Abstract
Though tyrosine kinase inhibitors have redefined the care of chronic myeloid leukemia (CML), these agents have not proved curative, likely due to resistance of the leukemia stem cells (LSC). While a number of potential therapeutic targets have emerged in CML, their expression in the LSC remains largely unknown. We therefore isolated subsets of CD34+ stem/progenitor cells from normal donors and from patients with chronic phase or blast crisis CML. These cell subsets were then characterized based on ability to engraft immunodeficient mice and expression of candidate therapeutic targets. The CD34+CD38− CML cell population with high aldehyde dehydrogenase (ALDH) activity was the most enriched for immunodeficient mouse engrafting capacity. The putative targets: PROTEINASE 3, SURVIVIN, and hTERT were expressed only at relatively low levels by the CD34+CD38−ALDHhigh CML cells, similar to the normal CD34+CD38−ALDHhigh cells and less than in the total CML CD34+ cells. In fact, the highest expression of these antigens was in normal, unfractionated CD34+ cells. In contrast, PRAME and WT1 were more highly expressed by all CML CD34+ subsets than their normal counterparts. Thus, ALDH activity appears to enrich for CML stem cells, which display an expression profile that is distinct from normal stem/progenitor cells and even the CML progenitors. Indeed, expression of a putative target by the total CD34+ population in CML does not guarantee expression by the LSC. These expression patterns suggest that PROTEINASE 3, SURVIVIN, and hTERT are not optimal therapeutic targets in CML stem cells; whereas PRAME and WT1 seem promising.
Keywords: chronic myeloid leukemia, CML, leukemia stem cell, WT1, PRAME
Introduction
The advent of tyrosine kinase inhibitors redefined the treatment of chronic myeloid leukemia (CML).[1] Complete cytogenetic response rates can now be seen in up to 90% of newly diagnosed patients in chronic phase (CP).[2] However, these “targeted” therapies have yet to prove curative. In fact, relapse is common after discontinuation of imatinib, even for patients in complete molecular remission at the time of cessation.[3, 4] This appears to be due to resistance of CML stem cells to the pro-apoptotic effects of imatinib[5, 6] and even newer tyrosine kinase inhibitors, such as dasatinib.[7] Indeed, primitive leukemic progenitors still can be readily detected in CML patients who have achieved complete cytogenetic remission on imatinib.[8] Blast crisis (BC) CML presents an even greater challenge, where in contrast to CP CML, tyrosine kinase inhibition rarely results in durable remission.[9] Hence, there has been a search for other therapeutic targets in CML, particularly on the leukemia stem cell (LSC), which in theory must be eradicated to achieve cure.[10–12]
A number of candidates have emerged as potential therapeutic targets in leukemia. Among the more promising are: the Wilm’s tumor gene (WT1), SURVIVIN, the preferentially expressed antigen of melanoma (PRAME), PROTEINASE 3 (PR3), and hTERT (the enzymatic component of telomerase). All of these are immunogenic, and each is over-expressed to varying degrees in many cancers, including CML.[13] Many of these candidate targets have been implicated in therapeutic resistance, including inhibition of apoptosis, and appear to correlate with prognosis.[13–15] There are ongoing vaccine trials targeting many of these antigens,[13, 16] as well as early phase clinical trials of pharmacologic inhibitors of telomerase[17] and SURVIVIN.[18] Presumably, any such new therapies will have curative potential only if their targets are actually expressed by the LSC. However, the expression of these putative targets in CML stem cells is largely unknown.
Indeed, existing data are limited regarding the precise characterization of CML stem cells; and expression of a gene by the differentiated leukemic bulk does not necessarily guarantee expression by the LSC. In fact, in many respects LSC more closely resemble normal hematopoietic stem cells (HSC) than their own differentiated leukemic progeny.[19–21] Nonetheless, it is expected that qualitative or quantitative expression of some genes must distinguish LSC from their normal counterparts. Accordingly, an optimal therapeutic target would not only be highly expressed by the LSC (and ideally their progeny as well); but it would also be absent or only minimally expressed in normal HSC to avoid unacceptable toxicity.[13]
LSC research to date has been impeded by the relative rarity of these cells, as well as the lack of a consensus on their exact phenotype. LSC are often phenotypically defined as simply the CD34+ leukemia cells or, occasionally, the more enriched CD34+CD38− subset; but even the CD34+CD38− cells are a heterogeneous population, of which the LSC constitute only a fraction.[19, 22–24] The CD34+CD38− population can be further refined for stem cells based on low side scatter and high aldehyde dehydrogenase (ALDH) activity.[23] ALDH, specifically the ALDH1A1 isoenzyme, mediates the biosynthesis of all-trans-retinoic acid, as well as the detoxification of a variety of compounds such as ethanol and active metabolites of cyclophosphamide;[25] and it is typically present at higher levels in adult stem cells, than in their differentiated progeny.[22–27] The fluorescently labeled ALDH1A1 substrate, Aldefluor, permits the isolation of viable normal and cancer stem cells.[22–24, 26–28]
Here we report that CML stem cells are characterized by high ALDH expression, and that these cells have a unique expression profile of putative targets as compared to both the more differentiated CML progenitors and normal HSC.
Methods
Patient and normal donor specimens
Bone Marrow and/or peripheral blood samples were obtained from a total of 14 patients with CML (10 CP and 4 myeloid BC). None of the BC CML patients had a duplicated Philadelphia chromosome. One of the BC CML patient samples was obtained at relapse from a patient who had been off of all treatment for several months, with imatinib-resistant disease at the time of relapse. All of the other patients were newly diagnosed and not yet treated at the time of sample collection. Demographic and clinical characteristics of the CML patients are displayed in Table I. A total of 15 normal samples (12 of which were used in the gene expression analyses and 3 of which were used for quantification of protein expression) were obtained as excess material from allogeneic bone marrow harvests. Informed consent was obtained from all patients and healthy donors prior to sample collection in accordance with the Declaration of Helsinski, under a research protocol approved by the Johns Hopkins Institutional Review Board.
Table I.
