Summary
Recent work on the PDZ-LIM protein family has revealed important activities at the cellular level, mediating signals between the nucleus and the cytoskeleton, with significant impact on organ development. We review and integrate current knowledge about the PDZ-LIM protein family and propose a new functional role, sequestering nuclear factors in the cytoplasm. Characterized by their PDZ and LIM domains, the PDZ-LIM family is comprised of evolutionarily conserved proteins found throughout the animal kingdom, from worms to humans. Combining two functional domains in one protein, PDZ-LIM proteins have wide-ranging and multi-compartmental cell functions during development and homeostasis while, in contrast, misregulation can lead to cancer formation and progression. New emerging roles include interactions with integrins, T-box transcription factors, and receptor tyrosine kinases. Facilitating the assembly of protein complexes, PDZ-LIM proteins can act as signal modulators, influence actin dynamics, regulate cell architecture and control gene transcription.
Keywords: PDZ-LIM, scaffold, actin cytoskeleton, organogenesis, T-box
Evolutionarily conserved building blocks for a heterogeneous protein family
The PDZ-LIM family is comprised of proteins with multiple functional domains and all share a PDZ domain combined with at least one LIM domain. As protein-protein interaction modules, PDZ and LIM domains act as scaffolds, binding to filamentous actin-associated proteins, a range of cytoplasmic signaling molecules, and nuclear proteins, allowing this family to carry out diverse functions during development and adulthood (1-17). PDZ-LIM proteins are conserved from invertebrates to vertebrates, indicating functional importance in all species (16,18). During the evolution of this protein family, a number of domain duplication and rearrangements occurred, generating various subfamilies, and individual proteins. The ALP gene subfamily (ALP, Clp-36/Elfin, Mystique and RIL) encodes proteins with a single PDZ and LIM domain at its N-terminal and C-terminal end, respectively. The Enigma subfamily of proteins (Enigma/LMP, Enigma Homolog and Cypher/Zasp) feature one N-terminal PDZ domain and three C-terminal LIM domains. The LIM only protein 7 (LMO7) contains one N-terminal PDZ domain, one C-terminal LIM domain, and an additional Calponin homology domain. The LIM kinase subfamily consists of two members (LIMK1 and 2), which encode two N-terminal LIM domains, one central PDZ domain and one C-terminal kinase domain. The individual PDZ and LIM domains of all PDZ-LIM encoding genes appear to be evolutionarily linked. The ALP and Enigma subfamilies have originated from a common ancestral gene with one N-terminal PDZ domain and four C-terminal LIM domains, similar to the worm C. elegans alp-1/eat-1 and Drosophila melanogaster tungus, respectively (16,18). In contrast, LMO7 and the LIMKs are the most distantly related genes. A current model for the molecular evolution of the PDZ-LIM genes and their diverse gene architecture has been recently described by te Velthuis et al. (18). Figure 1 shows the architecture of the PDZ-LIM protein subfamilies and their phylogenetic relationship.
Figure 1.
The PDZ-LIM protein family. (A) General architecture and domain structure of the Eat-1/Tungus, Enigma and ALP subfamilies of PDZ-LIM proteins. The PDZ domain is indicated by the blue box and the LIM domain is depicted by the purple boxes. (B) Phylogenetic tree of members of the PDZ-LIM protein family. The single LIM domain (ALP subfamily) and three LIM domain (Enigma subfamily) gene structures evolved from a common ancestral gene encoding four LIM domains as found in the C. elegans Alp-1/Eat-1 and D. melanogaster Tungus proteins. While the ALP/Enigma subfamilies and LMO7 and LIM kinases appear to have a common ancestral origin (16,18), LMO7 and LIMK proteins reveal a complex evolution and are not included in the phylogenetic tree. Available sequences from C. elegans (Ce), zebrafish (Dr), chicken (Gg), mouse (Mm), and human (Hs) were compiled using MacVector 7.0 software (Oxford Molecular Company, Madison, WI). C. elegans Alp-1, the earliest known PDZ-LIM protein, was used as the out-group. The new nomenclature proposed by GenBank is displayed next to the phylogenetic tree with the previously published names on the brackets to the right. GenBank accession numbers for all genes discussed are as follows: C. elegans alp-1, CAE52903. D. melanogaster tungus, NM_137217; Ldb3: Homo sapiens, NM_007078; Mus Musculus, NM_011918; Gallus gallus, XM_421495; Danio rerio, DQ012157; D. melanogaster, NM_145757. Pdlim5: Homo sapiens, NM_006457; Mus Musculus, NM_019808; Gallus gallus, AJ851689; Danio rerio, BC045922. Pdlim7: Homo sapiens, NM_005451, Mus musculus, NM_026131: Gallus gallus, NM_001005345; Danio rerio, NM_200840. Pdlim1: Homo sapiens, NM_020992; Mus musculus, NM_016861; Gallus gallus, XM_426503; Danio rerio, BC092978. Pdlim3: Homo sapiens, NM_014476; Mus musculus, NM_016798; Gallus gallus, NM_001001764; Danio rerio, NM_001042718. Pdlim4: Homo sapiens, NM_003687; Mus musculus, NM_019417; Gallus gallus, NM_204839; Danio rerio, NM_001042696. Pdlim2: Homo sapiens, NM_176871; Mus musculus, NM_145978; Danio rerio, NM_001042766. Pdlim2 sequence could not be identified in the Gallus gallus genome.
While both the PDZ and the LIM domains are often found in combination with other peptide motifs or domains, the co-evolution of the two invariant functional domains, PDZ and LIM, indicate high functional relevance and thus, is of particular interest and the focus of this review. All family members associate with the actin cytoskeleton and this property, organizing protein complexes at the cytoskeleton, may serve a range of unexpected, yet important biological roles.
The PDZ domain
Found in bacteria, yeast, and throughout the animal kingdom, the PDZ domain is one of the most common protein-protein binding domains (17,19). It is characterized by a highly conserved sequence of 80-90 amino acids consisting of six anti-parallel β-strands and two α-helices (19,20). PDZ domains can be found in proteins with only a PDZ domain or in proteins consisting of additional functional motifs (21). The PDZ domain provides a protein-binding interface, allowing the formation of multi-protein complexes with a variety of partners including, membrane-associated proteins, cytoplasmic signaling proteins and cytoskeletal proteins (17,19,21-25). PDZ domain interaction with actin-binding proteins, α-actinin, palladin or β-tropomyosin, localizes the PDZ-LIM proteins and other complexed proteins to filamentous actin, which points to a function in the organization and regulation of cell architecture (4-6,10,12-15,26-29).
The LIM domain
Similar to the PDZ domain, the LIM domain also acts as a modular protein binding interface identified in bacteria, yeast and animals (30,31). The LIM domain is approximately 55 amino acids long and characterized by highly conserved and spatially defined cysteine and histidine residues, which coordinate the binding of two zinc ions, thereby forming a two zinc-finger-like structure (17,30-35). The LIM domain can be found in proteins alone or in conjunction with other domains (30,31). The LIM domains bind highly diverse partners, ranging from signaling molecules, actin cytoskeletal components to transcription factors, which support cellular functions including, actin organization, integrin-dependent adhesion and signaling and cell-fate determination (1-4,6,17,30,31,36-40). Of note, while structurally very similar, individual LIM domains within the same protein bind different targets with high specificity. This enables complex protein interactions and opportunities for this family to be involved in many biological functions as indicated by expression in the brain, liver, kidney and bones (15,28,41).
