Abstract
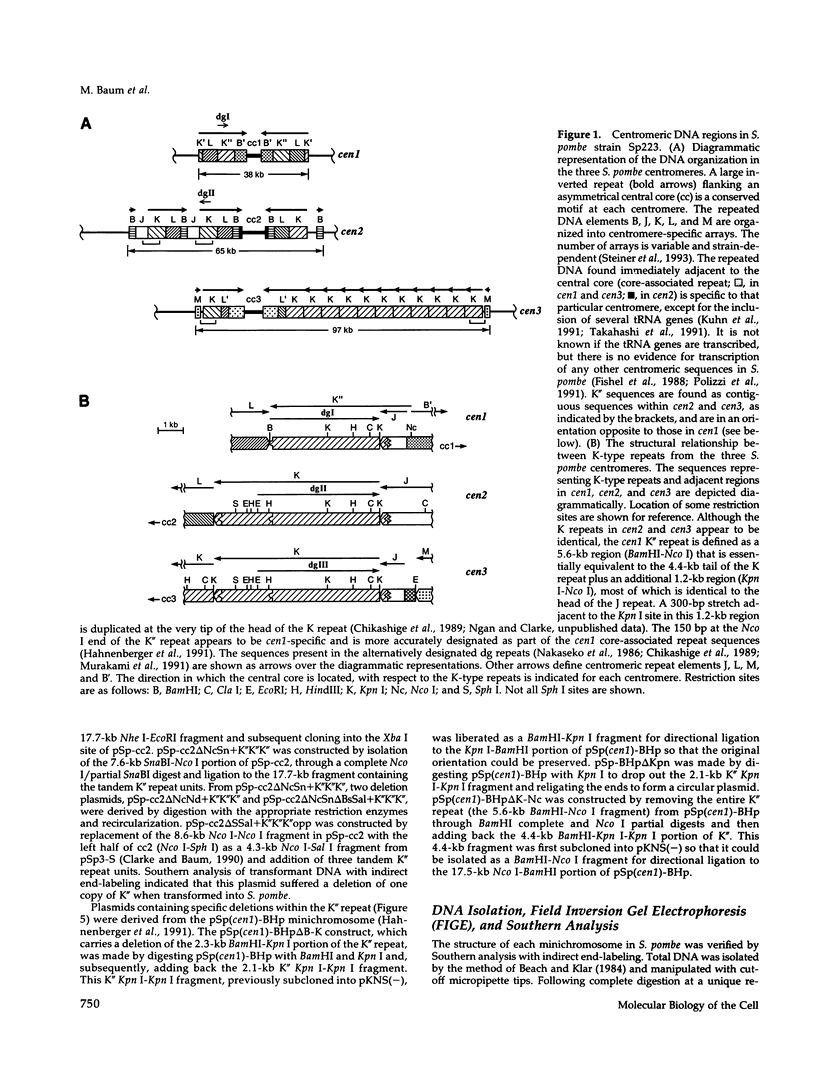
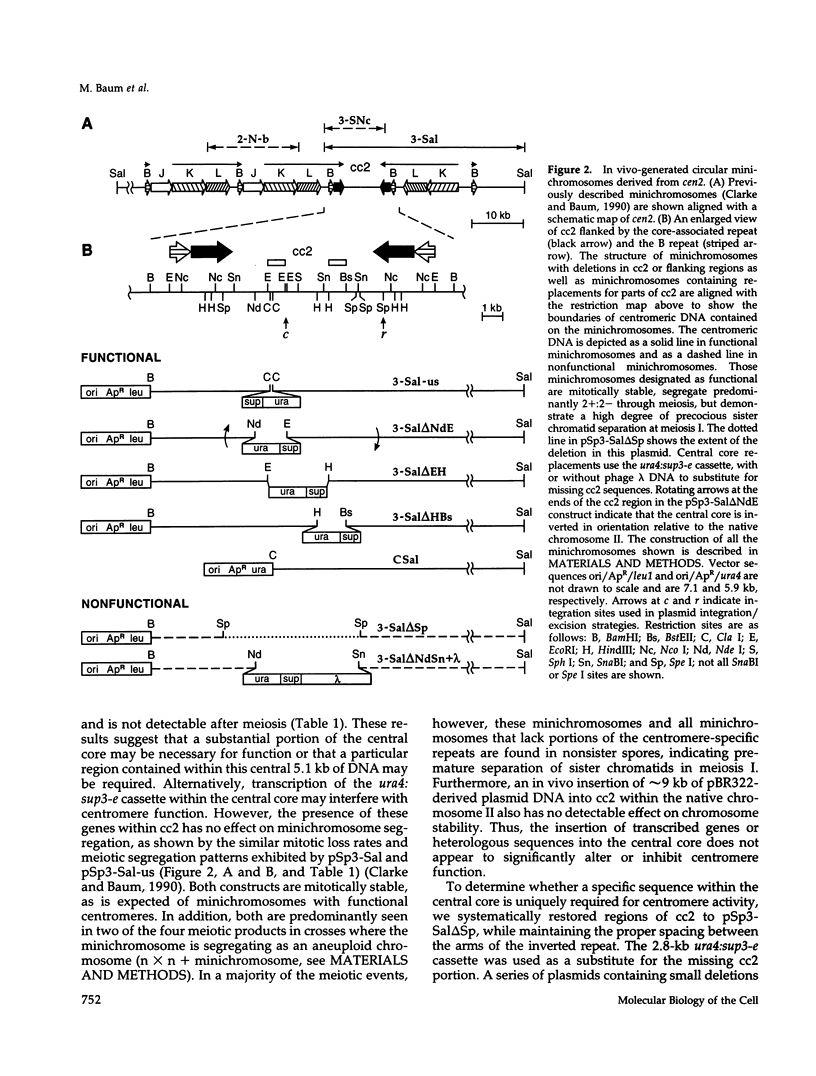
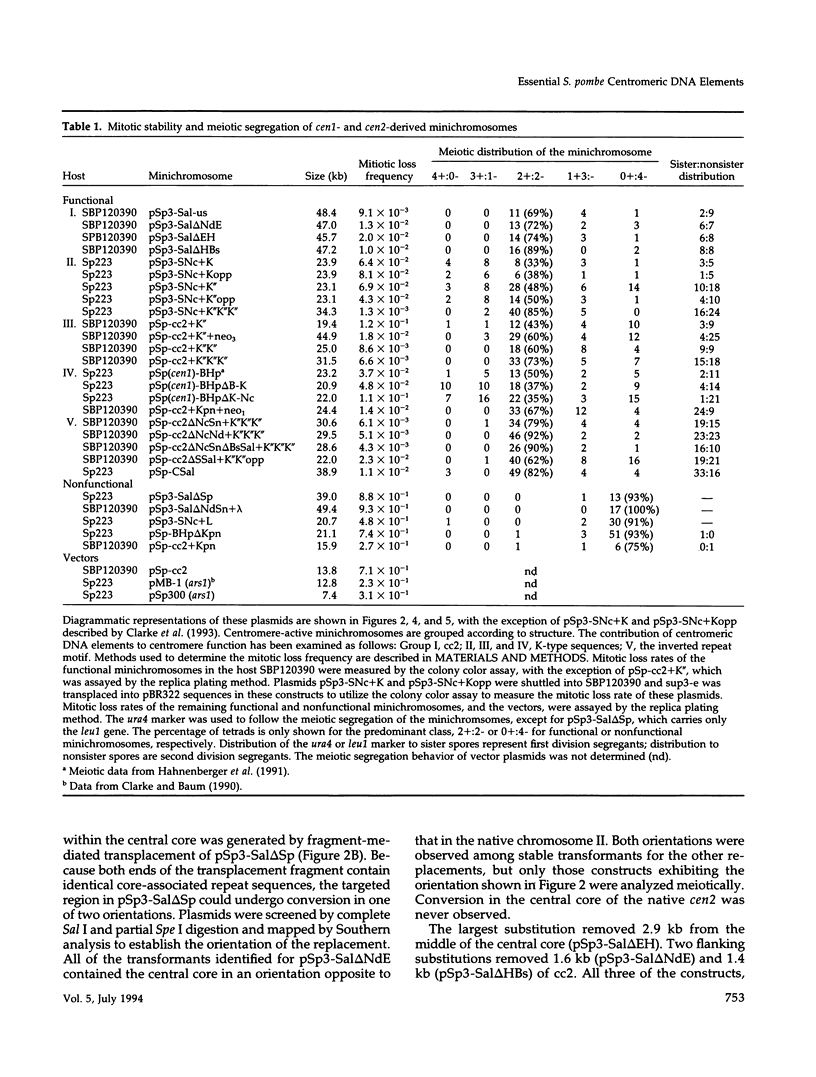
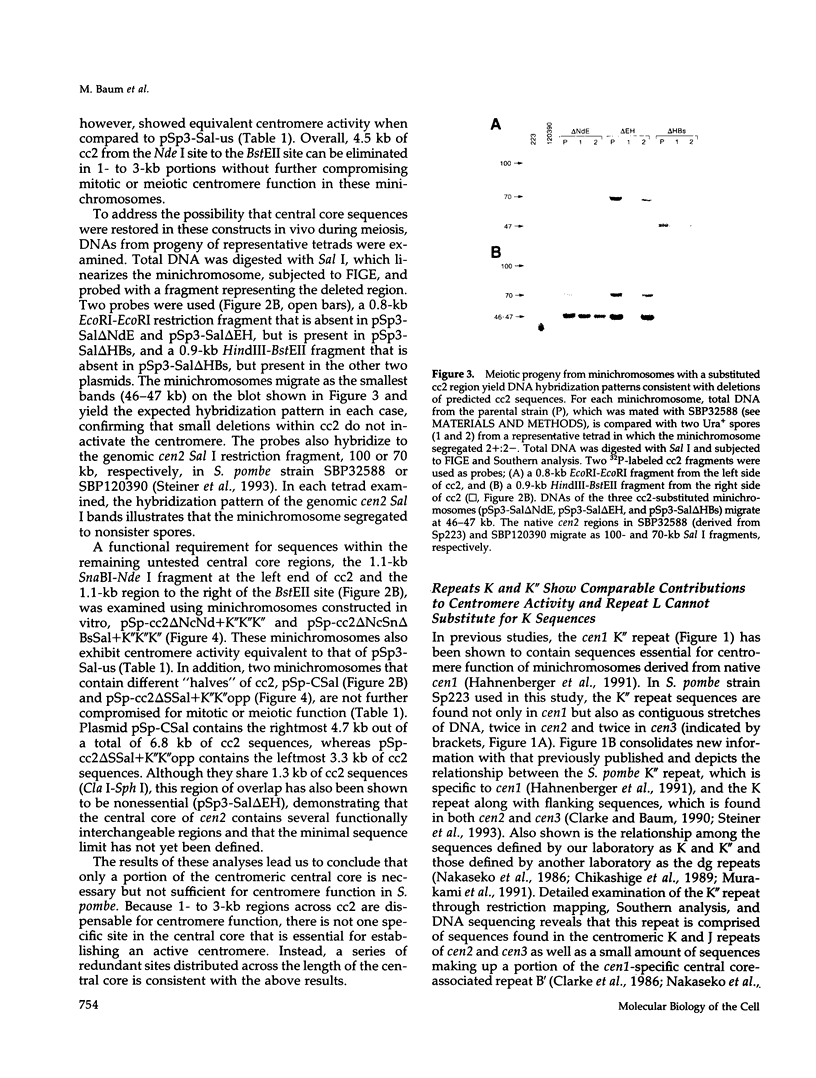
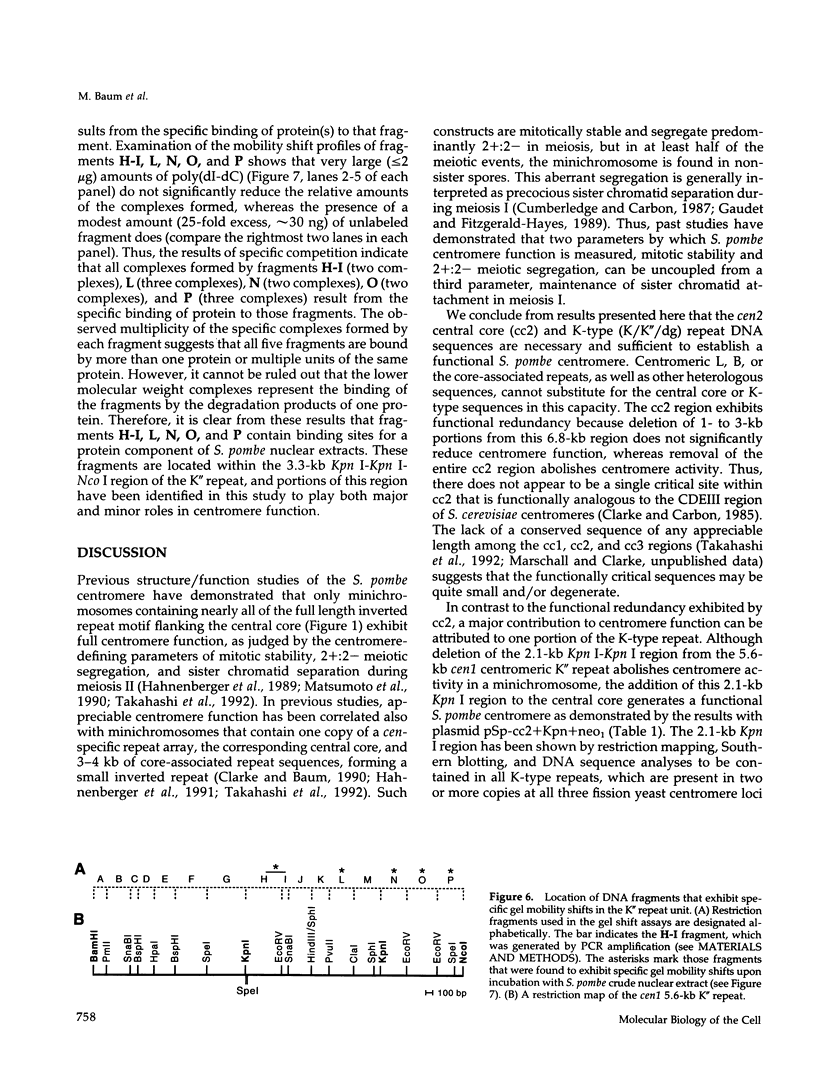
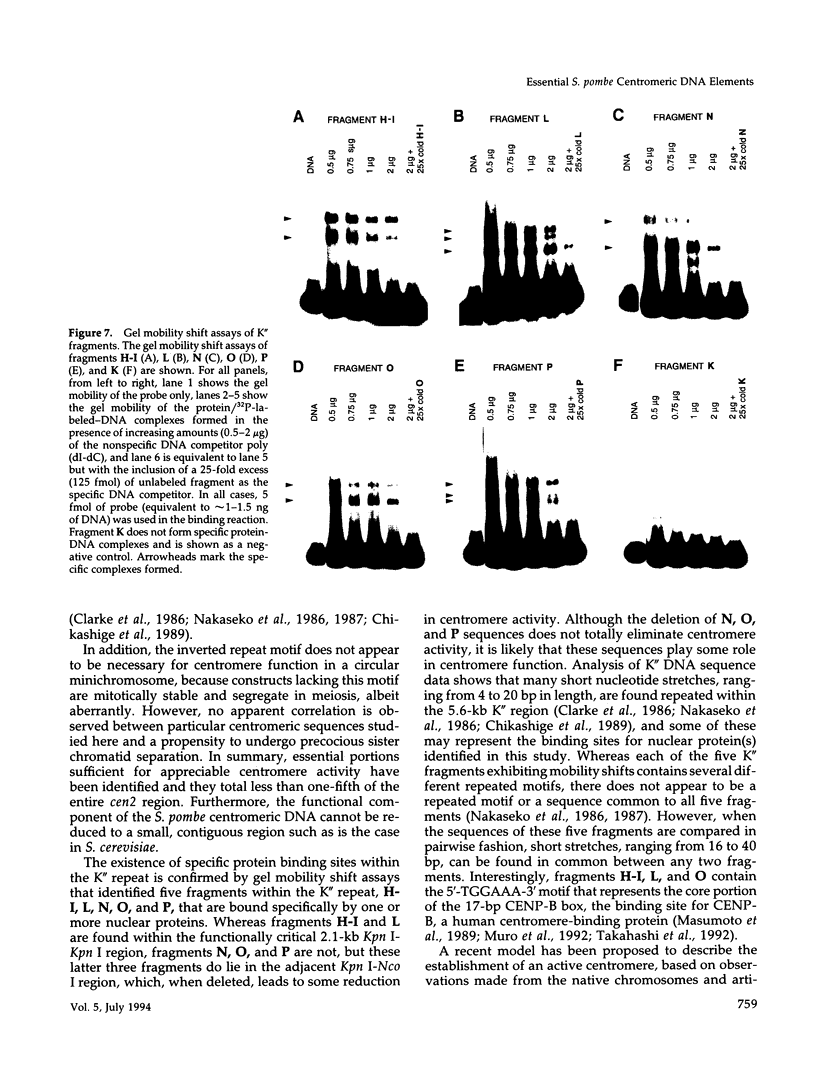
The DNA requirements for centromere function in fission yeast have been investigated using a minichromosome assay system. Critical elements of Schizosaccharomyces pombe centromeric DNA are portions of the centromeric central core and sequences within a 2.1-kilobase segment found on all three chromosomes as part of the K-type (K/K"/dg) centromeric repeat. The S. pombe centromeric central core contains DNA sequences that appear functionally redundant, and the inverted repeat motif that flanks the central core in all native fission yeast centromeres is not essential for centromere function in circular minichromosomes. Tandem copies of centromeric repeat K", in conjunction with the central core, exert an additive effect on centromere function, increasing minichromosome mitotic stability with each additional copy. Centromeric repeats B and L, however, and parts of the central core and its core-associated repeat are dispensable and cannot substitute for K-type sequences. Several specific protein binding sites have been identified within the centromeric K-type repeat, consistent with a recently proposed model for centromere/kinetochore function in S. pombe.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baker R. E., Fitzgerald-Hayes M., O'Brien T. C. Purification of the yeast centromere binding protein CP1 and a mutational analysis of its binding site. J Biol Chem. 1989 Jun 25;264(18):10843–10850. [PubMed] [Google Scholar]
