embryo shows abnormal palate-mandible fusion in the posterior palatal region. (A, D) At E14.5, the wild type palatal shelves have elevated to a position above the tongue and merged at the midline. (B, C, E, F) In the Nog-/- embryo at the comparable stage, the palatal shelves remain in a vertical position, showing progressive adhesion (E) and fusion (F) with the mandible. The asterisks denote a confluence of mesenchyme. The presence of the maxillary molar (arrows in A, B and C) marks the level of the sections. (G, H) Tgfβ3 is expressed in the MEE of the wild type palatal shelves (G); in the Nog-/- palate, Tgfβ3 expression is not affected in the MEE (black arrow in H), but is ectopically activated in the oral side epithelium (red arrows in H) where aberrant palate-mandible adhesion occurs. (I) TUNEL assay detects ectopic apoptotic periderm cells (arrows) in the oral side palatal epithelium prior to abnormal palate-mandible adhesion. No apoptotic cells are observed in the adjacent mandible epithelium. The yellow fluorescent dots in (I) represent autofluorescence from blood cells. The yellow line marks the basement membrane between palatal mesenchyme and the basal epithelial layer. M, Meckel's cartilage; T, tongue; PS, palatal shelf. Scale bars represent 100 μm.
Ectopic activation of BMP signaling in the palate epithelium recapitulates the abnormal palate-mandible fusion phenotype in Nog-/- mice
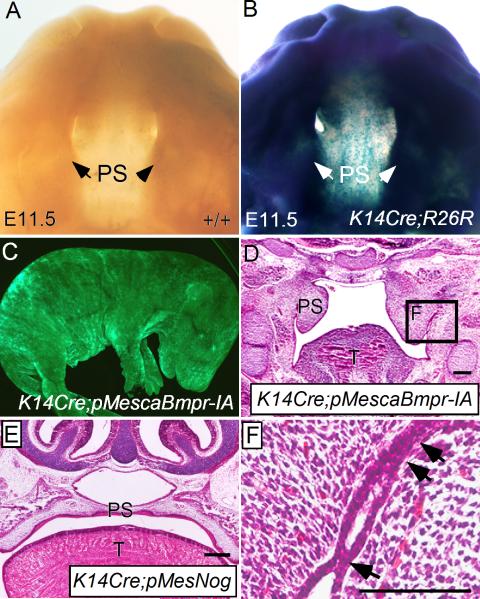
To test if ectopic activation of BMP signaling in the developing palatal epithelium could recapitulate the abnormal palate-mandible fusion phenotype observed in Nog-/- mice, we generated conditional transgenic mice that express a constitutively active form of BMP receptor-IA (pMescaBmpr-IA) or a constitutively active form of BMP receptor-IB (pMescaBmpr-IB) upon crossing to a Cre mouse line. These mutated forms of BMP receptors have been demonstrated to activate BMP signaling pathway in the absence of BMP ligands (Zou et al., 1997; Zhang et al., 2000). Using a K14-Cre mouse line (Andl et al., 2004; Xu et al., 2008), we ectopically expressed caBmpr-IA or caBmpr-IB in embryonic epithelium, confirmed by assessing expression of an Egfp transgene integrated into the transgenic vector (Fig. 7C). As shown in Fig. 7B, this K14-Cre transgenic allele was able to activate Rosa26 reporter expression uniformly in craniofacial region as early as E11.5, including the developing palatal shelves. We found that mice carrying the K14-Cre and caBmpr-IB double transgenic alleles (K14Cre;caBmpr-IB) did not develop a cleft palate defect (data not shown). However, these mice in fact developed severe ichthyosis-like skin phenotype postnatally, indicating a functional transgenicc allele (Yu, X. et al., unpublished results). The K14Cre;pMescaBmpr-IA mice, however, exhibited a cleft palate phenotype (data not shown). Histological analyses revealed an abnormal palate-mandible fusion in the posterior palate region at E14.5, resembling the phenotype found in Nog-/- mice (Fig. 7D, F). We have shown previously that transgenic overexpression of Noggin in the palatal mesenchyme causes a cleft palate defect (Xiong et al., 2009). Using the same conditional Noggin transgenic line (pMesNog), we overexpressed Noggin in the palatal epithelium by compounding the K14-Cre allele with the pMesNog conditional transgenic allele (Supplemental Fig. 1G). While mice carrying the K14Cre and pMesNog transgenic alleles exhibited an arrest of tooth development at the lamina/early bud stage (data not shown), they did not develop a cleft palate defect (Fig. 7E), consistent with the previous report that mice carrying the K14-Noggin transgenic allele survived to adulthood (Plikus et al., 2005). These results support the idea that repression of BMP activities by Noggin in the palatal epithelium is required for the maintenance of palatal epithelium integrity and is essential for normal palatogenesis.