PDZ-LIM proteins have diverse functions in heart and skeletal muscle development
As species evolved into larger and more complex organisms, so did the process of organogenesis. This required protein networks working together in a highly organized and regulated manner to insure correct temporal and spatial development. Therefore, it is not surprising that PDZ and LIM interaction modules have been selected to facilitate protein binding and subcellular localization in vital organs, such as the heart from zebrafish to human (6,11,42-47). Expression during cardiac development has been detected for many PDZ-LIM genes in overlapping domains (6,11,42-46). However, despite overlapping expression, specific heart defects are observed following knock-down/out of several PDZ-LIM genes (11,42,43,45,46). LMO7 knock-down in zebrafish results in conduction system defects, including arrhythmias (46). Zebrafish ldb3 (also known as Cypher/Zasp) morphants have an elongated atrium and a smaller, disorganized ventricle (45). Homozygous Ldb3 knock-out mice display right and left ventricular dilation (43). Similarly, mutations in human LDB3 have been identified in patients with dilated cardiomyopathy (47). Unlike Ldb3, Pdlim3 (also known as ALP) mutant mice exhibit only right ventricular dilation (42). The differing heart defects observed are likely due to the fact that Ldb3 localizes to the Z-lines while Pdlim3 localizes to the intercalated discs of cardiomyocytes (4,42,43). Knock-down of Pdlim7 (also known as Enigma/LMP) in zebrafish causes deficiencies at the atrio-ventricular (AV) boundary resulting in loss of valve tissue and looping defects producing a string-like heart (11). These knock-down/out studies strongly support distinct roles for PDZ-LIM proteins during heart development and maintenance.
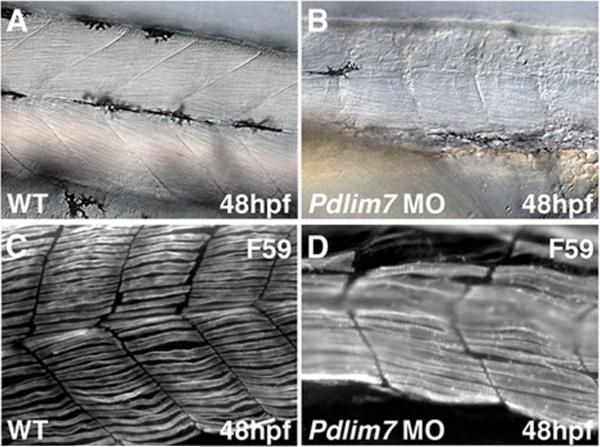
In addition to the heart, PDZ-LIM proteins show localization to the developing and adult skeletal musculature, suggesting a role not only in skeletal muscle organization, but also maintenance (4,5,9,11,14,28,42,43,45,46,48,49). In agreement with this notion, pdlim7 compromised zebrafish display severe defects in skeletal muscle (11). For example, Figure 2 shows disorganization of trunk slow muscle fibers resulting in contractile disfunction. Ldb3 mutations in the fruit fly D. melanogaster are lethal at the first larval instar, corresponding to the start of muscle contraction (5). In addition, knock-down of Ldb3 in zebrafish results in failure of somite compartmentalization and skeletal muscle organization (45). In the mouse, homozygous Ldb3 mutants do not survive past postnatal day five due to a combination of limb weakness, an inability to suckle and a gasping respiratory pattern (43). Unlike fruit flies and zebrafish, examination of the striated muscle in these mutant mice showed that Ldb3 is necessary for maintenance of the Z-line during muscle contraction, rather than formation of the Z-line structure (43). In contrast, Pdlim3 mutant mice do not display skeletal muscle defects, suggesting that either this PDZ-LIM protein is not necessary for skeletal muscle development/maintenance or there are compensatory activities by other family members (42,50). The overlapping expression of both Pdlim3 and Ldb3 at the Z-line in skeletal muscle supports the hypothesis of functional redundancy (43). The evolutionary conservation of PDZ-LIM proteins indicates specific and important roles in organ development and maintenance, but also suggests that they have a critical function in basic morphogenetic processes. Future studies in the worm, fruit fly, and zebrafish, where individual protein function appears to be less masked by redundancy but also the generation of compound PDZ-LIM mutant mice will hopefully uncover the full range of biological properties.
Figure 2.
PDZ-LIM proteins are required for muscle development and maintenance. High magnification example of zebrafish muscle phenotype after knock-down of pdlim7 with a splice-inhibiting morpholino (MO) antisense oligonucleotide (11). (A-B) Brightfield microscopy representing a lateral view of wild-type (A), and pdlim7 MO injected embryos (B) at 48 hpf. (C-D) Fluorescent microscopy showing a lateral view of wild-type (C), and pdlim7 MO injected embryos (D) at 48 hpf with F59 antibody (Developmental Studies Hybridoma Bank, University of Iowa) to detect slow muscle fibers. In comparison to the wild-type, the pdlim7 morphants exhibit elongated and severely disorganized slow muscle fibers. hpf=hours post-fertilization.
PDZ-LIM proteins function with integrins and during tumorgenesis via actin regulation
One of the most basic cellular processes is cell migration. During directed migration, a cell adheres to either the extracellular matrix (ECM) or neighboring cells via transmembrane receptors, such as integrins. The cell must also break and rebuild these contacts in order to make progress towards its destination (51). Therefore, regulating expression and modification of cell adhesion molecules is critical for proper cell migration.
The PDZ domain of Pdlim1 (also known as Clp-36/Elfin), Pdlim3, Pdlim5 (also known as ENH), LMO7, and Ldb3 binds to α-actinin at adherens junctions (5,9,13-15,52,53), a site of integrin localization (5,26). In D. melanogaster, Ldb3 and integrins were shown to genetically interact (5). Loss of Ldb3 results in muscle detachment following contraction, suggesting Ldb3 regulates or strengthens the association of integrins with the actin cytoskeleton. These findings are in agreement with in vitro data indicating a role for Ldb3 in cell adhesion. Pdlim2 (also known as Mystique) co-localizes with α-actinin and β1-integrin at cytoskeleton focal contacts in breast epithelial cells (26). Knock-down of Pdlim2 causes loss of adhesion and migration while over-expression results in enhanced cell adhesion (54). Similarly, knock-down of Pdlim1 in a trophoblast-derived choriocarcinoma cell line (BeWo) results in failure of stress fiber and focal adhesion formation (52). Pdlim4 (also known as RIL) functions in regulating stress fiber formation and turnover by increasing the ability of α-actinin to associate with actin filaments (27). The functional interactions with integrins suggest that PDZ-LIM proteins may participate in focal adhesion signaling cascades, communicating the extracellular milieu to intracellular regulatory pathways and as a result modifying the actin cytoskeleton. These studies with different family members indicate an overall role for PDZ-LIM proteins in cell-cell, cell-matrix interaction and migration, all functions essential for organ formation and maintenance.
However, when PDZ-LIM proteins become misregulated, rather than supporting the maintenance of an organ, they can actually damage it by promoting cancer cell invasion and metastasis (55-57). The hallmarks of tumor cell invasion and the initial steps in metastasis are uncontrolled cell motility and proliferation (58). Loss of cell-cell contact as a function of disruption of adherens junctions by misregulation of integrins and E-cadherin is thought to contribute to cancer cell invasion (59,60). LMO7 localizes to adherens junctions in epithelial cells where it directly binds the filamentous actin binding proteins, afadin and α-actinin. LMO7 connects the cell-cell adhesion molecules, nectin and E-cadherin via α-actinin, thus stabilizing the nectin and E-cadherin based adherens junctions (53). Loss of LMO7 in mice causes the formation of irregular epithelial lesions in the respiratory bronchioles and alveolar ducts early in adulthood, which later develops into lung cancer (55). In addition, LIMK1 is activated during mitosis by cyclin-dependent kinases (CDKs), which results in inactivation of cofilins and consequently actin cytoskeleton reorganization (61,62). Over-expression of LIMK1 in human breast cancer cells leads to increased cell proliferation and cell invasion by up-regulating the urokinase-type plasminogen activator (uPA) system (56). Similarly, enhanced levels of LIMK1 in prostate cancer cells result in increased cell invasion while reduction causes cells to arrest at the G2/M phase of the cell cycle (57). Furthermore, searching the NCBI Gene Expression Omnibus (GEO) for microarray profiles in cancer samples produces a number of data sets showing differential regulation of PDZ-LIM genes in cancer. Undoubtedly, proper regulation of PDZ-LIM proteins is key to preventing uncontrolled actin reorganization, proliferation, and cell motility. A deeper knowledge of how PDZ-LIM proteins function in basic cellular processes will be essential in understanding their role in tissue homeostasis and disease progression, such as cancer.