- Beach D. H., Klar A. J. Rearrangements of the transposable mating-type cassettes of fission yeast. EMBO J. 1984 Mar;3(3):603–610. doi: 10.1002/j.1460-2075.1984.tb01855.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Cai M. J., Davis R. W. Purification of a yeast centromere-binding protein that is able to distinguish single base-pair mutations in its recognition site. Mol Cell Biol. 1989 Jun;9(6):2544–2550. doi: 10.1128/mcb.9.6.2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carle G. F., Frank M., Olson M. V. Electrophoretic separations of large DNA molecules by periodic inversion of the electric field. Science. 1986 Apr 4;232(4746):65–68. doi: 10.1126/science.3952500. [DOI] [PubMed] [Google Scholar]
- Chikashige Y., Kinoshita N., Nakaseko Y., Matsumoto T., Murakami S., Niwa O., Yanagida M. Composite motifs and repeat symmetry in S. pombe centromeres: direct analysis by integration of NotI restriction sites. Cell. 1989 Jun 2;57(5):739–751. doi: 10.1016/0092-8674(89)90789-7. [DOI] [PubMed] [Google Scholar]
- Clarke L., Amstutz H., Fishel B., Carbon J. Analysis of centromeric DNA in the fission yeast Schizosaccharomyces pombe. Proc Natl Acad Sci U S A. 1986 Nov;83(21):8253–8257. doi: 10.1073/pnas.83.21.8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke L., Baum M. P. Functional analysis of a centromere from fission yeast: a role for centromere-specific repeated DNA sequences. Mol Cell Biol. 1990 May;10(5):1863–1872. doi: 10.1128/mcb.10.5.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke L., Baum M., Marschall L. G., Ngan V. K., Steiner N. C. Structure and function of Schizosaccharomyces pombe centromeres. Cold Spring Harb Symp Quant Biol. 1993;58:687–695. doi: 10.1101/sqb.1993.058.01.076. [DOI] [PubMed] [Google Scholar]