Figure 7.
Ectopic activation of BMP signaling in the palatal epithelium recapitulates the palate-mandible fusion phenotype observed in Nog-/- mice (A) An E11.5 wild type embryonic head shows negative staining of β-galactosidase. (B) An E11.5 K14Cre;R26R embryonic head shows staining of β-galactosidase in the craniofacial region, including the palatal shelves. (C) An E17.5 K14Cre;pMescaBmpr-IA embryo shows GFP expression throughout the entire embryo. (D) An E14.5 K14Cre;pMescaBmpr-IA embryo exhibits abnormal palate-mandible fusion and failure in palatal shelf elevation in the posterior portion. (E) An E16.5 K14Cre;pMesNog embryo shows a normally formed palate. (F) Higher magnification of the defined area in (D), showing an abnormal palate-mandible adhesion/fusion sites (arrows). T, tongue; PS, palatal shelf. Scale bars represent 100 μm.
Discussion
In this paper, we report the expression pattern of the BMP antagonist Noggin in the mouse developing palate, and show that inactivation of Noggin causes a cleft palate formation. We demonstrate that both cell proliferation and cell apoptosis are deregulated in the Nog-/- palate. Particularly, we found abnormal cell apoptosis in periderm cells in the oral side of the posterior palate, which leads to aberrant palate-mandible fusion and blocks the normal palatal elevation. We further show that ectopic activation of BMP signaling in the palatal epithelium recapitulates the abnormal palate-mandible fusion phenotype observed in Nog-/- mice. Our studies thus demonstrate that repression of BMP activities by Noggin is required for palate epithelium integrity, which is essential for normal palate development.
Palatal epithelium integrity and palate development
Epithelial-mesenchymal interaction controls each step of palate development. Palatal epithelium thus plays an essential role in palate shelf outgrowth, elevation, fusion and further differentiation (Ferguson, 1988). In addition to these aspects, recent studies have demonstrated that the integrity of palatal epithelium is particularly required for palate elevation (Alappat et al., 2005; Casey et al., 2006; Jiang et al., 1998; Richardson et al., 2006; 2009; Xiong et al., 2009). In normal palate development, the palatal shelves may contact the adjacent tissues, including tongue and mandible. However, a clear epithelial boundary is always maintained at the contact interface, preventing abnormal adhesion and fusion between palate and these structures. In pathological conditions, when the epithelium integrity is disrupted, abnormal adhesion or fusion occurs. Abnormal adhesion is observed in Irf6 heterozygous mice: the palatal epithelium adheres to the mandible and the tongue; however, the palatal shelves still manage to elevate and fuse to form an intact structure despite the abnormal epithelial adhesion (Richardson et al., 2006; 2009). In the event that adhesion is further deregulated, with elimination of the epithelial seam, (i.e., when a mesenchymal continuity is formed between palatal shelves and adjacent structures), palatal shelf elevation is hampered, as observed in Irf6 homozygous mice (Richardson et al., 2006; 2009). Similar abnormal palate-mandible or palate-tongue fusion phenotypes have also been reported in Jagged2 mutant, Fgf10 mutant and Hand2 hypomorphic mice (Alappat et al., 2005; Casey et al., 2006; Xiong et al., 2009). In these animal models, ectopic cell apoptosis is a common cause of disruption in epithelium integrity. However, since adhesion/fusion always occurs between two epithelial surfaces, disruption of either surface could lead to abnormal adhesion/fusion. In Jagged2 mutants, the abnormal palate-tongue fusion phenotype results from an abnormal tongue epithelial differentiation, despite normally developed palatal shelves as assessed at histological, cellular, and molecular levels (Casey et al., 2006). In Hand2 hypomorphic and Fgf10 mutant mice, the abnormal palate-mandible fusion has been attributed to ectopic cell apoptosis within the palatal epithelium (Alappat et al., 2005; Xiong et al., 2009). As shown in this study, in E13.5 Noggin mutants, ectopic apoptotic cells were found in the oral side epithelium of the anterior palate and in the mandibular epithelium, but not in the posterior palate (Fig. 5G). Since the anterior palatal shelves do not make contact with the mandible, an abnormal palate-mandible fusion does not occur. However, in the posterior palate at E14.5, we observed ectopic cell death in the oral side palatal epithelium, but not in the mandibular epithelium, where abnormal fusion occurred (Fig. 6I). Here, aberrant apoptosis was initially detected in periderm cells in the abnormal fusion region prior to making contact with the mandibular epithelium. A similar phenotype was also observed in Hand2 hypomorphic mice (Xiong et al., 2009). During normal palate development, periderm cells in the MEE region are removed prior to palatal shelf contact and subsequent fusion (Fitchett and Hay, 1989), and artificial removal of periderm cells causes degradation of the basal layer cells in the MEE (Cuervo and Covarrubias, 2004). Together, these results support an essential role for periderm cells in the maintenance of palatal epithelial integrity.