PDZ-LIM proteins retain an evolutionary role: scaffolds bringing signaling molecules to the actin cytoskeleton
In addition to interacting with integrins, PDZ-LIM proteins have been shown to bind several other signaling molecules via their LIM domains (1-4,36,37,41,63). In 1994, Wu and Gill first identified human PDLIM7 to bind the endocytic code of the insulin receptor (InsR), which is thought to become accessible after ligand binding and activation. The interaction requires LIM domain 3 of PDLIM7 and functions during endocytosis of the InsR (36). LIM domain 2 of Pdlim7 binds to the carboxy-terminus of the Ret receptor, anchoring the Ret/ptc2 complex to the proper subcellular localization (2,36,37). However, LIM domain 1 of Pdlim7 does not interact with either signaling molecule and presumably has a yet unidentified binding partner (1). Each of the three individual LIM domains of Pdlim5 is sufficient for interaction with activated Protein Kinase C (PKC), resulting in the translocation of Pdlim5 from the membrane to the cytosol (3). Similarly, the LIM domains of Ldb3 interact with PKC delivering the kinase to the actin cytoskeleton via its PDZ domain (4). Taken together, these studies strongly support a role as adapter proteins, recruiting signaling molecules to the actin cytoskeleton. Though, the functional significance of the interactions between the LIM domains of PDZ-LIM proteins and receptor tyrosine kinases, as well as other signaling molecules, remains elusive.
Uncovering a new role: PDZ-LIM proteins sequester nuclear proteins in the cytoplasm
In addition to localizing signaling molecules to the actin cytoskeleton, more recent studies demonstrate that the LIM domains of PDZ-LIM proteins also bind key nuclear proteins, re-localizing and tethering them to cytoplasmic sites (6-8,10,11,64). For example, Pdlim1 sequesters the predominantly nuclear kinase, Clik1, to actin stress fibers where the kinase is thought to alter the actin cytoskeleton (64). Human PDLIM5 binds Id2, a negative regulator of basic helix-loop-helix (bHLH) transcription factors, via LIM domain 1 (8). During neuronal differentiation, PDLIM5 is upregulated by retinoic acid, resulting in the sequestration of Id2 in the cytoplasm and loss of inhibitory effects on nuclear transcription factors (7,8). Blocking nuclear export of Id2 results in accumulation of the protein in the nucleus and enhanced repression of bHLH target genes, MyoD and E27, which would indicate that Id2 function is regulated by nucleo-cytoplasmic shuttling (7). In vitro studies show that Pdlim5 also binds Id1 and Id3, suggesting that Pdlim5 may function to sequester Id proteins in the cytoplasm; however, this remains to be confirmed (8). Our laboratory has shown that chicken Pdlim7 binds the C-terminus of the Tbx4 and Tbx5 transcription factors in a regulated fashion, tethering them to the actin cytoskeleton (6,10,44). Studies both in vitro and in vivo during zebrafish cardiogenesis where the amount of Pdlim7 is increased, thus lowering nuclear Tbx5 levels, result in loss of Tbx5 target gene expression, demonstrating that Pdlim7 is capable of regulating Tbx5 transcriptional activity (10,11). Taken together, these findings indicate a role for PDZ-LIM proteins in regulating nuclear activities by sequestering transcription factors in the cytoplasm. It will be of interest to determine what additional nuclear factors are interacting with PDZ-LIM proteins and potentially regulated in their activity by subcellular relocalization.
The T-box transcription factor family can localize to nuclear and cytoplasmic sites
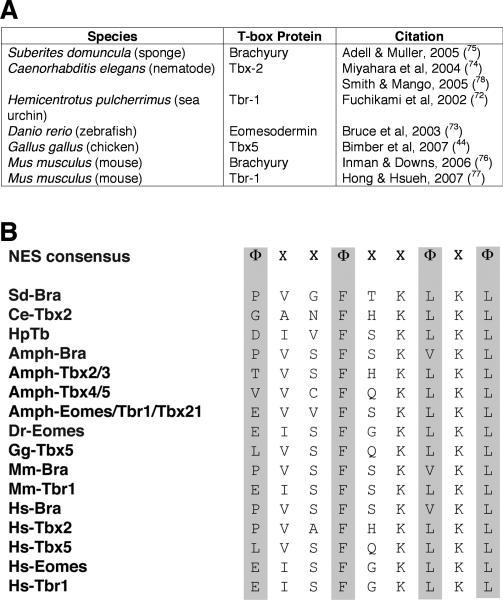
Based on studies from our laboratory, one intriguing possibility for additional nuclear binding partners of PDZ-LIM proteins is the T-box family of transcription factors. At least 22 different T-box genes have been reported in vertebrates, and like the PDZ-LIM gene family, they all evolved from a single primordial gene that underwent unequal crossing-over and gene duplication events (65-68). Like most transcription factors, T-box proteins contain nuclear localization signal (NLS) sequences (69,70). In addition, our group has characterized a functional nuclear export signal (NES) in chicken Tbx5, which provides a mechanism for how the transcription factor exits the nucleus to bind Pdlim7 in the cytoplasm of the cell (71). In fact, an NES motif is present within the N-terminal DNA binding domain of all T-box proteins from humans down to the simplest metazoan, the sponge (72-78). A current account of T-box proteins with documented in vivo cytoplasmic localization and conserved NES is compiled in Figure 3. In addition, Tbx3, Tbx5 and Brachyury all interact with the CRM1 export protein via the conserved NES, indicating that T-box transcription factors use the CRM1 nuclear export pathway (71). We have shown for Tbx5 that only in the presence of Pdlim7, it can dynamically shuttle between nuclear and cytoplasmic cell compartments (10). This relocalization function appears to have been adopted at the earliest stage of T-box protein evolution and retained in all family members; however, the functional roles for the protein shuttling remain elusive.
Figure 3.
T-box transcription factors can localize outside the nucleus using a conserved nuclear export signal (NES). (A) Table listing reports of T-box proteins localized to the cytoplasm ranging from the mouse to the sponge. (B) Alignment of the NES from T-box proteins known to localize to the cytoplasm and their orthologs in the cephalochordate amphioxus and humans, demonstrating an evolutionarily conserved nuclear export mechanism. The NES consensus sequence was derived from proteins that utilize the CRM1 nuclear export pathway, where Φ represents hydrophobic residues (L, I, F, V, M, P) and X represents any amino acid. GenBank accession numbers for all sequences are as follows: Brachyury: Suberites domuncula AJ_544242; Branchiostoma floridae X_91903; Mus musculus NP_033335; Homo sapiens NM_003181. Tbx2: C. elegans Q_19691; Branchiostoma floridae (Tbx2/3) AF_262563; Homo sapiens NM_005994. Tbx5: Gallus gallus NM_204173; Branchiostoma floridae (Tbx4/5) AF_262564; Homo sapiens NM_000192. Tbr-1: Hemicentrotus pulcherrimus AB_048760; Branchiostoma floridae (Eomesdermin /Tbr1/Tbx21) AF_262568; Mus musculus NP_033348; Homo sapiens NM_006593. Eomesdermin: Danio rerio AF_329830; Homo sapiens NM_005442.
