Abstract
The SMRT (Silencing Mediator of Retinoid and Thyroid hormone receptors) corepressor mediates gene repression by nuclear receptors and other transcriptional factors. The SMRT protein serves as a key nucleating core that organizes the assembly of a larger corepressor complex. We report here that SMRT interacts with itself to form a protein dimer, and that Erk2, a mitogen-activated protein (MAP) kinase, disrupts this SMRT self-dimerization in vitro and in vivo. Notably Erk2 phosphorylation also results in a re-organization of the overall corepressor complex, characterized by a reduced sedimentation coefficient, partial release of HDAC3, TBL-1, and TBLR-1, and inhibition of transcriptional repression. We propose that SMRT dimers form the central platform on which additional corepressor components assemble, and that kinase signaling modifies the architecture, composition, and function of this complex. These observations contribute to our understanding of how the SMRT corepressor complex assembles and is regulated during cell proliferation and differentiation.
Keywords: SMRT, corepressor complex, dimer, MAP kinases, Erk
1. INTRODUCTION
The SMRT protein was originally identified based on its ability to mediate repression by thyroid hormone receptors (TRs) and retinoic acid receptors (RARs) [Chen and Evans, 1995, Sande and Privalsky, 1996]. Subsequently SMRT, and its closely-related paralog NCoR, were shown to serve as corepressors for additional members of the nuclear receptor family, as well as a much wider variety of transcription factors, such as PLZF, BCL-6, NF-κB, ETO-1/2, and c-Myb [Hörlein et al., 1995, Jepsen and Rosenfeld, 2002, Lazar, 2003, Lee et al., 2001, Moehren et al., 2004, Ordentlich et al., 2001, Perissi et al., Privalsky, 2004, Seol et al., 1996, Stanya and Kao, 2009, Zamir et al., 1996]. Notably, SMRT and NCoR do not appear to possess intrinsic repressive properties, but instead are thought to function by recruiting additional polypeptides that mediate the actual molecular events required to inhibit gene transcription [Jepsen and Rosenfeld, 2002, Lazar, 2003, Lee et al., 2001, Moehren et al., 2004, Ordentlich et al., 2001, Perissi et al., Privalsky, 2004, Stanya and Kao, 2009]. Histone deacetylases (HDACs) were among the earliest of these SMRT-tethered auxiliary proteins to be identified [Guenther et al., 2000, Heinzel et al., 1997, Huang et al., 2000, Jones et al., 2001, Kao et al., 2000, Li et al., 2000, Li et al., 2002, Nagy et al., 1997, Wen et al., 2000]. SMRT and NCoR are therefore viewed as molecular platforms that nucleate assembly of the overall corepressor complex, interact through additional contact surfaces with their specific transcription factor partners, and thereby tether the functional corepressor complex to specific target genes. In fact, the corepressor complexes formed in cells are quite large, with estimated sizes of 1.5 to 2 MDa [Guenther et al., 2000, Li et al., 2000]. When purified by coimmunoprecipitation methodologies, these SMRT complexes have been shown to contain an assortment of associated proteins, including HDAC3, TBL1/TBLR1 (ubiquitin ligases implicated in the release of the corepressor complex from target genes on their activation), and GPS2 (a multifunctional protein that helps recruit TBL1/TBLR1) [Guenther et al., 2000, Guenther and Lazar, 2003, Li et al., 2000, Wen et al., 2000, Yoon et al., 2003, Zhang et al., 2002]. Still additional corepressor components have been identified by various means, such as mSin3A/B, HDACs 1, 2, 4, and 5, SHARP, MeCP2, KAP-1, JMJD2A, and an assortment of ATP-dependent chromatin remodeling proteins [Jepsen and Rosenfeld, 2002, Lazar, 2003, Lee et al., 2001, Moehren et al., 2004, Ordentlich et al., 2001, Perissi et al., Privalsky, 2004, Stanya and Kao, 2009]. Multiple forms of corepressor complex are likely to exist in cells (e.g. [Downes et al., 2000, Jones et al., 2001, Kao et al., 2000]); the stoichiometry of the corepressor components that comprise these complexes, and what defines the ability of SMRT and NCoR to assemble into different complexes, have remained incompletely resolved.