Differential cellular and gene expression responses to the absence of Noggin along the A-P axis of developing palate
It has been well documented that the developing secondary palatal shelf is an asymmetric structure along the anterior-posterior axis (Zhang et al., 2002; Alappat et al., 2005; Yu et al., 2005; He et al., 2008). The anterior and posterior portions of the palatal shelves differ not only at the cellular level, but also at the molecular level (reviewed in Hilliard, 2005; Okano et al., 2006; Gritli-Linde, 2007). Noggin is expressed in the oral side palatal epithelium in both anterior and posterior palate shelves and Noggin deficiency leads to ectopic activation of BMP/Smad signaling in these structures. Associated with this enhanced/ectopic BMP/Smad signaling is ectopic cell death in both anterior and posterior palatal epithelium. However, it is interesting to note that the timing of the apoptosis occurring varies along the A-P axis. In the anterior palatal epithelium of Nog-/- embryo, ectopic cell death was detected as early as E13.5, while in the posterior palate, ectopic cell death was not detected until E14.5, suggesting that molecular mechanisms underlying the cell death are different along the A-P axis. This idea is supported by the fact that Tgfβ3, which is required for cell death in the MEE during normal fusion of palate, is ectopically expressed in the posterior palatal epithelium, but not in the anterior counterpart. These results suggest that the ectopic Tgfβ3 expression may mediate the cell death in the posterior palate. The ectopic cell apoptosis found in the anterior palatal epithelium in Noggin mutant is mediated by a different mechanism.
In addition, the palatal mesenchymal cells of the anterior and posterior palate also respond differentially to the loss of Noggin. At the cellular level, the anterior palatal mesenchyme exhibits a down-regulated cell proliferation rate; while in the posterior palate, the cell proliferation rate is not affected (Fig. 5E). In terms of gene expression, Bmp2 expression was down-regulated in the anterior palatal mesenchyme, but remained unaltered in the posterior palatal mesenchyme (Fig. 4). It has been shown previously that Bmp2 acts downstream of Bmp4 and Shh to stimulate cell proliferation in the anterior palatal mesenchyme (Zhang et al., 2002). This down-regulation of Bmp2 expression appears to be responsible for the reduction in Smad1/5/8 phosphorylation in the anterior palatal mesenchyme of Noggin mutant, which contributes to the reduced cell proliferation rate. While Smad1/5/8 phosphorylation was enhanced in the posterior palatal mesehcyme in the absence of Noggin, the cell proliferation rate was unaltered, consistent with the previous finding that neither BMP2 nor BMP4 regulate cell proliferation in the posterior palatal mesenchyme (Zhang et al., 2002; Hilliard et al., 2005). In the posterior palate of Noggin mutant, Bmp2 expression was not changed in the mesenchyme, but was upregulated in the oral side epithelium. Although we cannot rule out the possibility that the expression and/or activity of other BMPs is upregulated in the posterior palate in the absence of Noggin, the upregulated Bmp2 expression in the oral side epithelium could count for, at least partially, the ectopic Smad1/5/8 phosphorylation in the oral side mesenchyme of the Nog-/- posterior palate.