T-box transcription factors interact with actin-associated PDZ-LIM proteins
Combining the hypotheses that all T-box proteins are shuttling factors and that PDZ-LIM proteins sequester nuclear proteins outside the nucleus, it would be further conceivable that these protein interactions may not be limited to Pdlim7 and Tbx4 or Tbx5. For example, during mouse heart development, Brachyury protein is detected in the cytoplasm temporally corresponding to actin associated localization of Pdlim3 (42,76). In addition, C. elegans TBX-2 is expressed in a filamentous pattern in pharyngeal muscle cells, a cell type that also expresses the actin-associated primordial PDZ-LIM protein ALP-1 (16,78). Of note, the C. elegans pharynx has contracting properties and may be considered an early pumping organ, preceding the vertebrate heart. T-box genes are expressed in a wide variety of tissues and are found in the earliest metazoans (68). PDZ-LIM genes are expressed in several of the same tissues as T-box genes and while the combination of the PDZ and LIM domain in a single protein appears to have evolved after T-box proteins, the LIM domain is found in all eukaryotes (30). Thus, the potential exists that these T-box and PDZ-LIM genes and the interactions between respective encoded protein domains may have co-evolved, supporting a functional relationship between these two seemingly unrelated protein families.
While direct binding has only been documented for Pdlim7 and Tbx4 or Tbx5, there is reason to believe that additional T-box and PDZ-LIM proteins may be interacting and thus functioning together during heart development (6,42-44,79-88). In the mouse, Tbx1, Tbx2, and Tbx20 are expressed together with Pdlim3 and Ldb3 at the same stages in the outflow tract (OFT) (42,43,79-83). Other examples include coexpression of Tbx2, Tbx20, Pdlim3, Pdlim7 and Ldb3 at similar stages in the atrioventricular canal (AVC) of the developing heart (42-44,79,81,82,84-87), and Tbx20, Pdlim3, Pdlim7 and Ldb3 coexpression in the developing right ventricle (6,42,43,85,88). The overlapping expression of multiple T-box and PDZ-LIM mRNAs and/or proteins in several cardiac tissues, support the hypothesis that T-box and PDZ-LIM proteins work together during cardiac development.
In agreement with the similarities in expression, knock-down/out studies of T-box and PDZ-LIM proteins result in heart defects in the same structures (11,42,43,80,89-94). For instance, OFT defects were observed following loss of Tbx1, Tbx2, Tbx3 and Pdlim3 (42,80,89-92). Tbx20, Pdlim3, and Ldb3 mutant mice all display right ventricular dilated cardiomyopathy (42,43,93). In the zebrafish, tbx5 mutant and pdlim7 morphant embryos both exhibit heart-looping defects as well as AV boundary and valve defects, which in these cases are related to changes in Tbx5/Pdlim7 protein interactions (11,94). Re-analysis of T-box and PDZ-LIM mutants, specifically focusing on the respective T-box or PDZ-LIM protein localization and function is warranted to gain new insights into the relationship these proteins have during development.
Conclusions, future perspectives and open questions: Regulation of protein interactions
We have reviewed the evolutionary history, structure, function and binding partners of the PDZ-LIM protein family. PDZ-LIM proteins function in skeletal muscle organization and maintenance by stabilizing the actin cytoskeleton and regulating cell adhesion via their interactions with α-actinin and integrins, respectively. While PDZ-LIM proteins have overlapping roles during skeletal/cardiac muscle development, they also possess more exclusive functions, such as in valve formation. This is likely due in part to specific interactions with other proteins. The full range of PDZ-LIM binding partners and the functional consequences of these interactions remain poorly understood, but hold great promise for exciting new discoveries.
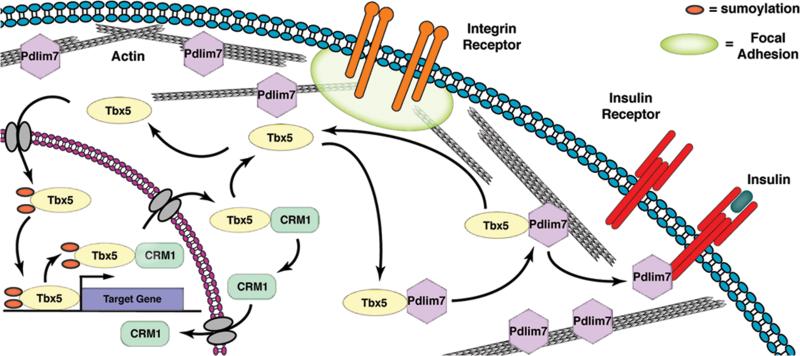
Recently, our laboratory was the first to show in vivo evidence of Pdlim7 regulating nppa and tbx2b gene expression via its interaction with the transcription factor Tbx5 (11). Pdlim7 morphant zebrafish fail to form an AV boundary due to the expansion of the Tbx5 target genes, nppa and tbx2b, which results in loss of valve specific tissue. In contrast, tbx5 null zebrafish display excessive valve-like tissue with reduced nppa and no tbx2b gene expression. The changes of nppa and tbx2b expression in vivo correspond with our in vitro findings that Pdlim7 can repress Tbx5 activity by actin-associated sequestration (10). In Figure 4, we propose a model for the novel function of PDZ-LIM proteins in regulating nucleo-cytoplasmic functions using Tbx5 and Pdlim7 as examples. Dynamic and regulated interaction of Pdlim7 and Tbx5 leads to the modulation of Tbx5 levels in the nucleus or cytoplasm and in turn influences target gene transcription. Therefore, PDZ-LIM proteins may serve to fine-tune levels of transcription factors in the nucleus to achieve appropriate gene regulation.
Figure 4.
Model for the diverse and multi-compartmental functions of PDZ-LIM proteins. PDZ-LIM proteins are engaged in dynamic interactions with cellular proteins. For example, Pdlim7 facilitates shuttling of Tbx5 between nucleus and actin cytoskeleton. Export of Tbx5 from the nucleus is promoted by CRM1, and protein subcellular localization and function possibly regulated by reversible sumoylation. Besides tethering Tbx5 to the actin cytoskeleton, Pdlim7 can interact with other cellular proteins, including the activated InsR. Pdlim7 uses the same domain to bind either Tbx5 or InsR and this competitive interaction may provide a mechanism for Tbx5 release from filamentous actin and relocation to the nucleus. Transmembrane integrin receptors anchor the actin cytoskeleton to focal adhesions and transmit information from the extracellular environment. Several PDZ-LIM proteins have been localized to focal adhesions, providing the possibility that Pdlim7 could similarly engage with proteins clustered in focal adhesions including integrin receptors to sense extracellular stress.
However, it is also possible that sequestering nuclear proteins to the actin cytoskeleton may provide certain nuclear factors with yet unknown functions at these cytoplasmic sites. For example, Pdlim1 sequesters the nuclear kinase, Clik1, in the cytoplasm where Clik1 kinase disrupts the periodic staining pattern of Pdlim1 on actin, suggesting the nuclear kinase plays a role in the cytoplasm altering actin stress fibers (64). It is also plausible that the binding of specific nuclear proteins by PDZ-LIM proteins in turn regulates the function of PDZ-LIM proteins on the actin cytoskeleton. Based on the fact that nuclear proteins localize to non-nuclear compartments via interaction with cytoplasmic proteins such as PDZ-LIM proteins, we must reanalyze their functions accordingly.
Regardless of whether the interactions between PDZ-LIM and specific nuclear proteins are required for transcriptional regulation or additional non-nuclear functions, the interactions need to be highly regulated. Coexpression of both, Pdlim7 and Tbx5 in vitro or in vivo is not sufficient for binding; however, the interaction and relocalization of the transcription factor seems to be linked to the induction of differentiation (10,44). Tbx5 utilizes the CRM1 export pathway in order to interact with Pdlim7 at the actin cytoskeleton (Fig. 4), but what triggers Tbx5 to interact with CRM1 or become exported remains to be determined. Pdlim7 binds both Tbx5 and the InsR via its third LIM domain in a competitive manner (Undesser, Camarata and Simon unpublished data). Thus, one idea could be that insulin signaling regulates Pdlim7/Tbx5 interactions. For example, in the presence of insulin, the InsR can bind to Pdlim7 LIM domain 3, thus releasing Tbx5 to relocalize to the nucleus (Fig. 4).