- Clarke L., Carbon J. The structure and function of yeast centromeres. Annu Rev Genet. 1985;19:29–55. doi: 10.1146/annurev.ge.19.120185.000333. [DOI] [PubMed] [Google Scholar]
- Cooke C. A., Bazett-Jones D. P., Earnshaw W. C., Rattner J. B. Mapping DNA within the mammalian kinetochore. J Cell Biol. 1993 Mar;120(5):1083–1091. doi: 10.1083/jcb.120.5.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottarel G., Shero J. H., Hieter P., Hegemann J. H. A 125-base-pair CEN6 DNA fragment is sufficient for complete meiotic and mitotic centromere functions in Saccharomyces cerevisiae. Mol Cell Biol. 1989 Aug;9(8):3342–3349. doi: 10.1128/mcb.9.8.3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumberledge S., Carbon J. Mutational analysis of meiotic and mitotic centromere function in Saccharomyces cerevisiae. Genetics. 1987 Oct;117(2):203–212. doi: 10.1093/genetics/117.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding R., McDonald K. L., McIntosh J. R. Three-dimensional reconstruction and analysis of mitotic spindles from the yeast, Schizosaccharomyces pombe. J Cell Biol. 1993 Jan;120(1):141–151. doi: 10.1083/jcb.120.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw W. C., Rothfield N. Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma. 1985;91(3-4):313–321. doi: 10.1007/BF00328227. [DOI] [PubMed] [Google Scholar]
- Fishel B., Amstutz H., Baum M., Carbon J., Clarke L. Structural organization and functional analysis of centromeric DNA in the fission yeast Schizosaccharomyces pombe. Mol Cell Biol. 1988 Feb;8(2):754–763. doi: 10.1128/mcb.8.2.