Patient Characteristics
| Sample # |
Disease Phase |
Age | Sex | Race |
BCR- ABL FISH |
BCR- ABL PCR* |
Other chromosomal abnormalities |
|---|---|---|---|---|---|---|---|
| 1 | chronic | 39 | Male | Hispanic | 96.5% | 144 | None |
| 2 | chronic | 53 | Male | African-American | 95.5% | 533 | None |
| 3 | chronic | 43 | Female | White | 81.5% | 645 | None |
| 4 | chronic | 54 | Male | African-American | 73.5% | 902 | None |
| 5 | chronic | 48 | Male | Hispanic | 93.0% | 531 | None |
| 6 | chronic | 59 | Male | White | 94.5% | 379 | Inv 2 |
| 7 | chronic | 50 | Female | White | 83.5% | 857 | None |
| 8 | chronic | 62 | Female | White | 96.5% | 562 | None |
| 9 | chronic | 25 | Male | White | 96.0% | 1495 | t(9;16) |
| 10 | chronic | 68 | Male | White | 96.5% | 345 | None |
| 11 | blast | 52 | Male | White | 70.5% | 615 | Inv 3 & t(1;10) |
| 12 | blast | 62 | Female | White | 82.0% | 433 | +8 |
| 13 | blast | 64 | Female | African-American | 93.5% | 808 | del 12p† |
| 14 | blast | 37 | Female | African-American | 87.5% | 195 | t(2;13) |
copies of BCR-ABL per 1,000 copies ABL
subclone present in 5/20 metaphases with additional abnormalities of +7, +8, and +21
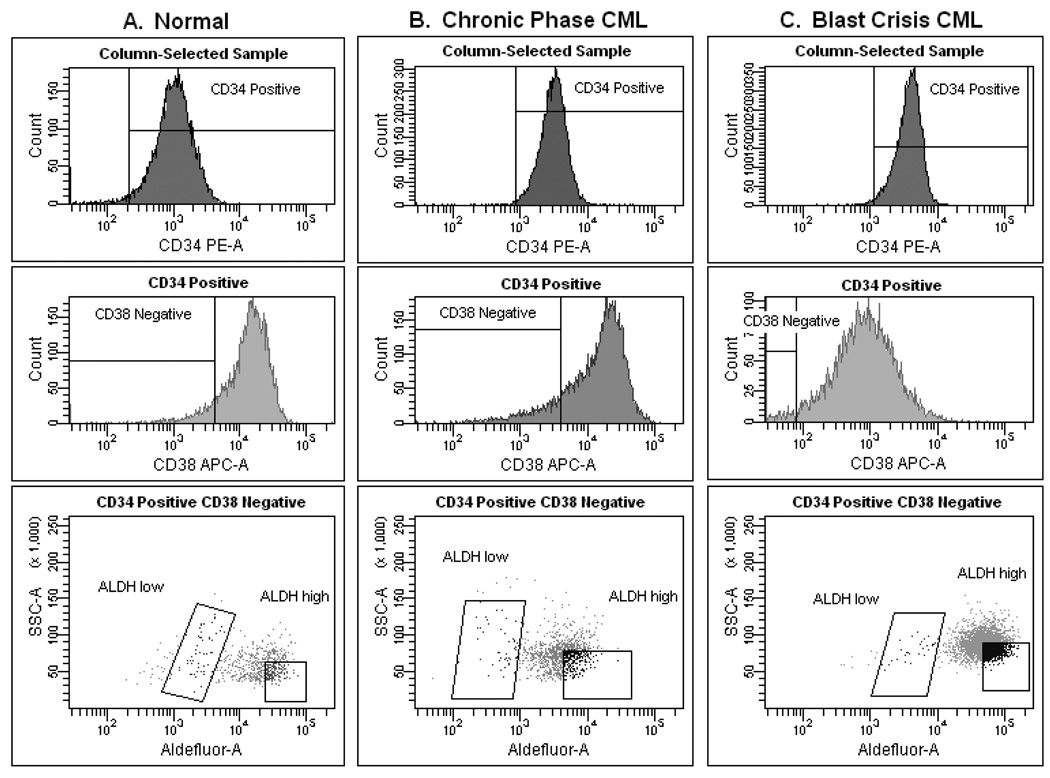
Mononuclear cells were isolated from fresh samples by Ficoll-Paque (GE Healthcare Life Sciences, Piscataway, NJ, USA) density (=1.077) centrifugation. CD34+ cells were selected by Miltenyi Biotec (Auburn, CA, USA) microbeads (binding the class II CD34 epitope) and column per manufacturer’s instructions. Next, ALDH1A1 activity was assessed in the CD34+ cells by staining with Aldefluor (Aldagen, Durham, NC, USA) per manufacturer’s guidelines; this was followed by labeling with monoclonal phycoerythrin (PE)-conjugated anti-CD34 (binding the class III CD34 epitope) and allophycocyanin (APC)-conjugated anti-CD38 (all antibodies purchased from BD Biosciences, San Jose, CA, USA). Cells were then sorted using a FACSAria (BD Biosciences) into 3 fractions: CD34+ total (unfractionated), CD34+CD38−ALDHlow, and (low side scatter) CD34+CD38−ALDHhigh. Representative examples of the staining profile and gating strategy for a normal sample, a CP CML sample, and a BC CML sample are depicted in Figure 1. Purity of the sorted cell fractions was >98% on reanalysis.
Figure 1. Staining profiles and gating strategies.
Representative staining patterns and gating strategies are depicted for a normal sample (A), a chronic phase CML sample (B), and a blast crisis CML sample (C). Following Ficoll-Paque density centrifugation and CD34+-selection via magnetic bead and column, each sample was stained (for CD34, CD38, and ALDH) and then sorted. The CD34+ total, CD34+CD38−ALDHlow, and (low side scatter) CD34+CD38−ALDHhigh populations were collected and analyzed from each sample. Given the variation across samples, attempts were made to isolate equivalent populations based on relative size and staining.
Fluorescence in situ hybridization (FISH)
Isolation of leukemic cells was confirmed by FISH for BCR-ABL on cytospins of 2×104 cells from each sorted cell fraction, fixed in 3:1 Methanol: Glacial Acetic acid (Sigma-Aldrich, St. Louis, MO, USA). FISH was performed by the Johns Hopkins Cytogenetics Core, using the Vysis LSI BCR-ABL Dual Color, Dual Fusion translocation probe (Abbot Molecular, Des Plaines, IL, USA) per manufacturer’s instructions and a fluorescence microscope with a triple-band pass filter for DAPI, Spectrum Orange, and Spectrum Green.