It was shown previously that application of exogenous Noggin protein to the globular process of the frontonasal mass of developing chick embryo induces clefts of the primary palate (Ashique et al., 2002). However, in Nog-/- mice, the absence of Noggin leads to a complete lack of the primary palate structure (Fig. 1J). These observations indicate that BMP homeostasis is also essential for development of the primary palate. There are many lines of evidence showing requirement of BMP signaling and BMP homeostasis in palate development (Matzuk et al., 1995; Zhang et al., 2002; Dudas et al., 2004; Liu et al., 2005; Xiong et al., 2009). Interestingly, while overexpression of Noggin in the palatal mesenchyme leads to cleft palate formation (Xiong et al., 2009), Noggin overexpression in the palatal epithelium does not cause a cleft palate defect (Fig. 7E, and Supplemental Fig. 1F), despite of the fact that K14Cre;pMesNog mice show an arrest of tooth development at the lamina/early bud stage (data not shown). This is likely due to different amounts of transgenic Noggin produced in the palatal mesenchyme and the palatal epithelium. The different responses of the developing palate and tooth to Noggin overexpression in the epithelium could be attributed to differential sensitivity to altered BMP signaling.
Bmp4 is unable to compensate for the reduction of Bmp2 expression in the anterior palate in vivo
While it remains to be determined how Bmp2 expression is differentially regulated in the developing palate along the A-P axis in the Noggin mutant, it is interesting to note that Bmp4 expression is unaltered. BMPs, including BMP2, BMP4 and BMP7, have been shown to function as mitogens in developing craniofacial structures (Barlow and Francis-West, 1997; Wang et al., 1999; Ashique et al., 2002; Zhang et al., 2002). Bmp2 and Bmp4 are expressed in a partially overlapping pattern in the anterior palate mesenchyme. Although exogenously applied BMP2 or BMP4 can equally induce cell proliferation in the anterior palatal mesenchyme in vitro (Zhang et al., 2002; Hilliard et al., 2005), the unaltered Bmp4 expression in the Nog-/- palate suggests that Bmp4 fails to compensate for the reduction of Bmp2 expression, in terms of activating BMP/Smad signaling and regulating cell proliferation, in the anterior palatal mesenchyme in vivo. Similar results have also been previously demonstrated in an in vitro study in which blocking Shh signaling did not affect Bmp4 expression but eliminated Bmp2 expression and subsequently inhibited cell proliferation in the anterior palatal mesenchyme (Zhang et al., 2002). Thus, Bmp4 appears to maintain Msx1 expression in the mesenchyme and Shh expression in the MEE, while Bmp2 is primarily responsible for the regulation of cell proliferation within the anterior palatal mesenchyme. This hypothesis is supported by the fact that expression pattern of pSmad1/5/8 in the developing palate is almost identical to that of Bmp2 in both the anterior and posterior portion (Fig. 3; Fig. 4). In addition, we also observed a significantly reduced Shox2 expression in the anterior palatal mesenchyme of Noggin mutants. We have also demonstrated previously that BMP activities are not sufficient to induce ectopic Shox2 expression, but are necessary for Shox2 expression in the anterior palatal mesenchyme (Yu et al., 2005). Given the fact that Bmp4 expression remains unaltered in the Nog-/- anterior palate, it cannot maintain Shox2 expression at the wild type level. BMP4 and BMP2 indeed have differential potency in activating BMP/Smad signaling (Upton et al., 2008), and BMP2/BMP4 heterodimers are more potent than homodimers of any of the proteins in activating BMP/Smad signaling. This functional difference is likely due to different binding affinities between specific BMP ligands and receptors, or availability of BMP receptors in specific cell context. Currently, three type I receptors (BMPR-IA, BMPR-IB, and ActRIa) and three type II receptors (BMPR-II; ActRIIA, and ActRIIB) have been identified to bind to BMP ligands with different affinities (Sieber et al., 2009). Expression of BMP receptors and their functions in palate development warrant future investigation.