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner M. M., Revzin A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 1981 Jul 10;9(13):3047–3060. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet A., Fitzgerald-Hayes M. Mutations in CEN3 cause aberrant chromosome segregation during meiosis in Saccharomyces cerevisiae. Genetics. 1989 Mar;121(3):477–489. doi: 10.1093/genetics/121.3.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haaf T., Warburton P. E., Willard H. F. Integration of human alpha-satellite DNA into simian chromosomes: centromere protein binding and disruption of normal chromosome segregation. Cell. 1992 Aug 21;70(4):681–696. doi: 10.1016/0092-8674(92)90436-g. [DOI] [PubMed] [Google Scholar]
- Hahnenberger K. M., Baum M. P., Polizzi C. M., Carbon J., Clarke L. Construction of functional artificial minichromosomes in the fission yeast Schizosaccharomyces pombe. Proc Natl Acad Sci U S A. 1989 Jan;86(2):577–581. doi: 10.1073/pnas.86.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahnenberger K. M., Carbon J., Clarke L. Identification of DNA regions required for mitotic and meiotic functions within the centromere of Schizosaccharomyces pombe chromosome I. Mol Cell Biol. 1991 Apr;11(4):2206–2215. doi: 10.1128/mcb.11.4.2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieter P., Mann C., Snyder M., Davis R. W. Mitotic stability of yeast chromosomes: a colony color assay that measures nondisjunction and chromosome loss. Cell. 1985 Feb;40(2):381–392. doi: 10.1016/0092-8674(85)90152-7. [DOI] [PubMed] [Google Scholar]