Another potential means for regulating PDZ-LIM/nuclear protein interactions is post-translational modification. For example, Pdlim5 binds Id2 and PKC via its LIM domains; therefore, activated PKC could phosphorylate Id2, Pdlim5 or both and in so doing alter the binding of the two proteins (3). In C. elegans pharyngeal muscle cells, TBX-2 protein displays cytoplasmic localization (78). In addition, normal TBX-2 function in the worm requires sumoylation at two consensus sites (95). Both SUMO sites are conserved in chicken Tbx5 and as compared to its predicted size (58kDa), the protein migrates at a higher molecular weight (75-80kDa) on a Western blot, a size difference close to the addition of two 10kDa SUMO proteins (10). The addition of SUMO groups has been shown to control nucleo-cytoplasmic shutting of other proteins (96,97). Therefore, future studies focusing on post-translational modification will provide a greater appreciation for how nucleo-cytoplasmic shuttling is regulated (Fig. 4).
The actin associated PDZ-LIM proteins were initially described as binding to signaling molecules during muscle development and maintenance. More recent studies reveal a novel role for this protein family in regulating transcriptional activity by sequestering nuclear proteins in the cytoplasm. For example, our laboratory has shown that PDZ-LIM regulation of T-box proteins is necessary for proper heart formation. In contrast, misregulation of PDZ-LIM and T-box proteins has been implicated in congenital heart disease and cancer formation and progression. We have only begun to scratch the surface of discovering the roles of PDZ-LIM proteins and the functional consequences of their interactions with nuclear factors. Could it be that PDZ-LIM proteins mediate information flow from extracellular to intracellular to nuclear compartments? Future studies focusing on such questions will provide essential new insights into how these dynamic protein networks relate to tissue and organ development and disease.
Acknowledgements
To keep the focus of this review we had to be selective concerning citations and apologize for the potential omission of other work. We thank Dr. Teng-Leong Chew for critical reading of the manuscript and helpful suggestions. This work is supported by NRSA Fellowship F31HL090031 (J. Krcmery), American Heart Association Pre- and Postdoctoral Fellowships (T. Camarata and A. Kulisz, respectively), and NIH grant R01HL085834 (H.-G. Simon).
References
- 1.Wu R, Durick K, Songyang Z, Cantley LC, Taylor SS, Gill GN. Specificity of LIM domain interactions with receptor tyrosine kinases. J Biol Chem. 1996;271(27):15934–41. doi: 10.1074/jbc.271.27.15934. [DOI] [PubMed] [Google Scholar]
- 2.Durick K, Wu RY, Gill GN, Taylor SS. Mitogenic signaling by Ret/ptc2 requires association with enigma via a LIM domain. J Biol Chem. 1996;271(22):12691–4. doi: 10.1074/jbc.271.22.12691. [DOI] [PubMed] [Google Scholar]
- 3.Kuroda S, Tokunaga C, Kiyohara Y, Higuchi O, Konishi H, Mizuno K, Gill GN, Kikkawa U. Protein-protein interaction of zinc finger LIM domains with protein kinase C. J Biol Chem. 1996;271(49):31029–32. doi: 10.1074/jbc.271.49.31029. [DOI] [PubMed] [Google Scholar]
- 4.Zhou Q, Ruiz-Lozano P, Martone ME, Chen J. Cypher, a striated muscle-restricted PDZ and LIM domain-containing protein, binds to alpha-actinin-2 and protein kinase C. J Biol Chem. 1999;274(28):19807–13. doi: 10.1074/jbc.274.28.19807. [DOI] [PubMed] [Google Scholar]
- 5.Jani K, Schock F. Zasp is required for the assembly of functional integrin adhesion sites. J Cell Biol. 2007;179(7):1583–97. doi: 10.1083/jcb.200707045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krause A, Zacharias W, Camarata T, Linkhart B, Law E, Lischke A, Miljan E, Simon HG. Tbx5 and Tbx4 transcription factors interact with a new chicken PDZ-LIM protein in limb and heart development. Dev Biol. 2004;273(1):106–20. doi: 10.1016/j.ydbio.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 7.Kurooka H, Yokota Y. Nucleo-cytoplasmic shuttling of Id2, a negative regulator of basic helix-loop-helix transcription factors. J Biol Chem. 2005;280(6):4313–20. doi: 10.1074/jbc.M412614200. [DOI] [PubMed] [Google Scholar]
- 8.Lasorella A, Iavarone A. The protein ENH is a cytoplasmic sequestration factor for Id2 in normal and tumor cells from the nervous system. Proc Natl Acad Sci U S A. 2006;103(13):4976–81. doi: 10.1073/pnas.0600168103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pomies P, Macalma T, Beckerle MC. Purification and characterization of an alpha-actinin-binding PDZ-LIM protein that is up-regulated during muscle differentiation. J Biol Chem. 1999;274(41):29242–50. doi: 10.1074/jbc.274.41.29242. [DOI] [PubMed] [Google Scholar]
- 10.Camarata T, Bimber B, Kulisz A, Chew TL, Yeung J, Simon HG. LMP4 regulates Tbx5 protein subcellular localization and activity. J Cell Biol. 2006;174(3):339–48. doi: 10.1083/jcb.200511109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Camarata T, Krcmery J, Snyder D, Park S, Topczewski J, Simon HG. Pdlim7 (LMP4) regulation of Tbx5 specifies zebrafish atrio-ventricular boundary and valve formation. Dev Biol. doi: 10.1016/j.ydbio.2009.10.039. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brenman JE, Chao DS, Gee SH, McGee AW, Craven SE, Santillano DR, Wu Z, Huang F, Xia H, Peters MF. Interaction of nitric oxide synthase with the postsynaptic density protein PSD-95 and alpha1-syntrophin mediated by PDZ domains. Cell. 1996;84(5):757–67. doi: 10.1016/s0092-8674(00)81053-3. others. [DOI] [PubMed] [Google Scholar]
- 13.Nakagawa N, Hoshijima M, Oyasu M, Saito N, Tanizawa K, Kuroda S. ENH, containing PDZ and LIM domains, heart/skeletal muscle-specific protein, associates with cytoskeletal proteins through the PDZ domain. Biochem Biophys Res Commun. 2000;272(2):505–12. doi: 10.1006/bbrc.2000.2787. [DOI] [PubMed] [Google Scholar]
- 14.Xia H, Winokur ST, Kuo WL, Altherr MR, Bredt DS. Actinin-associated LIM protein: identification of a domain interaction between PDZ and spectrin-like repeat motifs. J Cell Biol. 1997;139(2):507–15. doi: 10.1083/jcb.139.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vallenius T, Luukko K, Makela TP. CLP-36 PDZ-LIM protein associates with nonmuscle alpha-actinin-1 and alpha-actinin-4. J Biol Chem. 2000;275(15):11100–5. doi: 10.1074/jbc.275.15.11100. [DOI] [PubMed] [Google Scholar]