The SMRT complex is subject to multiple modes of regulation. Binding of hormone agonists by many nuclear receptors, for example, causes the release of SMRT (and its associated auxiliary protein entourage) from the receptor and permits subsequent coactivator recruitment, thereby switching these receptors from transcriptional repressors to activators [Chen and Evans, 1995, Glass and Rosenfeld, 2000, Hörlein et al., 1995, Sande and Privalsky, 1996, Perissi et al., 2004, Zamir et al., 1996]. A variety of kinase signaling pathways also converge on the SMRT corepressor complex and modulate its activity [Privalsky, 2001, Privalsky, 2004]. For example, stimulation of the epidermal growth factor (EGF) receptor activates a downstream MAP kinase cascade that induces phosphorylation of SMRT, causing its release from its nuclear receptor partners, its redistribution from nucleus to cytoplasm, and derepression of its target genes [Hong et al., 1998, Hong and Privalsky, 2000, Jonas and Privalsky, 2004]. This same phenomenon contributes to the pro-differentiation and anti-neoplastic effects of arsenic trioxide in the treatment of acute promyelocytic leukemia [Hong et al., 2001] and may contribute to anti-androgen resistance in prostate cancer [Eisold et al., 2009]
We have recently reported that the subcellular redistribution of SMRT, and the release of this corepressor from its nuclear receptor partners are regulated by distinct tiers of this MAPK cascade [Jonas et al., 2007]. The top two tiers, MEKK1 and MEK1, were found to be the primary mediators of SMRT relocalization to the cytoplasm, whereas the bottom tier kinase, Erk2, proved to be the principal effector of the inhibition of the SMRT/nuclear receptor interaction [Jonas et al., 2007]. We now report that SMRT self-associates in vitro and in vivo to form SMRT:SMRT dimers, and that phosphorylation of SMRT by Erk2 not only induces release of the corepressor complex from its nuclear receptor partners, but also results in a reorganization of the corepressor complex itself. We suggest that this MAPK modulation of SMRT dimer formation is a mechanism through which the function of the SMRT complex can be regulated by proproliferative signals in normal and neoplastic cells.
2. MATERIALS AND METHODS
2.1. Molecular clones and cell culture
The origins of the pUC18, pCH110, pADH-Gal4-17-mer, pSG5-Gal4DBD, pCMV5-FLAG-ΔMEKK1, and pCMV-HA-MEK1 (R4F) plasmids were previously described [Hong et al., 1998, Hong and Privalsky, 2000, Wong and Privalsky, 1998]. The pSG5-HA-Erk2 (L75P/S153D) vector, which encodes a constitutively-active form of Erk2 [Emrick et al., 2001], was created by QuikChange mutagenesis of a wild-type Erk2 clone (Stratagene, La Jolla CA). The pCMV-sGFP-SMRTα (1-2470) expression vector was created by inserting HindIII-XhoI fragments from the parental full-length SMRT cloning vector into HindIII-XhoI digested pCMV-sGFP. The pSG6-Gal4DBD vector was created by inserting a synthetic oligonucleotide (Biosource International, Camarillo, CA) encoding an expanded multiple cloning site into pSG5-Gal4DBD. The pSG6-Gal4DBD-SMRTα expression vector was created by inserting a parental full-length SMRTα (1-2470) into pSG6-Gal4DBD. The pSG5-Myc-SMRTα and pSG5-HA-SMRTα constructs, used for mammalian cell expression or in vitro translation, were created using polymerase chain reaction to introduce approximate restriction sites (generally HindIII and XhoI) on the ends of the corresponding open reading frames and by ligating the DNA products into the pSG5-Myc or pSG5-HA vectors. A similar approach, using XbaI and XhoI sites, was used to generate the pGEX-MP constructs employed for expression of GST-fusion proteins in E. coli. Base substitution mutations, designed to abolish MAPK sites within the SMRT sequence, were created by using QuikChange.
CV-1 cells (Jensen et al., 1964) were the generous gift of Dr. K. R. Yamamoto (University of California at San Francisco) and were propagated in Dulbecco's modified Eagle medium (DMEM) formulated with high glucose, L-glutamine, and pyridoxine hydrochloride (Invitrogen, Carlsbad, CA) and supplemented with 5% heat inactivated fetal bovine serum (HIFBS; Hyclone, Logan, UT); cells were maintained at 37°C in a humidified 5% CO2 atmosphere. Transient transfections were performed using Effectene reagent, and the manufacturer's protocol (Qiagen, Valencia, CA).