- Hyman A. A., Middleton K., Centola M., Mitchison T. J., Carbon J. Microtubule-motor activity of a yeast centromere-binding protein complex. Nature. 1992 Oct 8;359(6395):533–536. doi: 10.1038/359533a0. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W. D., Philippsen P. Purification of a protein binding to the CDEI subregion of Saccharomyces cerevisiae centromere DNA. Mol Cell Biol. 1989 Dec;9(12):5585–5593. doi: 10.1128/mcb.9.12.5585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W., Lechner J., Carbon J. Isolation and characterization of a gene (CBF2) specifying a protein component of the budding yeast kinetochore. J Cell Biol. 1993 May;121(3):513–519. doi: 10.1083/jcb.121.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klar A. J., Miglio L. M. Initiation of meiotic recombination by double-strand DNA breaks in S. pombe. Cell. 1986 Aug 29;46(5):725–731. doi: 10.1016/0092-8674(86)90348-x. [DOI] [PubMed] [Google Scholar]
- Kuhn R. M., Clarke L., Carbon J. Clustered tRNA genes in Schizosaccharomyces pombe centromeric DNA sequence repeats. Proc Natl Acad Sci U S A. 1991 Feb 15;88(4):1306–1310. doi: 10.1073/pnas.88.4.1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner J., Carbon J. A 240 kd multisubunit protein complex, CBF3, is a major component of the budding yeast centromere. Cell. 1991 Feb 22;64(4):717–725. doi: 10.1016/0092-8674(91)90501-o. [DOI] [PubMed] [Google Scholar]
- Masumoto H., Masukata H., Muro Y., Nozaki N., Okazaki T. A human centromere antigen (CENP-B) interacts with a short specific sequence in alphoid DNA, a human centromeric satellite. J Cell Biol. 1989 Nov;109(5):1963–1973. doi: 10.1083/jcb.109.5.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S., Klar A., Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Murakami S., Matsumoto T., Niwa O., Yanagida M. Structure of the fission yeast centromere cen3: direct analysis of the reiterated inverted region. Chromosoma. 1991 Dec;101(4):214–221. doi: 10.1007/BF00365153. [DOI] [PubMed] [Google Scholar]