NOD/SCID-IL2Rγnull (NOG) mouse transplants
For a subset of CML patients (4 CP and 4 BC) from whom sufficient cellular yields of the isolated CD34 subsets were obtained, NOG mouse transplantation was used as a functional assay for stem cells. Following irradiation with 300cGy (via Cesium irradiator), 3 mice per cell fraction were injected (via tail vein) with 104 –105 cells; for any given sample, equal cell numbers from all fractions were transplanted. Mice were sacrificed >3 months later, and bone marrow was harvested at necropsy. The harvested mouse bone marrow was treated with RBC lysis buffer (Sigma-Aldrich) and then stained with an APC-conjugated monoclonal antibody against human CD45 (BD Biosciences). Cells were then analyzed on the FACSAria cell sorter and the human CD45+ population (if present) collected and cytospun onto slides for analysis of BCR-ABL by FISH. A sample fraction was considered to have successfully engrafted if at least 1 transplanted mouse had ≥ 0.1% human CD45+ cells detectable, with a cytospin of those cells positive for BCR-ABL by FISH. All mouse research was performed under a protocol approved by the Johns Hopkins Animal Care and Use Committee and complied with National Institutes of Health and American Veterinary Medical Association guidelines.
Reverse transcription polymerase chain reaction (RT-PCR)
Aliquots of 104 – 5×105 cells from the three CD34+ subsets isolated from 10 CP CML patients, 4 BC CML patients, and 12 normal donors were pelleted, flash frozen, and stored at −80C until assay. Aliquots of the K562 cell line (American Type Culture Collection, Manassas, VA, USA) were similarly pelleted and frozen, for use as a control between assay runs. RNA was extracted with the RNeasy kit (Quiagen) per manufacturer’s instructions. Expression of BCR-ABL, SURVIVIN, PR3, PRAME, hTERT, and WT1 mRNA was quantified by real-time, one-step RT-PCR using sequence-specific probes per the QuantiTect Probe RT-PCR kit (Qiagen, Valencia, CA, USA). Standard curves were generated using plasmids for each candidate and validated on fixed aliquots of K562 cells. Taqman primers were obtained from Applied Biosystems (Foster City, CA, USA; GAPDH 4326317, hTERT 162669, SURVIVIN 153353, WT1 240913, PRAME 196132, and PR3 1957752) and Ipsogen (Marseilles, France; BCR-ABL IPPF-70). mRNA expression of each putative target was also measured in 104 cell aliquots of the K562 cell line as a control. Candidate target expression was normalized to 104 copies of GAPDH mRNA. All RT-PCR reactions were performed in duplicate and run on the ABI 7500 machine (Applied Biosystems).
Protein quantification
Cells were fixed in 2% paraformalin, then permeabilized in 0.1% Triton-X-100 in PBS (Sigma-Aldrich), and then single-stained with unconjugated rabbit monoclonal antibody against PR3 or WT1 (Santa Cruz Biotechnology, Santa Cruz, CA, USA) or unconjugated rabbit polyclonal antibody against PRAME (Abcam, Cambridge, MA, USA). Surface membrane expression of PRAME was also analyzed on fresh cells (neither fixed nor permeabilized).[29] Cells were then secondarily stained with PE-conjugated goat anti-rabbit antibodies (BD Biosciences) and analyzed on the FACSAria. Due to limited quantities of CML samples, these experiments were performed only on normal donor samples and the K562 cell line.
Statistical considerations
mRNA expression of each of the six genes (PROTEINASE 3, SURVIVIN, hTERT, WT1, PRAME, and BCR-ABL) was quantified (per 104 copies of GAPDH) from all 3 cell fractions (CD34+ total, CD34+CD38−ALDHlow, and CD34+CD38−ALDHhigh) and compared among normal, CP CML, and BC CML. To account for the correlation among the three cell fractions obtained from the same patient, generalized estimating equations (GEE) were used (assuming a compound symmetry correlation structure) for model estimation and hypothesis testing of mean differences based on a Chi-squared statistic. The vector of expression from the cell fractions among patients was modeled as a function of clinical phase of CML (normal, CP, BC), cell fraction, and the interaction of the two. A log (base 10) transformation was applied to all expression data. A similar statistical approach was used to examine BCR-ABL FISH data (expressed as percentage of cells with the BCR-ABL fusion gene), whereby GEE was used to model the FISH percentage of cell fractions for corresponding hypothesis tests of equal means among cell fractions and between clinical phases. All reported p-values are by Chi-squared test, with values <0.05 considered significant.
Correlation in expression between the CD34+ total and CD34+CD38−ALDHhigh groups was explored using Spearman rank for BCR-ABL, WT1, and PRAME (reported p-values are also by Chi-squared test). PR3, SURVIVIN, and hTERT expression were further examined for mean differences between the CD34+ total and CD34+CD38−ALDHhigh cell fractions within each of CP and BC CML by Chi-squared test.
Results
The CD34+CD38−ALDHhigh cell fraction is highly enriched for CML stem cells
CD34+ cell fractions were isolated from 10 patients with CP CML and 4 patients with BC CML. CP CML staining patterns were quite reproducible and closely mirrored that of normal samples, whereas staining patterns of BC CML were more variable (Figure 1). The majority of the cells (on average >90%) from all fractions of all CML patients were positive for BCR-ABL by FISH, with the exception of the CD34+CD38−ALDHhigh fraction of one CP CML patient (which was only 39.5% BCR-ABL FISH-positive) (Table II). The CD34+CD38−ALDHhigh cells constituted <5% of the total CD34+ cells and about 20% of the total CD34+CD38− population (and typically, <0.1% of the total mononuclear cells). There were no statistically significant differences in the mean percentage of BCR-ABL FISH-positive cells between CP and BC CML or between the CD34+ cell subsets within each disease phase.
Table II.