2.2. Co-immunoprecipitation Assays
CV-1 cells (8 × 105 cells/plate in a 10cm plate) were transfected with various combinations of Myc-SMRTα, HA-SMRTα, a constitutively active Erk2 construct, or appropriate amounts of equivalent empty vectors using the Effectene protocol described above. Cells were collected 48 h after transfection and lysed by a 30-min incubation at 4°C in 1ml of immunoprecipitation buffer consisting of phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, and 1.5 mM KH2PO4) plus 1 mM EDTA, 1.5 mg/ml iodoacetamide, 100 μM Na3VO4, 0.5% Triton X-100, 20 mM glycerophosphate, 1 mM NaF, 0.2 mM phenylmethylsulfonyl fluoride, 1x Complete phosphatase inhibitor mixture I (EMD Biosciences, Inc., La Jolla, CA), and 1x Complete protease inhibitor mixture (Roche Applied Science, Mannheim, Germany). The cell lysates were cleared by centrifugation at 18,000 × G at 4°C. A 50-μl aliquot of each cell lysate was saved, and the remaining lysate was incubated at 4°C for 1 h with mouse anti-Myc monoclonal antiserum (diluted 1:200; Gamma One Laboratories, Lexington, KY). Next, 40 μl of a 50% slurry of protein G-Sepharose beads (GE Healthcare, Piscataway NJ) were added, and the samples were incubated overnight at 4°C on a rotator. The Sepharose beads and any proteins bound to them were collected by centrifugation at 1000 × G in a microcentrifuge at 4°C for 2 min. The beads were washed four times with 1 ml of immunoprecipitationbuffer, and any proteins remaining bound to the beads were then eluted by boiling in SDS sample buffer, were resolved by SDS-PAGE using a Tris acetate 3-12% gradient gel system; and were visualized by immunoblotting using mouse anti-HA monoclonal antibody (diluted 1:1000; Gamma One Laboratories, Lexington, KY), horseradish peroxidase-conjugated goat anti-mouse IgG antibody (diluted 1:2000; Bio-Rad, Hercules CA), and the ECL Plus Western blot detection system (GE Healthcare, Piscataway NJ). The resulting chemiluminescent signal was detected and quantified using a Fluorchem 8900 digitaldetection system (Alpha Innotech, San Leandro, CA).
2.3. Glycerol gradient fractionation
CV-1 cells (8 × 105 cells/plate in a 10cm plate) were transfected with pSG5-Myc-SMRTα, a constitutively-active Erk2 construct, or appropriate amounts of equivalent empty vectors using the Effectene protocol described above. Cells were collected 48 h after transfection and lysed and cleared as described in section 2.2. The detergent-solubilized supernatants were subjected to centrifugation in a linear glycerol gradient, consisting of 5-25% glycerol (5.25 ml total) in 10 mM Tris-HCl and 100 mM NaCl. Gradients were centrifuged at 151,000 × G at 4°C for 18h in a SW41 Ti rotor (Beckman), and fractions of 0.5 ml were collected from the bottom under gravity flow. An aliquot of each fraction was resolved by 3-12% gradient SDS-PAGE gels and analyzed by Western blotting with mouse monoclonal anti-Myc antibody (diluted 1:1000; Gamma One Laboratories) using the protocol in section 2.2.
2.4. In Vitro Kinase Assays
Appropriate GST-SMRT fusion proteins were expressed in Escherichia coli BL21 cells and were immobilized on glutathione-agarose beads as described previously [Hong et al., 1998, Wong and Privalsky, 1998]. GST-SMRT proteins were then incubated with 500 Units of purified Erk2 (New England Biolabs)) for 18 hours at 30°C in 50 μl of 1x Erk2 kinase buffer (50 mM Tris-HCl, 10 mM MgCl2, 1 mM EGTA, 2 mM dithiothreitol, 0.01% Brij 35, pH 7.5) containing 1 mM ATP, 1x Complete phosphatase inhibitor mixture I (EMD Biosciences, Inc., La Jolla, CA), and 1x Complete protease inhibitor mixture (Roche Applied Science, Mannheim, Germany). The kinase reactions were terminated by washing the beads by centrifugation four times with 1ml of ice-cold HEMG buffer (4 mM HEPES [pH 7.8], 100 mM KCl, 0.2 mM EDTA, 5 mM MgCl2, 0.1% NP-40, 10% glycerol, 1.5 mM dithiothreitol) per well. The washed beads with the bound proteins were then used in the in vitro protein-protein interaction assays in section 2.5.