BMPR-IA likely mediates overactive BMP signaling in disruption of palatal epithelial integrity
During development and disease, BMP signaling is involved in the regulation of multiple cellular processes, including proliferation and apoptosis. In the palatal epithelium of Noggin mutants, in which BMP signaling is overactive, we observed the simultaneous occurrence of elevated level of cell proliferation and excess apoptotic cells. This simultaneous elevation of cell proliferation rate and apoptosis was also observed in the palatal mesenchyme in mice bearing inactivation of Alk5 in neural crest cells and their derivatives (Dudas et al., 2006). In the posterior palate of Noggin mutant, the excess cell apoptosis in the palatal epithelium resulted in abnormal palate-mandible fusion. This phenotype was recapitulated by ectopic expression of a constitutively active form of BMPR-IA in the palatal epithelium. In contrast, ectopic expression of a constitutively active form of BMPR-IB did not produce any obvious palate defect. Since Bmp2 expression and Smad1/5/8 phosphorylation were elevated in the region where excess cell death and abnormal fusion occurred, it appears that overexpressed BMP2 acts through BMPR-IA and Smad-dependent pathway in this context, leading to the disruption of palatal epithelial integrity. This view is supported by the observations of high binding affinity of BMP2 to BMPR-IA (Knaus and Sebald, 2001), and Bmpr-IA but not Bmpr-IB is strongly expressed in the palatal epithelium during palatogenesis (Li, L. and Chen, Y.P., unpublished results).
In conclusion, our studies show an absolute requirement for Noggin in normal palate development. The absence of Noggin alters the levels of BMP signaling, causing aberrant cell proliferation and cell death in the developing palate. Repression of BMP activities by Noggin in the palatal epithelium is essential for the maintenance of epithelium integrity. Our results highlight the importance of fine-tuned BMP signaling in palate formation.
Supplementary Material
Acknowledgements
This work was supported by the NIH grants to Y.P.C., J.K., and Y.C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alappat SR, Zhang Z, Suzuki K, Zhang X, Liu H, Jiang R, Yamada G, Chen YP. The cellular and molecular etiology of the cleft secondary palate in Fgf10 mutant mice. Dev. Biol. 2005;277:102–113. doi: 10.1016/j.ydbio.2004.09.010. [DOI] [PubMed] [Google Scholar]
- Anderson RM, Stottmann RW, Choi M, Klingensmith J. Endogenous bone morphogenetic protein antagonists regulate mammalian neural crest generation and survival. Dev. Dyn. 2006;235:2507–2520. doi: 10.1002/dvdy.20891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andl T, Ahn K, Kairo A, Chu EY, Wine-Lee L, Reddy ST, Croft NJ, Cebra-Thomas JA, Metzger D, Chambon P, Lyons KM, Mishina Y, Seykora JT, Crenshaw EB, 3rd., Millar SE. Epithelial Bmpr1a regulates differentiation and proliferation in postnatal hair follicles and is essential for tooth development. Development. 2004;131:2257–2268. doi: 10.1242/dev.01125. [DOI] [PubMed] [Google Scholar]
- Bachiller D, Klingensmith J, Kemp C, Belo JA, Anderson RM, May SR, McMahon JA, McMahon AP, Harland RM, Rossant J, De Robertis EM. The organizer factors Chordin and Noggin are required for mouse forebrain development. Nature. 2000;403:658–661. doi: 10.1038/35001072. [DOI] [PubMed] [Google Scholar]
- Balemans W, Van Hul W. Extracellular regulation of BMP signaling in vertebrates: a cocktail of modulators. Dev. Biol. 2002;250:231–250. [PubMed] [Google Scholar]
- Barlow AJ, Francis-West PH. Ectopic application of recombinant BMP-2 and BMP-4 can change patterning of developing chick facial primordial. Development. 1997;124:391–398. doi: 10.1242/dev.124.2.391. [DOI] [PubMed] [Google Scholar]