Percentages of each sorted cell fraction positive for BCR-ABL by FISH in chronic phase (CP) and blast crisis (BC) CML.
| CD34+ total | CD34+CD38−ALDHlow | CD34+CD38−ALDHhigh | |
|---|---|---|---|
| CP CML mean | 94% | 92.8% | 85.2% |
| CP CML median | 97.5% | 95.5% | 94% |
| CP CML range | 70–99.5% | 79–99% | 39.5–100% |
| BC CML mean | 98.4% | 93.3% | 98.6% |
| BC CML median | 98.5% | 92.8% | 98.5% |
| BC CML range | 97–99.5% | 88–99.5% | 98.3–99% |
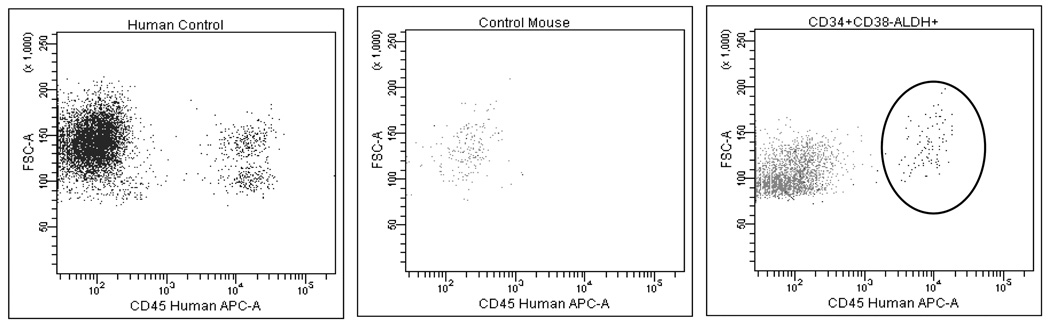
Stem cell function was defined by the ability to engraft immunodeficient mice.[30, 31] The transplanted CD34+CD38−ALDHhigh cell fraction from all 8 CML patients (4 CP and 4 BC) produced leukemic (by FISH for BCR-ABL) engraftment in irradiated NOG mice (median 3%, range 0.1 – 31%, of total RBC-lysed marrow cells). A representative example of an engrafted CML sample is shown in Figure 2 (right panel). Equal numbers of the total CD34+ cells also produced leukemic engraftment, but less efficiently; 7/8 patient samples engrafted, with a median population size of 0.1% (range 0.1–1.3% of total RBC-lysed marrow cells). The CD34+CD38−ALDHlow fraction engrafted in only 1 case, a BC CML sample. Normal CD34+CD38−ALDHhigh cells similarly produced reliable engraftment.
Figure 2. The CD34+CD38−ALDHhigh fraction was highly enriched for NOG mouse engrafting capacity.
Left panel: a human control, depicting a CD45+ cell population. Middle panel: an untransplanted NOG mouse control, lacking a detectable human CD45+ population. Right panel: a NOG mouse which engrafted with a detectable human CD45+ population after injection with the CD34+CD38−ALDHhigh fraction of a CP CML sample several months prior; 98% of the sorted human CD45+ cells from this sample were positive for BCR-ABL by FISH.
Expression of PR3, SURVIVIN, and hTERT by CML stem/progenitor cells parallels that in their normal counterparts
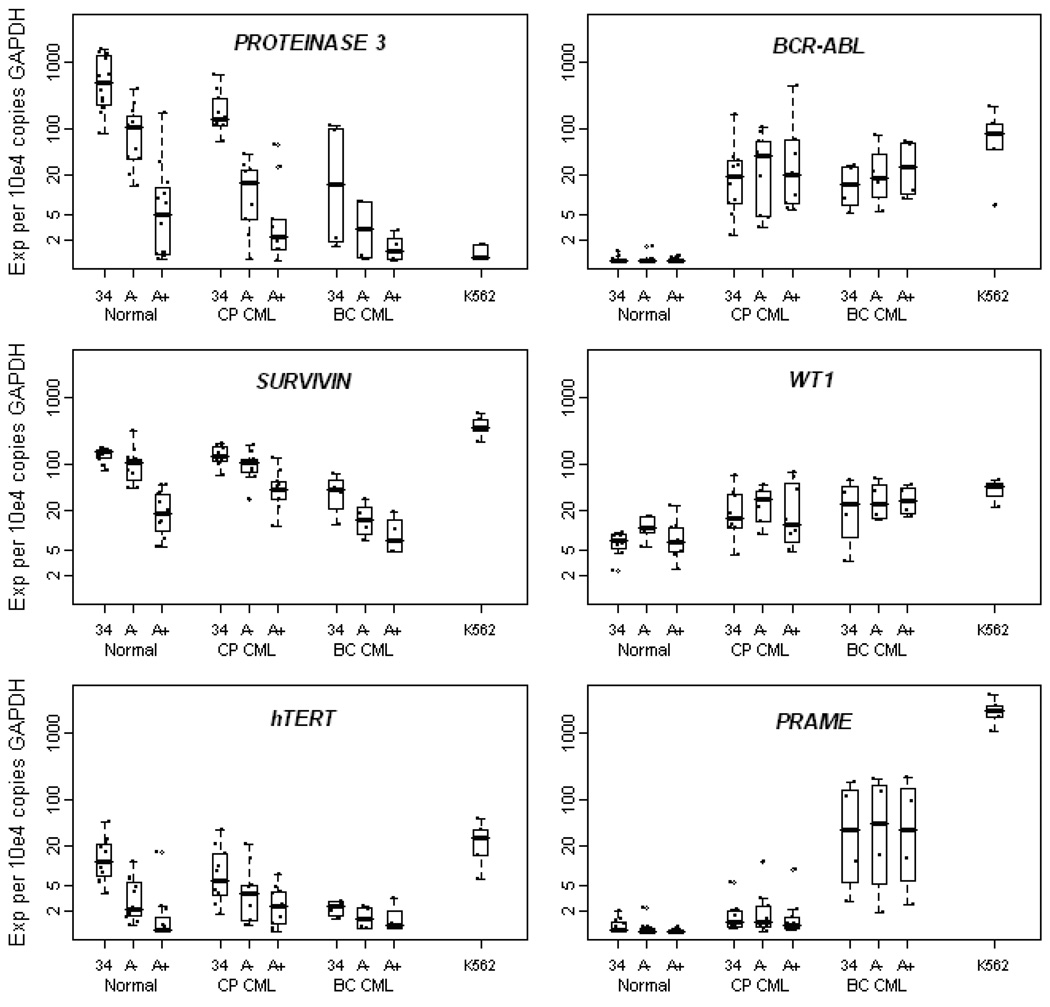
mRNA expression of the potential therapeutic targets and BCR-ABL was assessed by real time RT-PCR in each of the three CD34+ cell subsets isolated from the CML patients and normal donors (Figure 3). There was a trend toward higher BCR-ABL mRNA expression in the CD34+CD38−ALDHhigh fraction of both CP (p=0.0568) and BC CML (p=0.081). PR3 and SURVIVIN mRNA showed similar patterns of expression in normal cells, with levels lowest in the CD34+CD38−ALDHhigh cells, intermediate in the CD34+CD38−ALDHlow population, and highest in the total CD34+ fraction. Expression of PR3 and SURVIVIN mRNA in both CP and BC CML paralleled that detected in the normal samples; the expression of these antigens was actually highest of all in the normal CD34+ total fraction and decreased with clinical progression from CP to BC CML. hTERT mRNA was expressed at only very low levels in any of the CD34+ cell fractions, especially in the CD34+CD38−ALDHhigh cells of both the normal controls and CML patients. In CP CML, expression levels of PR3 (p=0.002), SURVIVIN (p=0.005), and hTERT (p=0.008) mRNA were significantly lower in the CD34+CD38−ALDHhigh fraction than in the total CD34+ fraction. These same trends were seen in BC CML, though only borderline significant for SURVIVIN (p=0.056) and not statistically significant for PR3 and hTERT.