- Muro Y., Masumoto H., Yoda K., Nozaki N., Ohashi M., Okazaki T. Centromere protein B assembles human centromeric alpha-satellite DNA at the 17-bp sequence, CENP-B box. J Cell Biol. 1992 Feb;116(3):585–596. doi: 10.1083/jcb.116.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray A. W., Szostak J. W. Construction of artificial chromosomes in yeast. Nature. 1983 Sep 15;305(5931):189–193. doi: 10.1038/305189a0. [DOI] [PubMed] [Google Scholar]
- Nakaseko Y., Adachi Y., Funahashi S., Niwa O., Yanagida M. Chromosome walking shows a highly homologous repetitive sequence present in all the centromere regions of fission yeast. EMBO J. 1986 May;5(5):1011–1021. doi: 10.1002/j.1460-2075.1986.tb04316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaseko Y., Kinoshita N., Yanagida M. A novel sequence common to the centromere regions of Schizosaccharomyces pombe chromosomes. Nucleic Acids Res. 1987 Jun 25;15(12):4705–4715. doi: 10.1093/nar/15.12.4705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman A. J., Lin R. J., Cheng S. C., Abelson J. Molecular consequences of specific intron mutations on yeast mRNA splicing in vivo and in vitro. Cell. 1985 Aug;42(1):335–344. doi: 10.1016/s0092-8674(85)80129-x. [DOI] [PubMed] [Google Scholar]
- Peterson J. B., Ris H. Electron-microscopic study of the spindle and chromosome movement in the yeast Saccharomyces cerevisiae. J Cell Sci. 1976 Nov;22(2):219–242. doi: 10.1242/jcs.22.2.219. [DOI] [PubMed] [Google Scholar]