- Brunet LJ, McMahon JA, McMahon AP, Harland RM. Noggin, cartilage morphogenesis, and joint formation in the mammalian skeleton. Nature. 1998;280:1455–1457. doi: 10.1126/science.280.5368.1455. [DOI] [PubMed] [Google Scholar]
- Casey LM, Lan Y, Cho ES, Maltby KM, Gridley T, Jiang R. Jag2-Notch1 signaling regulates oral epithelial differentiation and palate development. Dev. Dyn. 2006;235:1830–1844. doi: 10.1002/dvdy.20821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y, Jiang X, Ito Y, Bringas P, Jr., Han J, Rowitch DH, Soriano P, McMahon AP, Sucov HM. Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development. 2000;127:1671–1679. doi: 10.1242/dev.127.8.1671. [DOI] [PubMed] [Google Scholar]
- Chen D, Zhao M, Mundy GR. Bone Morphogenetic Proteins. Growth Factors. 2004;22:233–241. doi: 10.1080/08977190412331279890. [DOI] [PubMed] [Google Scholar]
- Cuervo R, Covarrubias L. Death is the major fate of medial edge epithelial cells and the cause of basal lamina degradation during palatogenesis. Development. 2004;131:15–24. doi: 10.1242/dev.00907. [DOI] [PubMed] [Google Scholar]
- Dudas M, Sridurongrit S, Nagy A, Okazaki K, Kaartinen V. Craniofacial defects in mice lacking BMP type I receptor Alk2 in neural crest cells. Mech. Dev. 2004;121:173–182. doi: 10.1016/j.mod.2003.12.003. [DOI] [PubMed] [Google Scholar]
- Dudas M, Kim J, Li WY, Nagy A, Larsson J, Karlsson S, Chai Y, Kaartinen V. Epithelial and ectomesenchymal role of the type I TGF-beta receptor ALK5 during facial morphogenesis and palatal fusion. Dev. Biol. 2006;296:298–314. doi: 10.1016/j.ydbio.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson MWJ. Palate development. Development. 1988;103(Suppl.):41–60. doi: 10.1242/dev.103.Supplement.41. [DOI] [PubMed] [Google Scholar]
- Ferguson MW, Honig LS. Epithelial-msenchymal interactions during vertebrate palatogenesis. In: Zimmerman EF, editor. Current Topics in Developmental Biology, Palate Development: Normal and Abnormal, Cellular and Molecular Aspects'. Academic Press; New York: 1984. pp. 137–164. [DOI] [PubMed] [Google Scholar]
- Fitchett JE, Hay ED. Medial edge epithelium transforms to mesenchyme after embryonic palatal shelves fues. Dev. Biol. 1989;131:455–474. doi: 10.1016/s0012-1606(89)80017-x. [DOI] [PubMed] [Google Scholar]
- Gazzerro E, Canalis E. Bone morphogenetic proteins and their antagonists. Rev. Endocr. Metab. Disord. 2006;7:51–65. doi: 10.1007/s11154-006-9000-6. [DOI] [PubMed] [Google Scholar]
- Gritli-Linde A. Molecular control of secondary palate development. Dev. Biol. 2007;301:309–326. doi: 10.1016/j.ydbio.2006.07.042. [DOI] [PubMed] [Google Scholar]
- Groppe J, Greenwald J, Wiater E, Rodriguez-Leon J, Economides AN, Kwiatkowski W, Affolter M, Vale WW, Belmonte JC, Choe S. Structural basis of BMP signalling inhibition by the cystine knot protein Noggin. Nature. 2002;420:636–642. doi: 10.1038/nature01245. [DOI] [PubMed] [Google Scholar]
- Hall BK. Cell-Cell Interactions in Craniofacial Growth and Development. In: Davidovitch Z, editor. Biological Mechanisms of Tooth Movement and Craniofacial Adaptation. Ohio State Univ.; Columbus: 1992. pp. 11–17. [Google Scholar]