Figure 3. mRNA expression of PROTEINASE 3, SURVIVIN, hTERT, BCR-ABL, WT1, and PRAME in CD34+ cell subsets of normal controls and CML patients.
Boxplots of log (base 10) mRNA expression (per 104 copies of GAPDH) of each candidate target by clinical phase (Normal, Chronic Phase (CP) CML, or Blast Crisis (BC) CML) and within phase by cell phenotype: CD34+ total (34), CD34+CD38−ALDHlow (A−), and CD34+CD38−ALDHhigh (A+). The K562 cell line is included as a control. Each dot refers to an individual sample. Horizontal lines indicate median expression.
PROTEINASE 3, SURVIVIN, and hTERT expression in CP and BC CML paralleled that in normal cells. Expression of each of these genes was highest in the normal CD34+ total cells and low in the CP and BC CML CD34+CD38−ALDHhigh cells. In contrast, BCR-ABL, WT1 (p=0.038 for CP CML and p=0.042 for BC CML vs. normal), and PRAME (p=0.056 for CP CML and p=0.032 for BC CML vs. normal) exhibited significantly higher expression in CML than in normal. BCR-ABL and WT1 did not differ significantly between cell fractions or between CP and BC. PRAME expression also did not differ significantly by cell fraction, but it did significantly increase from CP to BC (p=0.039).
WT1 and PRAME are over-expressed in CML
Mean WT1 mRNA expression was significantly higher in CP (p=0.038) and BC (p=0.042) CML than normal, with 2 to 5-fold higher expression in each CD34+ fraction versus the corresponding normal fraction (Figure 3). Mean expression of WT1 mRNA did not differ significantly between CP and BC CML (p=0.498). PRAME mRNA was expressed at very low to undetectable levels in all of the normal samples, especially the CD34+CD38−ALDHhigh population (Figure 3). All three CP CML CD34 cell fractions expressed PRAME mRNA levels on average that were >30-fold higher than the normal CD34+CD38−ALDHhigh cells and >4-fold higher than the corresponding normal fraction (p=0.056 for mean PRAME expression in CP CML vs. normal). PRAME mRNA was significantly more highly expressed in all BC CML CD34+ fractions; >50-fold on average higher than the corresponding CP CML fractions (p=0.039 for CP vs. BC CML) and >300-fold higher on average than any normal fraction (p=0.032 for BC CML vs. normal). Notably, there was a statistically significant correlation between mRNA expression in the CD34+ total fraction and that in the CD34+CD38−ALDHhigh fraction for both WT1 (Spearman’s rho=0.798; p<0.001) and PRAME (Spearman’s rho=0.806; p<0.001).
PR3, WT1, and PRAME protein expression mirrored mRNA expression
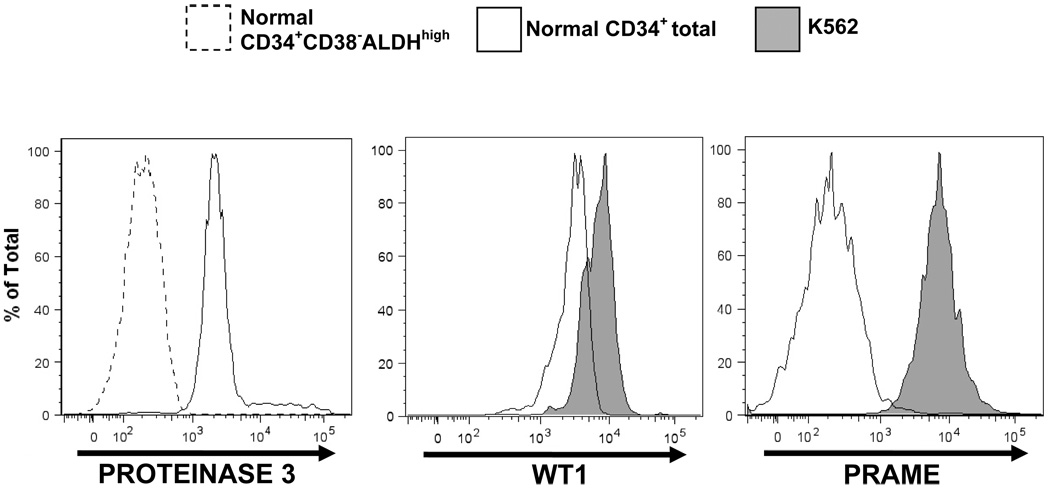
PR3 protein levels by flow cytometry paralleled mRNA expression; unfractionated normal CD34+ cells exhibited markedly higher pr3 expression than the normal CD34+CD38−ALDHhigh cell subset (Figure 4, left panel). WT1 protein expression also correlated with mRNA expression, with higher levels in K562 cells than in normal unfractionated CD34+ cells (Figure 4, middle panel). Likewise, PRAME protein expression correlated with mRNA levels. Intense PRAME surface membrane staining was seen in the K562 cells, substantially higher than that in normal unfractionated CD34+ cells (Figure 4, right panel).