- Polizzi C., Clarke L. The chromatin structure of centromeres from fission yeast: differentiation of the central core that correlates with function. J Cell Biol. 1991 Jan;112(2):191–201. doi: 10.1083/jcb.112.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- Steiner N. C., Hahnenberger K. M., Clarke L. Centromeres of the fission yeast Schizosaccharomyces pombe are highly variable genetic loci. Mol Cell Biol. 1993 Aug;13(8):4578–4587. doi: 10.1128/mcb.13.8.4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Murakami S., Chikashige Y., Funabiki H., Niwa O., Yanagida M. A low copy number central sequence with strict symmetry and unusual chromatin structure in fission yeast centromere. Mol Biol Cell. 1992 Jul;3(7):819–835. doi: 10.1091/mbc.3.7.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Murakami S., Chikashige Y., Niwa O., Yanagida M. A large number of tRNA genes are symmetrically located in fission yeast centromeres. J Mol Biol. 1991 Mar 5;218(1):13–17. doi: 10.1016/0022-2836(91)90867-6. [DOI] [PubMed] [Google Scholar]
- Waye J. S., Willard H. F. Nucleotide sequence heterogeneity of alpha satellite repetitive DNA: a survey of alphoid sequences from different human chromosomes. Nucleic Acids Res. 1987 Sep 25;15(18):7549–7569. doi: 10.1093/nar/15.18.7549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willard H. F. Centromeres of mammalian chromosomes. Trends Genet. 1990 Dec;6(12):410–416. doi: 10.1016/0168-9525(90)90302-m. [DOI] [PubMed] [Google Scholar]
- Zinkowski R. P., Meyne J., Brinkley B. R. The centromere-kinetochore complex: a repeat subunit model. J Cell Biol. 1991 Jun;113(5):1091–1110. doi: 10.1083/jcb.113.5.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]