- 16.McKeown CR, Han HF, Beckerle MC. Molecular characterization of the Caenorhabditis elegans ALP/Enigma gene alp-1. Dev Dyn. 2006;235(2):530–8. doi: 10.1002/dvdy.20633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.te Velthuis AJ, Bagowski CP. PDZ and LIM domain-encoding genes: molecular interactions and their role in development. ScientificWorldJournal. 2007;7:1470–92. doi: 10.1100/tsw.2007.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.te Velthuis AJ, Isogai T, Gerrits L, Bagowski CP. Insights into the molecular evolution of the PDZ/LIM family and identification of a novel conserved protein motif. PLoS ONE. 2007;2(2):e189. doi: 10.1371/journal.pone.0000189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sheng M, Sala C. PDZ domains and the organization of supramolecular complexes. Annu Rev Neurosci. 2001;24:1–29. doi: 10.1146/annurev.neuro.24.1.1. [DOI] [PubMed] [Google Scholar]
- 20.Fanning AS, Anderson JM. PDZ domains: fundamental building blocks in the organization of protein complexes at the plasma membrane. J Clin Invest. 1999;103(6):767–72. doi: 10.1172/JCI6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jelen F, Oleksy A, Smietana K, Otlewski J. PDZ domains - common players in the cell signaling. Acta Biochim Pol. 2003;50(4):985–1017. [PubMed] [Google Scholar]
- 22.Kim E, Niethammer M, Rothschild A, Jan YN, Sheng M. Clustering of Shaker-type K+ channels by interaction with a family of membrane-associated guanylate kinases. Nature. 1995;378(6552):85–8. doi: 10.1038/378085a0. [DOI] [PubMed] [Google Scholar]
- 23.Kornau HC, Schenker LT, Kennedy MB, Seeburg PH. Domain interaction between NMDA receptor subunits and the postsynaptic density protein PSD-95. Science. 1995;269(5231):1737–40. doi: 10.1126/science.7569905. [DOI] [PubMed] [Google Scholar]
- 24.Niethammer M, Kim E, Sheng M. Interaction between the C terminus of NMDA receptor subunits and multiple members of the PSD-95 family of membrane-associated guanylate kinases. J Neurosci. 1996;16(7):2157–63. doi: 10.1523/JNEUROSCI.16-07-02157.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sato T, Irie S, Kitada S, Reed JC. FAP-1: a protein tyrosine phosphatase that associates with Fas. Science. 1995;268(5209):411–5. doi: 10.1126/science.7536343. [DOI] [PubMed] [Google Scholar]
- 26.Loughran G, Healy NC, Kiely PA, Huigsloot M, Kedersha NL, O'Connor R. Mystique is a new insulin-like growth factor-I-regulated PDZ-LIM domain protein that promotes cell attachment and migration and suppresses Anchorage-independent growth. Mol Biol Cell. 2005;16(4):1811–22. doi: 10.1091/mbc.E04-12-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vallenius T, Scharm B, Vesikansa A, Luukko K, Schafer R, Makela TP. The PDZ-LIM protein RIL modulates actin stress fiber turnover and enhances the association of alpha-actinin with F-actin. Exp Cell Res. 2004;293(1):117–28. doi: 10.1016/j.yexcr.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 28.Guy PM, Kenny DA, Gill GN. The PDZ domain of the LIM protein enigma binds to beta-tropomyosin. Mol Biol Cell. 1999;10(6):1973–84. doi: 10.1091/mbc.10.6.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maeda M, Asano E, Ito D, Ito S, Hasegawa Y, Hamaguchi M, Senga T. Characterization of interaction between CLP36 and palladin. FEBS J. 2009;276(10):2775–85. doi: 10.1111/j.1742-4658.2009.07001.x. [DOI] [PubMed] [Google Scholar]
- 30.Kadrmas JL, Beckerle MC. The LIM domain: from the cytoskeleton to the nucleus. Nat Rev Mol Cell Biol. 2004;5(11):920–31. doi: 10.1038/nrm1499. [DOI] [PubMed] [Google Scholar]
- 31.Bach I. The LIM domain: regulation by association. Mech Dev. 2000;91(1-2):5–17. doi: 10.1016/s0925-4773(99)00314-7. [DOI] [PubMed] [Google Scholar]
- 32.Perez-Alvarado GC, Miles C, Michelsen JW, Louis HA, Winge DR, Beckerle MC, Summers MF. Structure of the carboxy-terminal LIM domain from the cysteine rich protein CRP. Nat Struct Biol. 1994;1(6):388–98. doi: 10.1038/nsb0694-388. [DOI] [PubMed] [Google Scholar]
- 33.Kosa JL, Michelsen JW, Louis HA, Olsen JI, Davis DR, Beckerle MC, Winge DR. Common metal ion coordination in LIM domain proteins. Biochemistry. 1994;33(2):468–77. doi: 10.1021/bi00168a011. [DOI] [PubMed] [Google Scholar]
- 34.Dawid IB, Breen JJ, Toyama R. LIM domains: multiple roles as adapters and functional modifiers in protein interactions. Trends Genet. 1998;14(4):156–62. doi: 10.1016/s0168-9525(98)01424-3. [DOI] [PubMed] [Google Scholar]
- 35.Jurata LW, Gill GN. Structure and function of LIM domains. Curr Top Microbiol Immunol. 1998;228:75–113. doi: 10.1007/978-3-642-80481-6_4. [DOI] [PubMed] [Google Scholar]
- 36.Wu RY, Gill GN. LIM domain recognition of a tyrosine-containing tight turn. J Biol Chem. 1994;269(40):25085–90. [PubMed] [Google Scholar]
- 37.Durick K, Gill GN, Taylor SS. Shc and Enigma are both required for mitogenic signaling by Ret/ptc2. Mol Cell Biol. 1998;18(4):2298–308. doi: 10.1128/mcb.18.4.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmeichel KL, Beckerle MC. The LIM domain is a modular protein-binding interface. Cell. 1994;79(2):211–9. doi: 10.1016/0092-8674(94)90191-0. [DOI] [PubMed] [Google Scholar]
- 39.Arber S, Caroni P. Specificity of single LIM motifs in targeting and LIM/LIM interactions in situ. Genes Dev. 1996;10(3):289–300. doi: 10.1101/gad.10.3.289. [DOI] [PubMed] [Google Scholar]
- 40.Feuerstein R, Wang X, Song D, Cooke NE, Liebhaber SA. The LIM/double zinc-finger motif functions as a protein dimerization domain. Proc Natl Acad Sci U S A. 1994;91(22):10655–9. doi: 10.1073/pnas.91.22.10655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boden SD, Liu Y, Hair GA, Helms JA, Hu D, Racine M, Nanes MS, Titus L. LMP-1, a LIM-domain protein, mediates BMP-6 effects on bone formation. Endocrinology. 1998;139(12):5125–34. doi: 10.1210/endo.139.12.6392. [DOI] [PubMed] [Google Scholar]
- 42.Pashmforoush M, Pomies P, Peterson KL, Kubalak S, Ross J, Jr., Hefti A, Aebi U, Beckerle MC, Chien KR. Adult mice deficient in actinin-associated LIM-domain protein reveal a developmental pathway for right ventricular cardiomyopathy. Nat Med. 2001;7(5):591–7. doi: 10.1038/87920. [DOI] [PubMed] [Google Scholar]
- 43.Zhou Q, Chu PH, Huang C, Cheng CF, Martone ME, Knoll G, Shelton GD, Evans S, Chen J. Ablation of Cypher, a PDZ-LIM domain Z-line protein, causes a severe form of congenital myopathy. J Cell Biol. 2001;155(4):605–12. doi: 10.1083/jcb.200107092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bimber B, Dettman RW, Simon HG. Differential regulation of Tbx5 protein expression and sub-cellular localization during heart development. Dev Biol. 2007;302(1):230–42. doi: 10.1016/j.ydbio.2006.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van der Meer DL, Marques IJ, Leito JT, Besser J, Bakkers J, Schoonheere E, Bagowski CP. Zebrafish cypher is important for somite formation and heart development. Dev Biol. 2006;299(2):356–72. doi: 10.1016/j.ydbio.2006.07.032. [DOI] [PubMed] [Google Scholar]