Figure 4. Protein staining by flow cytometry mirrors mRNA expression for PROTEINASE 3, WT1, and PRAME.
Left panel: PROTEINASE 3 staining was markedly higher in normal CD34+ total cells (solid curve) than in normal CD34+CD38−ALDHhigh cells (dotted curve). Middle panel: WT1 staining was higher in the K562 cell line (shaded) than in normal CD34+ total cells (solid curve). Right panel: PRAME staining was substantially higher in the K562 cell line (shaded) than in normal CD34+ total cells (solid curve).
Discussion
The CD34+CD38−ALDHhigh cell population was highly enriched for CML stem cells, as determined by its ability to engraft immunodeficient mice. Equal numbers of unfractionated CD34+ CML cells produced about 30-fold less chimerism, and little to no engraftment was generated by the CD34+CD38−ALDHlow CML cells. Most importantly, the CD34+CD38−ALDHhigh CML stem cells exhibited a unique expression pattern, distinct from both their own leukemic progeny and the normal hematopoietic stem/progenitor cells.
Expression of PR3, SURVIVIN, and hTERT was highest in the more differentiated CD34+ total cells of both CML and normal samples, with levels lowest in BC CML. This pattern is consistent with markers of differentiation. Indeed, PR3, the antigen for which this pattern was most prominent, has been proposed as a marker of myeloid differentiation.[32] Furthermore, PR3, SURVIVIN, and hTERT were all expressed at lower levels in each of the CML CD34+ subsets analyzed (including the CD34+CD38−ALDHhigh fraction), than in the normal unfractionated CD34+ cells. Protein levels of PR3 reflected the mRNA expression levels, and the same has been reported previously for SURVIVIN[33] and hTERT.[34] These data suggest that efforts to target these antigens in CML stem cells may be compromised by substantial toxicity to normal hematopoietic cells. Additionally, these results illustrate that the CD34+ total fraction in CML is not necessarily representative of the LSC and underscore the importance of assessing target expression in refined LSC populations.
Conversely, given their more selective expression, including at the level of the LSC, WT1 and PRAME may be more suitable therapeutic targets in CML. WT1 appears to act downstream of BCR-ABL and has been implicated in inhibition of apoptosis;[14, 35] it has also been proposed as a marker for minimal residual disease.[36, 37] In contrast to the somewhat disappointing experience to date with vaccines against BCR-ABL, early promise has been seen with WT1 vaccines in myeloid leukemias.[38] Additionally, WT1 siRNA has been demonstrated to decrease proliferation and increase apoptosis of leukemic cell lines, including K562, supplying further evidence of a critical role for this antigen.[35, 39] It remains to be determined whether the degree of over-expression of WT1 in both CP and BC CML provides a sufficient therapeutic window to be exploited.
PRAME may be a superior target, given its minimal to null expression in the normal CD34+ cells, particularly the CD34+CD38−ALDHhigh subset. In fact, PRAME appears to have very limited expression in any normal adult tissue, other than testis.[13] Our results support existing data that PRAME expression increases from CP to advanced phase CML, suggesting a role in leukemic progression.[40] Furthermore, PRAME has been demonstrated to interfere with retinoic acid signaling[41] and may thus contribute to the inhibition of differentiation in advanced phase CML.[42] Though PRAME has been recommended as a vaccine target in CML,[12, 16] to our knowledge, there are yet no active clinical trials specifically targeting it.
Interestingly, investigators at our institution have developed a K562-based vaccine engineered to produce GM-CSF;[43] this vaccine has shown promise in early phase clinical trials against CML.[44] The K562 cell line, upon which the vaccine is based, expresses low levels of PR3, but high levels of both WT1 and PRAME, similar to the expression pattern seen in CML stem cells (Figure 3 & Figure 4). Thus, this vaccine may be capable of selectively delivering CML stem cell antigens to the immune system. Since both WT1 and PRAME mRNA expression were similar in all leukemic CD34+ cell fractions, the CD34+ total fraction could serve as a surrogate marker for the LSC with regard to these two targets, facilitating clinical screening of patients.
Yong et al previously reported on antigen expression by phenotypically defined populations of CML stem and progenitor cells.[12] They, too, found WT1 mRNA to be more highly expressed in CML stem cells than in normal hematopoietic stem/progenitor cells and PRAME mRNA to be over-expressed in advanced phase CML. Likewise, they also found substantially higher levels of PR3 mRNA expression in the differentiated CML and normal CD34+ subsets than in their more primitive counterparts.[12] Importantly, our results confirm these findings in functionally defined CML stem/progenitor populations and also demonstrate that protein expression of these putative therapeutic targets correlates with mRNA levels.
Despite the remarkable success of tyrosine kinase inhibitors against CML, these drugs do not seem to cure the disease.[3, 4] This appears to be due to the failure of these agents to reliably eliminate the CML stem cells.[5–7, 10, 45] The long term implications of the persistence of CML stem cells in patients with responsive disease remain unclear. Moreover, in a significant minority of patients with CML, including most with BC CML, the disease will not respond to tyrosine kinase inhibition or ultimately progresses in spite of it.[9, 11, 45] Thus, the search for novel therapies, especially those that target the rare CML stem cells, remains imperative. Although CML stem cells share many characteristics with their normal counterparts, WT1 and PRAME, like BCR-ABL, are selectively expressed by the CML stem/progenitor cells. These candidates thus hold promise as potential therapeutic targets in CML. Further work is warranted for their functional validation.
Acknowledgements
The authors would like to thank James P. Barber and Laura Morsberger for their expert technical assistance.