- He F, Xiong W, Yu X, Espinoza-Lewis R, Liu C, Gu S, Nishita M, Suzuki K, Yamada G, Minami Y, Chen YP. Wnt5a regulates directional cell migration and cell proliferation via Ror2-mediated non-canonical pathway in mammalian palate development. Development. 2008;135:3871–3879. doi: 10.1242/dev.025767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliard SA, Yu L, Gu S, Zhang Z, Chen YP. Regional regulation of palatal growth and patterning along the anterior-posterior axis in mice. J. Anat. 2005;207:655–667. doi: 10.1111/j.1469-7580.2005.00474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y, Yeo JY, Chytil A, Han J, Bringas P, Jr., Nakajima A, Shuler CF, Moses HL, Chai Y. Conditional inactivation of Tgfbr2 in cranial neural crest causes cleft palate and calvaria defects. Development. 2003;130:5269–5280. doi: 10.1242/dev.00708. [DOI] [PubMed] [Google Scholar]
- Jiang R, Lan Y, Chapman HD, Shawber C, Norton CR, Serreze DV, Weinmaster G, Gridley T. Defects in limb, craniofacial, and thymic development in Jagged2 mutant mice. Genes Dev. 1998;12:1046–1057. doi: 10.1101/gad.12.7.1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knaus P, Sebald W. Cooperativity of binding epitopes and receptor chains in the BMP/TGFbeta superfamily. Biol. Chem. 2001;382:1189–1195. doi: 10.1515/BC.2001.149. [DOI] [PubMed] [Google Scholar]
- Levi G, Mantero S, Barbieri O, Cantatore D, Paleari L, Beverdam A, Genova F, Robert B, Merlo GR. Msx1 and Dlx5 act independent in development of craniofacial skeleton, but converge on the regulation of Bmp signaling in palate formation. Mech. Dev. 2006;123:3–16. doi: 10.1016/j.mod.2005.10.007. [DOI] [PubMed] [Google Scholar]
- Liu W, Sun X, Braut A, Mishina Y, Behringer RR, Mina M, Martin JF. Distinct functions for Bmp signaling in lip and palate fusion in mice. Development. 2005;132:1453–1461. doi: 10.1242/dev.01676. [DOI] [PubMed] [Google Scholar]
- Lu H, Jin Y, Tipoe GL. Alteration in the expression of bone morphogenetic protein-2,3,4,5 mRNA during pathogenesis of cleft palate in BALB/c mice. Arch. Oral Biol. 2000;45:133–40. doi: 10.1016/s0003-9969(99)00118-1. [DOI] [PubMed] [Google Scholar]
- Matzuk MM, Kumar TR, Vassalli A, Bickenbach JR, Roop DR, Jaenisch R, Bradley A. Functional analysis of activins during mammalian development. Nature. 1995;374:354–356. doi: 10.1038/374354a0. [DOI] [PubMed] [Google Scholar]
- McMahon JA, Takada S, Zimmerman LB, Fan CM, Harland RM, McMahon AP. Noggin-mediated antagonism of BMP signaling is required for growth and patterning of the neural tube and somite. Genes Dev. 1998;12:1438–1452. doi: 10.1101/gad.12.10.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie X. Differential expression of Bmp2, Bmp4 and Bmp3 in embryonic development of mouse anterior and posterior palate. Chin. Med. J. (Engl) 2005;118:1710–1716. [PubMed] [Google Scholar]
- Nie X, Luukko K, Kettunen P. BMP signaling in craniofacial development. Int. J. Dev. Biol. 2006;50:511–521. doi: 10.1387/ijdb.052101xn. [DOI] [PubMed] [Google Scholar]
- Okano J, Suzuki S, Shiota K. Regional heterogeneity in the developing palate: morphological and molecular evidence for normal and abnormal palatogenesis. Congenit. Anom.(Kyoto) 2006;46:49–54. doi: 10.1111/j.1741-4520.2006.00103.x. [DOI] [PubMed] [Google Scholar]
- Plikus MV, Zeichner-David M, Mayer J-A, Reyna J, Bringas P, Thewissen JGM, Snead ML, Chai Y, Chuong C-M. Morphoregulation of teeth: modulation the number, size, shape and differentiation by tuning Bmp activity. Evol. Dev. 2005;7:440–457. doi: 10.1111/j.1525-142X.2005.05048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice R, Spencer-Dene B, Connor EC, Gritli-Linde A, McMahon AP, Dickson C, Thesleff I, Rice DP. Disruption of Fgf10/Fgfr2b coordinated epithelial-mesenchymal interactions causes cleft palate. J. Clin. Invest. 2004;113:1692–1700. doi: 10.1172/JCI20384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson RJ, Dixon J, Malhotra S, Hardman MJ, Knowles L, Boot Handford RP, Shore P, Whitmarsh A, Dixon MJ. Irf6 is a key determinant of the keratinocyte proliferation-differentiation switch. Nat. Genet. 2006;38:1329–1334. doi: 10.1038/ng1894. [DOI] [PubMed] [Google Scholar]