2.5. In vitro protein-protein interaction assay
Glutathione S-transferase (GST)-corepressor protein fusions were produced in E.coli strain BL-21 cells transformed by the appropriate pGEX vector [Guan and Dixon, 1991]. The bacteria were lysed by sonication and the GST fusion proteins were bound to a glutathione-agarose beads as previously described [Guan and Dixon, 1991]. 35S-radiolabeled corepressor fragments were synthesized in vitro using a TnT coupled reticulocyte lysate system (Promega, Madison, WI). Each radiolabeled protein (5 μl of TnT product per reaction) was then incubated with the GST fusion protein of interest (typically 10 to 20 ng) immobilized on 20 μl of glutathione-agarose beads in a total volume of 200 μl of HEMG buffer (4 mM HEPES [pH 7.8], 100 mM KCl, 0.2 mM EDTA, 5 mM MgCl2, 0.1% NP-40, 10% glycerol, 1.5 mM dithiothreitol) at 4°C. All of the GST-proteins in a given experiment were used at the same concentration. The binding reactions were performed in 1.5 ml Eppendorf tubes on a rotating platform to ensure thorough mixing. After a 1 hour of incubation, the beads were washed by centrifugation four times with 1ml of ice-cold HEMG buffer, and any radiolabeled proteins remaining bound to the immobilized GST fusion proteins were eluted with 50 μl of 10 mM glutathione in 50mM Tris-HCl [pH 7.8]. The eluted proteins were resolved by SDS-PAGE and were visualized and quantified using a Storm PhosphorImager (Amersham Biosciences, Piscataway, NJ).
2.6. Chromatin Immunoprecipitation (ChIP) assays
CV-1 cells (8 × 105 cells/plate in a 10cm plate) were transfected with 300 ng/plate of pGL4CP-TK 2xGal4, 200 ng/plate of Gal4-DBD-SMRT, 1.5 μg/μl of HA-SMRT, and 500 ng/plate of a constitutively active Erk2 construct (or equivalent empty vector)using the Effectene protocol (Qiagen, Valencia, CA). Forty-eight hours after transfection, formalin was added to the tissue culture medium to a final concentration of 1%. After 10 minutes of incubation the reaction was terminated by adding 0.125 M glycine. The cells were rinsed twice with ice-cold PBS and scraped into 5 ml/plate of PBS containing 1x Complete protease inhibitor mixture (Roche Applied Science, Mannheim, Germany). The cell pellet was resuspended in 200 μl of cell lysis buffer (5 mM PIPES, pH 8.0, 85 mM KCl, 0.5% NP-40, 1x complete protease inhibitors) and homogenized by using a B Dounce homogenizer 10 times. The nuclei were pelleted by centrifugation in a micro centrifuge for 5 minutes at 2,200 × G at 4°C and were resuspended in 100 μl of nuclei lysis buffer (50 mM Tris-Cl, pH 8.1, 10 mM EDTA, 1% SDS, 1X Complete protease inhibitors). Chromatin was sonicated in a Heat-Systems Branson sonicator at 50% duty cycle, at setting 4, for 4 × 15 seconds, after which samples were centrifuged at 18,000 × G for 10 minutes. The supernatant was precleared by adding 1mg/ml of herring sperm DNA, 1 mg/ml of BSA, 10 μl of Staph A cells (Pansorbin, CalBiochem 507862), and incubating the samples on ice for 15 minutes. After the samples were centrifuged at 18,000 × G for 5 min, the pellets were discarded and 1/10 of each supernatant was saved as “input.” Two volumes of IP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-Cl, pH 8.1, 167 mM NaCl, 1X Complete protease inhibitors) were then added to each supernatant, together with either 5 μl of antisera directed against the HA or Myc epitope, or 5 μl of non-specific IgG, and the samples were incubated overnight at 4 °C with mixing. Next, 10 μl of blocked Staph A cells were added to each sample and the samples were incubated at 4 °C for 15 minutes. The Staph A pellets, together with any immunocomplexes bound by them, were collected by centrifugation, washed twice with 1.4 ml of 1x dialysis buffer (2 mM EDTA, 50 mM Tris-Cl, pH 8.1) and four times with 1.4 ml of IP wash buffer (100 mM Tris-Cl, pH 9.0, 500 mM LiCl, 1% NP-40, 1% Deoxycholic acid, 1X Complete protease inhibitors). The protein/DNA complexes were then eluted into 100 μl of elution buffer (50 mM NaHCO3, 1% SDS), the crosslinks were reversed by adding 12 μl of 5 M NaCl and by incubating the samples at 67°C overnight. The eluted DNA was purified using a QIAquick PCR purification kit (Qiagen), was eluted into water and was analyzed by PCR using primers specific for the introduced TK-DR4-Luc reporter (CTAGCAAAATAGGCTGTCCC and GAGTGGGTAGAATGGCGCTGG).