This work was supported by grants from the National Institutes of Health (PO1 CA15396, PO1 CA070970, and 5 T32 HL0075225); an American Society of Hematology Research Training Award for Fellows; and a Maryland Stem Cell Research Fund Post-Doctoral Fellowship grant (RFA-MD-07-3)
References
- 1.Druker BJ, Talpaz M, Resta DJ, et al. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med. 344:1031–1037. doi: 10.1056/NEJM200104053441401. [DOI] [PubMed] [Google Scholar]
- 2.Kantarjian H, Talpaz M, O'Brien S, et al. High-dose imatinib mesylate therapy in newly diagnosed Philadelphia chromosome-positive chronic phase chronic myeloid leukemia. Blood. 103:2873–2878. doi: 10.1182/blood-2003-11-3800. [DOI] [PubMed] [Google Scholar]
- 3.Cortes J, O'Brien S, Kantarjian H H. Discontinuation of imatinib therapy after achieving a molecular response. Blood. 104:2204–2205. doi: 10.1182/blood-2004-04-1335. [DOI] [PubMed] [Google Scholar]
- 4.Rousselot P, Huguet F, Rea D, et al. Imatinib mesylate discontinuation in patients with chronic myelogenous leukemia in complete molecular remission for more than 2 years. Blood. 109:58–60. doi: 10.1182/blood-2006-03-011239. [DOI] [PubMed] [Google Scholar]
- 5.Graham SM, Jorgensen HG, Allan E, et al. Primitive, quiescent, Philadelphia-positive stem cells from patients with chronic myeloid leukemia are insensitive to STI571 in vitro. Blood. 99:319–325. doi: 10.1182/blood.v99.1.319. [DOI] [PubMed] [Google Scholar]
- 6.Angstreich GR, Matsui W, Huff CA, et al. Effects of imatinib and interferon on primitive chronic myeloid leukaemia progenitors. Br J Haematol. 130:373–381. doi: 10.1111/j.1365-2141.2005.05606.x. [DOI] [PubMed] [Google Scholar]
- 7.Copland M, Hamilton A, Elrick LJ, et al. Dasatinib (BMS-354825) targets an earlier progenitor population than imatinib in primary CML but does not eliminate the quiescent fraction. Blood. 107:4532–4539. doi: 10.1182/blood-2005-07-2947. [DOI] [PubMed] [Google Scholar]
- 8.Bhatia R, Holtz M, Niu N, et al. Persistence of malignant hematopoietic progenitors in chronic myelogenous leukemia patients in complete cytogenetic remission following imatinib mesylate treatment. Blood. 101:4701–4707. doi: 10.1182/blood-2002-09-2780. [DOI] [PubMed] [Google Scholar]
- 9.Shah NP. Advanced CML: therapeutic options for patients in accelerated and blast phases. J Natl Compr Canc Netw. 6 Suppl 2:S31–S36. [PubMed] [Google Scholar]
- 10.Savona M, Talpaz M. Getting to the stem of chronic myeloid leukaemia. Nat Rev Cancer. 8:341–350. doi: 10.1038/nrc2368. [DOI] [PubMed] [Google Scholar]
- 11.Heaney NB, Holyoake TL. Therapeutic targets in chronic myeloid leukaemia. Hematol Oncol. 25:66–75. doi: 10.1002/hon.813. [DOI] [PubMed] [Google Scholar]
- 12.Yong AS, Keyvanfar K, Eniafe R, et al. Hematopoietic stem cells and progenitors of chronic myeloid leukemia express leukemia-associated antigens: implications for the graft-versus-leukemia effect and peptide vaccine-based immunotherapy. Leukemia. 22:1721–1727. doi: 10.1038/leu.2008.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greiner J, Dohner H, Schmitt M. Cancer vaccines for patients with acute myeloid leukemia--definition of leukemia-associated antigens and current clinical protocols targeting these antigens. Haematologica. 91:1653–1661. [PubMed] [Google Scholar]
- 14.Svensson E, Vidovic K, Lassen C, et al. Deregulation of the Wilms' tumour gene 1 protein (WT1) by BCR/ABL1 mediates resistance to imatinib in human leukaemia cells. Leukemia. 21:2485–2494. doi: 10.1038/sj.leu.2404924. [DOI] [PubMed] [Google Scholar]
- 15.Badran A, Yoshida A, Wano Y, et al. Expression of the antiapoptotic gene survivin in chronic myeloid leukemia. Anticancer Res. 23:589–592. [PubMed] [Google Scholar]
- 16.Greiner J, Schmitt M. Leukemia-associated antigens as target structures for a specific immunotherapy in chronic myeloid leukemia. Eur J Haematol. 80:461–468. doi: 10.1111/j.1600-0609.2008.01053.x. [DOI] [PubMed] [Google Scholar]
- 17.Phatak P, Burger AM. Telomerase and its potential for therapeutic intervention. Br J Pharmacol. doi: 10.1038/sj.bjp.0707374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakahara T, Takeuchi M, Kinoyama I, et al. YM155, a novel small-molecule survivin suppressant, induces regression of established human hormone-refractory prostate tumor xenografts. Cancer Res. 67:8014–8021. doi: 10.1158/0008-5472.CAN-07-1343. [DOI] [PubMed] [Google Scholar]
- 19.Bedi A, Zehnbauer BA, Collector MI, et al. BCR-ABL gene rearrangement and expression of primitive hematopoietic progenitors in chronic myeloid leukemia. Blood. 81:2898–2902. [PubMed] [Google Scholar]
- 20.Wang JC, Dick JE. Cancer stem cells: lessons from leukemia. Trends Cell Biol. 15:494–501. doi: 10.1016/j.tcb.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 21.Gal H, Amariglio N, Trakhtenbrot L, et al. Gene expression profiles of AML derived stem cells; similarity to hematopoietic stem cells. Leukemia. 20:2147–2154. doi: 10.1038/sj.leu.2404401. [DOI] [PubMed] [Google Scholar]
- 22.Bonnet D. Normal and leukaemic stem cells. Br J Haematol. 130:469–479. doi: 10.1111/j.1365-2141.2005.05596.x. [DOI] [PubMed] [Google Scholar]