- 46.Ott EB, van den Akker NM, Sakalis PA, Gittenberger-de Groot AC, Te Velthuis AJ, Bagowski CP. The lim domain only protein 7 is important in zebrafish heart development. Dev Dyn. 2008;237(12):3940–52. doi: 10.1002/dvdy.21807. [DOI] [PubMed] [Google Scholar]
- 47.Vatta M, Mohapatra B, Jimenez S, Sanchez X, Faulkner G, Perles Z, Sinagra G, Lin JH, Vu TM, Zhou Q. Mutations in Cypher/ZASP in patients with dilated cardiomyopathy and left ventricular non-compaction. J Am Coll Cardiol. 2003;42(11):2014–27. doi: 10.1016/j.jacc.2003.10.021. others. [DOI] [PubMed] [Google Scholar]
- 48.Passier R, Richardson JA, Olson EN. Oracle, a novel PDZ-LIM domain protein expressed in heart and skeletal muscle. Mech Dev. 2000;92(2):277–84. doi: 10.1016/s0925-4773(99)00330-5. [DOI] [PubMed] [Google Scholar]
- 49.Faulkner G, Pallavicini A, Formentin E, Comelli A, Ievolella C, Trevisan S, Bortoletto G, Scannapieco P, Salamon M, Mouly V. ZASP: a new Z-band alternatively spliced PDZ-motif protein. J Cell Biol. 1999;146(2):465–75. doi: 10.1083/jcb.146.2.465. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jo K, Rutten B, Bunn RC, Bredt DS. Actinin-associated LIM protein-deficient mice maintain normal development and structure of skeletal muscle. Mol Cell Biol. 2001;21(5):1682–7. doi: 10.1128/MCB.21.5.1682-1687.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. Cell migration: integrating signals from front to back. Science. 2003;302(5651):1704–9. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 52.Tamura N, Ohno K, Katayama T, Kanayama N, Sato K. The PDZ-LIM protein CLP36 is required for actin stress fiber formation and focal adhesion assembly in BeWo cells. Biochem Biophys Res Commun. 2007;364(3):589–94. doi: 10.1016/j.bbrc.2007.10.064. [DOI] [PubMed] [Google Scholar]
- 53.Ooshio T, Irie K, Morimoto K, Fukuhara A, Imai T, Takai Y. Involvement of LMO7 in the association of two cell-cell adhesion molecules, nectin and E-cadherin, through afadin and alpha-actinin in epithelial cells. J Biol Chem. 2004;279(30):31365–73. doi: 10.1074/jbc.M401957200. [DOI] [PubMed] [Google Scholar]
- 54.Healy NC, O'Connor R. Sequestration of PDLIM2 in the cytoplasm of monocytic/macrophage cells is associated with adhesion and increased nuclear activity of NF-kappaB. J Leukoc Biol. 2009;85(3):481–90. doi: 10.1189/jlb.0408238. [DOI] [PubMed] [Google Scholar]
- 55.Tanaka-Okamoto M, Hori K, Ishizaki H, Hosoi A, Itoh Y, Wei M, Wanibuchi H, Mizoguchi A, Nakamura H, Miyoshi J. Increased susceptibility to spontaneous lung cancer in mice lacking LIM-domain only 7. Cancer Sci. 2009;100(4):608–16. doi: 10.1111/j.1349-7006.2009.01091.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bagheri-Yarmand R, Mazumdar A, Sahin AA, Kumar R. LIM kinase 1 increases tumor metastasis of human breast cancer cells via regulation of the urokinase-type plasminogen activator system. Int J Cancer. 2006;118(11):2703–10. doi: 10.1002/ijc.21650. [DOI] [PubMed] [Google Scholar]
- 57.Davila M, Frost AR, Grizzle WE, Chakrabarti R. LIM kinase 1 is essential for the invasive growth of prostate epithelial cells: implications in prostate cancer. J Biol Chem. 2003;278(38):36868–75. doi: 10.1074/jbc.M306196200. [DOI] [PubMed] [Google Scholar]
- 58.Jiang P, Enomoto A, Takahashi M. Cell biology of the movement of breast cancer cells: Intracellular signalling and the actin cytoskeleton. Cancer Lett. 2009 doi: 10.1016/j.canlet.2009.02.034. [DOI] [PubMed] [Google Scholar]
- 59.Takeichi M. Cadherins in cancer: implications for invasion and metastasis. Curr Opin Cell Biol. 1993;5(5):806–11. doi: 10.1016/0955-0674(93)90029-p. [DOI] [PubMed] [Google Scholar]
- 60.Avraamides CJ, Garmy-Susini B, Varner JA. Integrins in angiogenesis and lymphangiogenesis. Nat Rev Cancer. 2008;8(8):604–17. doi: 10.1038/nrc2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sumi T, Matsumoto K, Nakamura T. Mitosis-dependent phosphorylation and activation of LIM-kinase 1. Biochem Biophys Res Commun. 2002;290(4):1315–20. doi: 10.1006/bbrc.2002.6346. [DOI] [PubMed] [Google Scholar]
- 62.Takahashi T, Koshimizu U, Abe H, Obinata T, Nakamura T. Functional involvement of Xenopus LIM kinases in progression of oocyte maturation. Dev Biol. 2001;229(2):554–67. doi: 10.1006/dbio.2000.9999. [DOI] [PubMed] [Google Scholar]
- 63.Sangadala S, Boden SD, Viggeswarapu M, Liu Y, Titus L. LIM mineralization protein-1 potentiates bone morphogenetic protein responsiveness via a novel interaction with Smurf1 resulting in decreased ubiquitination of Smads. J Biol Chem. 2006;281(25):17212–9. doi: 10.1074/jbc.M511013200. [DOI] [PubMed] [Google Scholar]
- 64.Vallenius T, Makela TP. Clik1: a novel kinase targeted to actin stress fibers by the CLP-36 PDZ-LIM protein. J Cell Sci. 2002;115(Pt 10):2067–73. doi: 10.1242/jcs.115.10.2067. [DOI] [PubMed] [Google Scholar]
- 65.Agulnik SI, Garvey N, Hancock S, Ruvinsky I, Chapman DL, Agulnik I, Bollag R, Papaioannou V, Silver LM. Evolution of mouse T-box genes by tandem duplication and cluster dispersion. Genetics. 1996;144(1):249–54. doi: 10.1093/genetics/144.1.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Simon H. T-box genes and the formation of vertebrate forelimb- and hindlimb specific pattern. Cell Tissue Res. 1999;296(1):57–66. doi: 10.1007/s004410051266. [DOI] [PubMed] [Google Scholar]
- 67.Horton AC, Mahadevan NR, Minguillon C, Osoegawa K, Rokhsar DS, Ruvinsky I, de Jong PJ, Logan MP, Gibson-Brown JJ. Conservation of linkage and evolution of developmental function within the Tbx2/3/4/5 subfamily of T-box genes: implications for the origin of vertebrate limbs. Dev Genes Evol. 2008;218(11-12):613–28. doi: 10.1007/s00427-008-0249-5. [DOI] [PubMed] [Google Scholar]
- 68.Naiche LA, Harrelson Z, Kelly RG, Papaioannou VE. T-box genes in vertebrate development. Annu Rev Genet. 2005;39:219–39. doi: 10.1146/annurev.genet.39.073003.105925. [DOI] [PubMed] [Google Scholar]
- 69.Collavoli A, Hatcher CJ, He J, Okin D, Deo R, Basson CT. TBX5 nuclear localization is mediated by dual cooperative intramolecular signals. J Mol Cell Cardiol. 2003;35(10):1191–5. doi: 10.1016/s0022-2828(03)00231-1. [DOI] [PubMed] [Google Scholar]
- 70.Zaragoza MV, Lewis LE, Sun G, Wang E, Li L, Said-Salman I, Feucht L, Huang T. Identification of the TBX5 transactivating domain and the nuclear localization signal. Gene. 2004;330:9–18. doi: 10.1016/j.gene.2004.01.017. [DOI] [PubMed] [Google Scholar]
- 71.Kulisz A, Simon HG. An evolutionarily conserved nuclear export signal facilitates cytoplasmic localization of the Tbx5 transcription factor. Mol Cell Biol. 2008;28(5):1553–64. doi: 10.1128/MCB.00935-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fuchikami T, Mitsunaga-Nakatsubo K, Amemiya S, Hosomi T, Watanabe T, Kurokawa D, Kataoka M, Harada Y, Satoh N, Kusunoki S. T-brain homologue (HpTb) is involved in the archenteron induction signals of micromere descendant cells in the sea urchin embryo. Development. 2002;129(22):5205–16. doi: 10.1242/dev.129.22.5205. others. [DOI] [PubMed] [Google Scholar]