- Richardson RJ, Dixon J, Jiang R, Dixon MJ. Integration of IRF6 and Jagged2 signalling is essential for controlling palatal adhesion and fusion competence. Hum. Mol. Genet. 2009;18:2632–42. doi: 10.1093/hmg/ddp201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber C, Kopf J, Hiepen C, Knaus P. Recent advances in BMP receptor signaling. Cytokine Growth Factor Rev. 2009;20:343–355. doi: 10.1016/j.cytogfr.2009.10.007. [DOI] [PubMed] [Google Scholar]
- St. Amand TR, Zhang Y, Semina EV, Zhao X, Hu YP, Nguyen L, Murray JC, Chen YP. Antagonistic signals between BMP4 and FGF8 define the expression of Pitx1 and Pitx2 in mouse tooth-forming anlage. Dev. Biol. 2000;217:323–332. doi: 10.1006/dbio.1999.9547. [DOI] [PubMed] [Google Scholar]
- Stottmann RW, Anderson RM, Klingensmith J. The BMP antagonists Chordin and Noggin have essential but redundant roles in mouse mandibular outgrowth. Dev. Biol. 2001;240:457–473. doi: 10.1006/dbio.2001.0479. [DOI] [PubMed] [Google Scholar]
- Taya Y, Okane S, Ferguson MWJ. Pathogenesis of cleft palate in TGF-β3 knockout mice. Development. 1999;126:3869–3879. doi: 10.1242/dev.126.17.3869. [DOI] [PubMed] [Google Scholar]
- Upton PD, Long L, Trembath RC, Morrell NW. Functional characterization of bone morphogenetic protein binding sites and Smad1/5 activation in human vascular cells. Mol. Pharmacol. 2008;73:539–552. doi: 10.1124/mol.107.041673. [DOI] [PubMed] [Google Scholar]
- Wang Y-H, Rutherford B, Uphot W, Mina M. Effects of BMP-7 on mouse tooth mesenchyme and chick mandibular mesenchyme. Dev. Dyn. 1999;216:320–335. doi: 10.1002/(SICI)1097-0177(199912)216:4/5<320::AID-DVDY2>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Xiong W, He F, Morikawa Y, Yu X, Zhang Z, Lan Y, Jiang R, Cserjesi P, Chen YP. Hand2 is required in the epithelium for palatogenesis in mice. Dev. Biol. 2009;330:131–141. doi: 10.1016/j.ydbio.2009.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Han J, Ito Y, Bringas P, Jr., Deng C, Chai Y. Ectodermal Smad4 and p38 MAPK are functionally redundant in mediating TGF-β/BMP signaling during tooth and palate development. Dev. Cell. 2008;15:322–329. doi: 10.1016/j.devcel.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Gu S, Alappat S, Song Y, Yan M, Zhang X, Zhang G, Jiang Y, Zhang Z, Zhang Y, Chen YP. Shox2-deficient mice exhibit a rare type of incomplete clefting of the secondary palate. Development. 2005;132:4397–4406. doi: 10.1242/dev.02013. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Song Y, Zhao X, Zhang X, Fermin C, Chen YP. Rescue of cleft palate in Msx1-deficient mice by transgenic Bmp4 reveals a network of BMP and Shh signaling in the regulation of mammalian palatogenesis. Development. 2002;129:4135–4146. doi: 10.1242/dev.129.17.4135. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Yu X, Zhang Y, Geronimo B, Lovlie A, Fromm SH, Chen YP. Targeted misexpression of constitutively active BMP receptor-IB causes bifurcation and duplication and posterior transformation of digit in mouse limb. Dev. Biol. 2000;220:154–167. doi: 10.1006/dbio.2000.9637. [DOI] [PubMed] [Google Scholar]
- Zimmerman LB, De Jesus-Escobar JM, Harland RM. The Spemann organizer signal noggin binds and inactivates bone morphogenetic protein 4. Cell. 1996;86:599–606. doi: 10.1016/s0092-8674(00)80133-6. [DOI] [PubMed] [Google Scholar]
- Zou H, Wieser R, Massagué J, Niswander L. Distinct roles of type I bone morphogenetic protein receptors in the formation and differentiation of cartilage. Genes Dev. 1997;11:2191–2203. doi: 10.1101/gad.11.17.2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.