3. RESULTS
3.1. SMRT proteins self-associate in vivo and in vitro
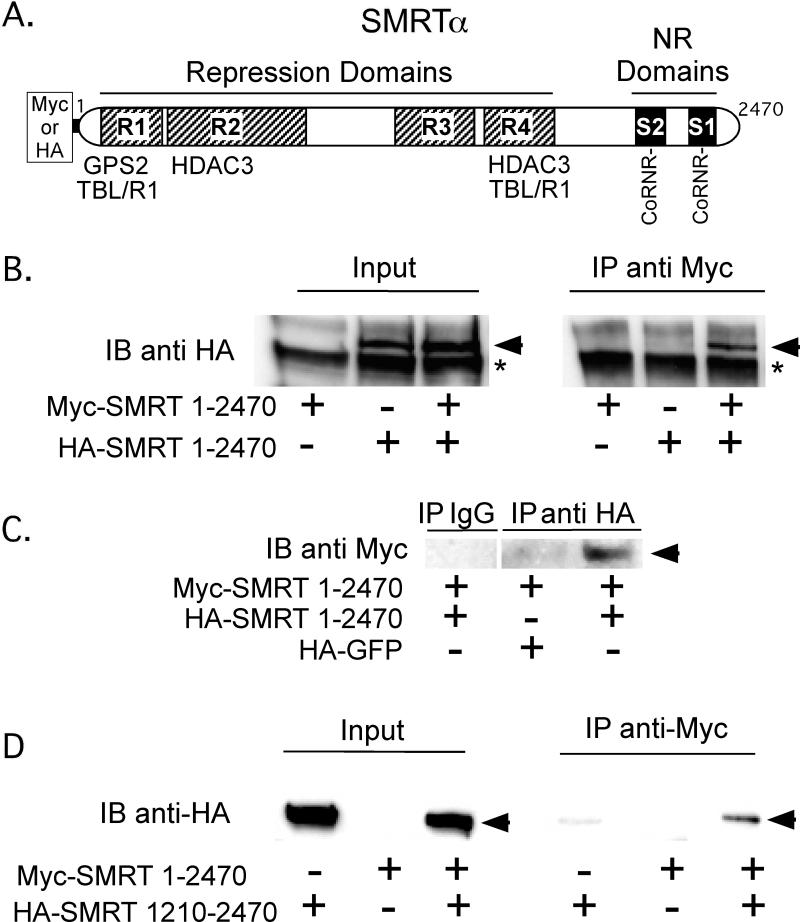
Although a variety of proteins are known to assemble into the SMRT corepressor complex, the stoichiometry of the individual components has not been fully determined. The very large overall size of the holo-complex, estimated as 1.5 to 2.0 MDa, suggested to us that there might be multiple copies of one or more of the proteins that comprise it [Guenther et al., 2000, Li et al., 2000]. We were particularly interested in determining if SMRT, the central platform that nucleates the overall corepressor complex, could itself self-associate to form higher order structures. To examine this question, we simultaneously introduced into CV1 cells two distinct epitope-tagged versions of this corepressor, Myc-SMRTα and HA-SMRTα (Figure 1A). The cells were lysed, and the Myc-SMRTα protein was immunoprecipitated using anti-Myc monoclonal antibody. The immuoprecipitates were then resolved by SDS-PAGE and any HA-SMRT that associated with the Myc-SMRT was detected by immunoblotting using anti-HA epitope antiserum (Figure 1B). Approximately 5-10% of the input HA-SMRT in the whole cell lysates (detected by anti-HA antibodies in lanes 2 and 3) immunoprecipitated with the Myc-SMRTα under these conditions (lane 6), whereas little or no HA-SMRTα was observed in the absence of the Myc-SMRT construct (lane 5) or in the absence of the HA-SMRTα construct (lanes 1 and 4). Comparable results were observed performing the experiment reciprocally by immunoprecipitating HA-SMRTα and immunoblotting against Myc-SMRTα (Figure 1C); Repeating these experiments with different HA-tagged subdomains of SMRTα demonstrated that amino acids 1210 to 2470 of SMRTα were sufficient to interact with the full-length Myc-tagged SMRTα, indicating at least one of the interaction surfaces participating in this self-association was within the C-terminal half of the corepressor (Figure 1D). The anti-HA and anti-Myc antibodies do not cross-react with the opposing epitopes (Figure 1, and data not shown).