- 23.Christ O, Lucke K, Imren S, et al. Improved purification of hematopoietic stem cells based on their elevated aldehyde dehydrogenase activity. Haematologica. 92:1165–1172. doi: 10.3324/haematol.11366. [DOI] [PubMed] [Google Scholar]
- 24.Pierre-Louis O, Clay D, Brunet de la GP, et al. Dual SP/ALDH Functionalities Refine the Human Hematopoietic Lin(−)CD34(+)CD38(−) Stem/Progenitor Cell Compartment. Stem Cells. 27:2552–2562. doi: 10.1002/stem.186. [DOI] [PubMed] [Google Scholar]
- 25.Brodsky RA, Jones RJ. Aplastic anaemia. Lancet. 365:1647–1656. doi: 10.1016/S0140-6736(05)66515-4. [DOI] [PubMed] [Google Scholar]
- 26.Matsui W, Wang Q, Barber JP, et al. Clonogenic multiple myeloma progenitors, stem cell properties, and drug resistance. Cancer Res. 68:190–197. doi: 10.1158/0008-5472.CAN-07-3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones RJ, Gocke CD, Kasamon YL, et al. Circulating clonotypic B cells in classic Hodgkin lymphoma. Blood. 113:5920–5926. doi: 10.1182/blood-2008-11-189688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones RJ, Barber JP, Vala MS, et al. Assessment of aldehyde dehydrogenase in viable cells. Blood. 85:2742–2746. [PubMed] [Google Scholar]
- 29.Proto-Siqueira R, Figueiredo-Pontes LL, Panepucci RA, et al. PRAME is a membrane and cytoplasmic protein aberrantly expressed in chronic lymphocytic leukemia and mantle cell lymphoma. Leuk Res. 30:1333–1339. doi: 10.1016/j.leukres.2006.02.031. [DOI] [PubMed] [Google Scholar]
- 30.Sirard C, Lapidot T, Vormoor J, et al. Normal and leukemic SCID-repopulating cells (SRC) coexist in the bone marrow and peripheral blood from CML patients in chronic phase, whereas leukemic SRC are detected in blast crisis. Blood. 87:1539–1548. [PubMed] [Google Scholar]
- 31.Lapidot T, Sirard C, Vormoor J, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 367:645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 32.Dengler R, Munstermann U, al-Batran S, et al. Immunocytochemical and flow cytometric detection of proteinase 3 (myeloblastin) in normal and leukaemic myeloid cells. Br J Haematol. 89:250–257. doi: 10.1111/j.1365-2141.1995.tb03297.x. [DOI] [PubMed] [Google Scholar]
- 33.Badran A, Yoshida A, Wano Y, et al. Expression of the anti-apoptotic gene survivin in myelodysplastic syndrome. Int J Oncol. 22:59–64. [PubMed] [Google Scholar]
- 34.Poole JC, Andrews LG, Tollefsbol TO. Activity, function, and gene regulation of the catalytic subunit of telomerase (hTERT) Gene. 269:1–12. doi: 10.1016/s0378-1119(01)00440-1. [DOI] [PubMed] [Google Scholar]
- 35.Elmaagacli AH, Koldehoff M, Peceny R, et al. WT1 and BCR-ABL specific small interfering RNA have additive effects in the induction of apoptosis in leukemic cells. Haematologica. 90:326–334. [PubMed] [Google Scholar]
- 36.Tamaki H, Ogawa H. Monitoring of minimal residual disease using WT1 assay for patients with chronic myeloid leukemia who undergo allogeneic stem cell transplantation. Bone Marrow Transplant. 34:653–654. doi: 10.1038/sj.bmt.1704620. [DOI] [PubMed] [Google Scholar]
- 37.Cilloni D, Saglio G. WT1 as a universal marker for minimal residual disease detection and quantification in myeloid leukemias and in myelodysplastic syndrome. Acta Haematol. 112:79–84. doi: 10.1159/000077562. [DOI] [PubMed] [Google Scholar]
- 38.Rusakiewicz S, Molldrem JJ. Immunotherapeutic peptide vaccination with leukemia-associated antigens. Curr Opin Immunol. 18:599–604. doi: 10.1016/j.coi.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 39.Glienke W, Maute L, Koehl U, Esser R, Milz E, Bergmann L. Effective treatment of leukemic cell lines with wt1 siRNA. Leukemia. 21:2164–2170. doi: 10.1038/sj.leu.2404878. [DOI] [PubMed] [Google Scholar]
- 40.Radich JP, Dai H, Mao M, et al. Gene expression changes associated with progression and response in chronic myeloid leukemia. Proc Natl Acad Sci U S A. 103:2794–2799. doi: 10.1073/pnas.0510423103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Epping MT, Wang L, Edel MJ, Carlee L, Hernandez M, Bernards R. The human tumor antigen PRAME is a dominant repressor of retinoic acid receptor signaling. Cell. 122:835–847. doi: 10.1016/j.cell.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 42.Oehler VG, Guthrie KA, Cummings CL, et al. The preferentially expressed antigen in melanoma (PRAME) inhibits myeloid differentiation in normal hematopoietic and leukemic progenitor cells. Blood. 114:3299–3308. doi: 10.1182/blood-2008-07-170282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Borrello I, Sotomayor EM, Cooke S, Levitsky HI. A universal granulocyte-macrophage colony-stimulating factor-producing bystander cell line for use in the formulation of autologous tumor cell-based vaccines. Hum Gene Ther. 10:1983–1991. doi: 10.1089/10430349950017347. [DOI] [PubMed] [Google Scholar]
- 44.Smith BD, Kasamon YL, Kowalski J, et al. K562/GM-CSF immunotherapy reduces tumor burden in chronic myeloid leukemia patients with residual disease on imatinib mesylate. Clin Cancer Res. 16:338–347. doi: 10.1158/1078-0432.CCR-09-2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Irvine DA, Heaney NB, Holyoake TL. Optimising chronic myeloid leukaemia therapy in the face of resistance to tyrosine kinase inhibitors--a synthesis of clinical and laboratory data. Blood Rev. 24:1–9. doi: 10.1016/j.blre.2009.11.002. [DOI] [PubMed] [Google Scholar]