- 73.Bruce AE, Howley C, Zhou Y, Vickers SL, Silver LM, King ML, Ho RK. The maternally expressed zebrafish T-box gene eomesodermin regulates organizer formation. Development. 2003;130(22):5503–17. doi: 10.1242/dev.00763. [DOI] [PubMed] [Google Scholar]
- 74.Miyahara K, Suzuki N, Ishihara T, Tsuchiya E, Katsura I. TBX2/TBX3 transcriptional factor homologue controls olfactory adaptation in Caenorhabditis elegans. J Neurobiol. 2004;58(3):392–402. doi: 10.1002/neu.10299. [DOI] [PubMed] [Google Scholar]
- 75.Adell T, Muller WE. Expression pattern of the Brachyury and Tbx2 homologues from the sponge Suberites domuncula. Biol Cell. 2005;97(8):641–50. doi: 10.1042/BC20040135. [DOI] [PubMed] [Google Scholar]
- 76.Inman KE, Downs KM. Localization of Brachyury (T) in embryonic and extraembryonic tissues during mouse gastrulation. Gene Expr Patterns. 2006;6(8):783–93. doi: 10.1016/j.modgep.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 77.Hong CJ, Hsueh YP. Cytoplasmic distribution of T-box transcription factor Tbr-1 in adult rodent brain. J Chem Neuroanat. 2007;33(3):124–30. doi: 10.1016/j.jchemneu.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 78.Smith PA, Mango SE. Role of T-box gene tbx-2 for anterior foregut muscle development in C. elegans. Dev Biol. 2007;302(1):25–39. doi: 10.1016/j.ydbio.2006.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Habets PE, Moorman AF, Clout DE, van Roon MA, Lingbeek M, van Lohuizen M, Campione M, Christoffels VM. Cooperative action of Tbx2 and Nkx2.5 inhibits ANF expression in the atrioventricular canal: implications for cardiac chamber formation. Genes Dev. 2002;16(10):1234–46. doi: 10.1101/gad.222902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vitelli F, Morishima M, Taddei I, Lindsay EA, Baldini A. Tbx1 mutation causes multiple cardiovascular defects and disrupts neural crest and cranial nerve migratory pathways. Hum Mol Genet. 2002;11(8):915–22. doi: 10.1093/hmg/11.8.915. [DOI] [PubMed] [Google Scholar]
- 81.Stennard FA, Costa MW, Elliott DA, Rankin S, Haast SJ, Lai D, McDonald LP, Niederreither K, Dolle P, Bruneau BG. Cardiac T-box factor Tbx20 directly interacts with Nkx2-5, GATA4, and GATA5 in regulation of gene expression in the developing heart. Dev Biol. 2003;262(2):206–24. doi: 10.1016/s0012-1606(03)00385-3. others. [DOI] [PubMed] [Google Scholar]
- 82.Christoffels VM, Hoogaars WM, Tessari A, Clout DE, Moorman AF, Campione M. T-box transcription factor Tbx2 represses differentiation and formation of the cardiac chambers. Dev Dyn. 2004;229(4):763–70. doi: 10.1002/dvdy.10487. [DOI] [PubMed] [Google Scholar]
- 83.Xu H, Morishima M, Wylie JN, Schwartz RJ, Bruneau BG, Lindsay EA, Baldini A. Tbx1 has a dual role in the morphogenesis of the cardiac outflow tract. Development. 2004;131(13):3217–27. doi: 10.1242/dev.01174. [DOI] [PubMed] [Google Scholar]
- 84.Lincoln J, Alfieri CM, Yutzey KE. Development of heart valve leaflets and supporting apparatus in chicken and mouse embryos. Dev Dyn. 2004;230(2):239–50. doi: 10.1002/dvdy.20051. [DOI] [PubMed] [Google Scholar]
- 85.Plageman TF, Jr., Yutzey KE. Differential expression and function of Tbx5 and Tbx20 in cardiac development. J Biol Chem. 2004;279(18):19026–34. doi: 10.1074/jbc.M314041200. [DOI] [PubMed] [Google Scholar]
- 86.Yamagishi T, Nakajima Y, Nishimatsu S, Nohno T, Ando K, Nakamura H. Expression of tbx20 RNA during chick heart development. Dev Dyn. 2004;230(3):576–80. doi: 10.1002/dvdy.20076. [DOI] [PubMed] [Google Scholar]
- 87.Shelton EL, Yutzey KE. Tbx20 regulation of endocardial cushion cell proliferation and extracellular matrix gene expression. Dev Biol. 2007;302(2):376–88. doi: 10.1016/j.ydbio.2006.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Takeuchi JK, Ohgi M, Koshiba-Takeuchi K, Shiratori H, Sakaki I, Ogura K, Saijoh Y, Ogura T. Tbx5 specifies the left/right ventricles and ventricular septum position during cardiogenesis. Development. 2003;130(24):5953–64. doi: 10.1242/dev.00797. [DOI] [PubMed] [Google Scholar]
- 89.Jerome LA, Papaioannou VE. DiGeorge syndrome phenotype in mice mutant for the T-box gene, Tbx1. Nat Genet. 2001;27(3):286–91. doi: 10.1038/85845. [DOI] [PubMed] [Google Scholar]
- 90.Lindsay EA, Vitelli F, Su H, Morishima M, Huynh T, Pramparo T, Jurecic V, Ogunrinu G, Sutherland HF, Scambler PJ. Tbx1 haploinsufficieny in the DiGeorge syndrome region causes aortic arch defects in mice. Nature. 2001;410(6824):97–101. doi: 10.1038/35065105. others. [DOI] [PubMed] [Google Scholar]
- 91.Harrelson Z, Kelly RG, Goldin SN, Gibson-Brown JJ, Bollag RJ, Silver LM, Papaioannou VE. Tbx2 is essential for patterning the atrioventricular canal and for morphogenesis of the outflow tract during heart development. Development. 2004;131(20):5041–52. doi: 10.1242/dev.01378. [DOI] [PubMed] [Google Scholar]
- 92.Bakker ML, Boukens BJ, Mommersteeg MT, Brons JF, Wakker V, Moorman AF, Christoffels VM. Transcription factor Tbx3 is required for the specification of the atrioventricular conduction system. Circ Res. 2008;102(11):1340–9. doi: 10.1161/CIRCRESAHA.107.169565. [DOI] [PubMed] [Google Scholar]
- 93.Stennard FA, Costa MW, Lai D, Biben C, Furtado MB, Solloway MJ, McCulley DJ, Leimena C, Preis JI, Dunwoodie SL. Murine T-box transcription factor Tbx20 acts as a repressor during heart development, and is essential for adult heart integrity, function and adaptation. Development. 2005;132(10):2451–62. doi: 10.1242/dev.01799. others. [DOI] [PubMed] [Google Scholar]
- 94.Garrity DM, Childs S, Fishman MC. The heartstrings mutation in zebrafish causes heart/fin Tbx5 deficiency syndrome. Development. 2002;129(19):4635–45. doi: 10.1242/dev.129.19.4635. [DOI] [PubMed] [Google Scholar]
- 95.Roy Chowdhuri S, Crum T, Woollard A, Aslam S, Okkema PG. The T-box factor TBX-2 and the SUMO conjugating enzyme UBC-9 are required for ABa-derived pharyngeal muscle in C. elegans. Dev Biol. 2006;295(2):664–77. doi: 10.1016/j.ydbio.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 96.Salinas S, Briancon-Marjollet A, Bossis G, Lopez MA, Piechaczyk M, Jariel-Encontre I, Debant A, Hipskind RA. SUMOylation regulates nucleo-cytoplasmic shuttling of Elk-1. J Cell Biol. 2004;165(6):767–73. doi: 10.1083/jcb.200310136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kindsmuller K, Groitl P, Hartl B, Blanchette P, Hauber J, Dobner T. Intranuclear targeting and nuclear export of the adenovirus E1B-55K protein are regulated by SUMO1 conjugation. Proc Natl Acad Sci U S A. 2007;104(16):6684–9. doi: 10.1073/pnas.0702158104. [DOI] [PMC free article] [PubMed] [Google Scholar]