Figure 1. The SMRT corepressor, when expressed in cells, self-associates.
A. A schematic of the full-length SMRTα protein is shown. Repression domains (R1 to R4) and nuclear receptor (NR) interaction domains (S1 and S2) are indicated, as are the binding sites for GPS2, TBL1, TBLR1, and HDAC2, the CoRNR box nuclear receptor contact motifs, and the Myc or HA epitope tags employed for the co-immunoprecipitation experiments. B. SMRTα expressed in CV-1 cells self-associates. HA-tagged full-length SMRTα, Myc-tagged full-length SMRTα, or both were expressed by transient transfection of CV-1 cells. Cell lysates were then either analyzed directly by SDS-PAGE/immunoblot using anti-HA antibodies (left panel), or were first immunoprecipitated using anti-Myc antibodies and the immunoprecipitates were analyzed by SDS-PAGE/immunoblot using anti-HA antibodies (right panel). HA-tagged SMRTα is indicated by an arrowhead; * indicates a non-specific background protein. C. A reciprocal coimmunoprecipitation confirms SMRT self-association The experiment in panel B was repeated, but immunoprecipitating with anti-HA sera and immunoblotting with anti-Myc sera; HA-GFP was employed as a negative control. D. The C-terminal portion of the SMRTα protein is sufficient to mediate an interaction with full-length SMRTα. The protocol in panel B was repeated using a HA-tagged SMRTα (1210-2470) in place of full-length HA-SMRTα.
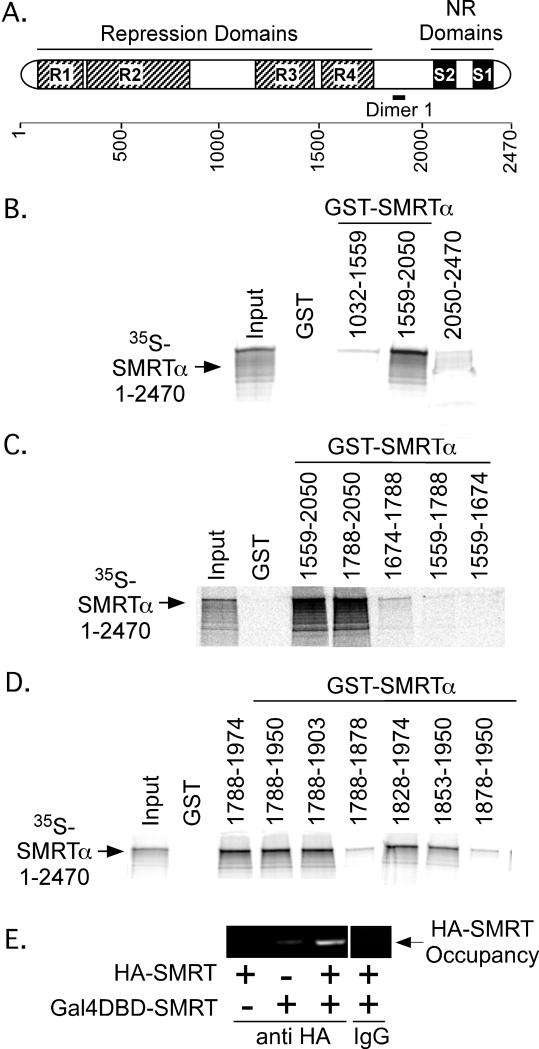
To confirm our co-immunoprecipitation results were due to direct protein-protein interactions, and to better map the surfaces involved, we turned to a GST-pulldown protocol, examining the ability of 35S-radiolabeled full-length SMRTα (generated by in vitro translation) to interact with GST-fusions representing different subdomains of SMRT (purified from recombinant E. coli) (Figure 2A). The 35S-labeled full-length SMRTα was able to efficiently bind to a GST-SMRTα (1559-2050) construct in this assay, but not to GST-alone, to GST-SMRTα (1032-1559), or to GST-SMRTα (2050-2440) constructs (Figure 2B). Analysis of additional subdomains within this region revealed that GST-SMRTα (1559-2050), GST-SMRTα (1788-2050), GST-SMRTα (1788-1974), GST-SMRTα (1788-1950), GST-SMRTα (1788-1903), GST-SMRTα (1828-1974), and GST-SMRTα (1853-1950) all retained a significant interaction with full-length SMRTα, whereas removal of yet-additional N or C-terminal sequences [i.e. GST-SMRTα (1674-1788), GST-SMRTα (1559-1788), GST-SMRTα (1559-1674), GST-SMRTα (1788-1878), or GST-SMRTα (1878-1950)] severely reduced or eliminated this association (Figure 2C and 2D). We conclude that amino acids 1788-1903 (denoted “Dimer 1” in Fig. 1A) are sufficient to promote a self-association with full-length SMRTα, with the core interaction surface within this domain likely mapping to 1853-1903. The same region of SMRTα also interacted with 35S-labeled full length SMRTω (an alternatively spliced version of SMRTα bearing an additional receptor interaction domain) and with 35S-labeled NCoRω (a SMRT paralog sharing 50% amino acid identity to SMRTω), but NCoRω did not interact with NCoRω (data not shown).
Figure 2. A self-association domain is located within the C-terminal third of SMRTα between amino acids 1788-1903.
A. A SMRTα schematic is presented; the deduced self-association domain within the SMRT C-terminal region (“Dimer 1”) is indicated. B. The C-terminal SMRTα dimerization domain lies between amino acids 1559-2050. Immobilized, GST-fusions containing different portions of SMRTα were incubated with full-length 35S-radiolabeled SMRTα. Radiolabled SMRTα protein remaining bound after washing was eluted and analyzed by SDS-PAGE/phosphorimager scanning. Representative phosphorimager scans are shown. Input lane represents 20% of the total 35S-protein employed in each incubation. C. and D. Further mapping localizes the C-terminal domain SMRTα dimerization sequences to amino acids 1788-1903. The protocol in panel B was repeated using additional GST-SMRTα truncations. E. SMRTα retains the ability to self-associate when bound to chromatin. Full-length SMRTα, either fused to a Gal4-DNA Binding Domain (DBD) or to an HA-tag, was expressed singularly or in combination in CV-1 cells together with a luciferase reporter containing GAL 17-mer binding sites. Empty GAL4DBD or HA-tag vectors were used as negative controls. After 48 hrs, the cells were subjected to a chromatin immunoprecipitation/PCR protocol using anti-HA antibodies (or non-immune IgG) and primers flanking the GAL-17 mer binding site; the resulting PCR products were analyzed by gel electrophoresis and ethidium bromide staining.
To determine if SMRTα is also capable of self-association in a chromatin context, we fused SMRTα to a GAL4-DNA binding domain (DBD) and introduced this construct, together with a full-length HA-tagged SMRT and a GAL4 17-mer reporter, into CV1 cells by transient transfection. Using chromatin immunoprecipitation, we determined that the GAL4DBD-SMRTα fusion was able to recruit full-length HA-tagged SMRTα to the GAL17-mer reporter, whereas little or no recruitment was observed in the absence of the GALDBD construct (Figure 2E and data not shown).
3.2. The C-terminal third of SMRTα interacts with amino acids 290-427 within the N-terminal third of SMRTα
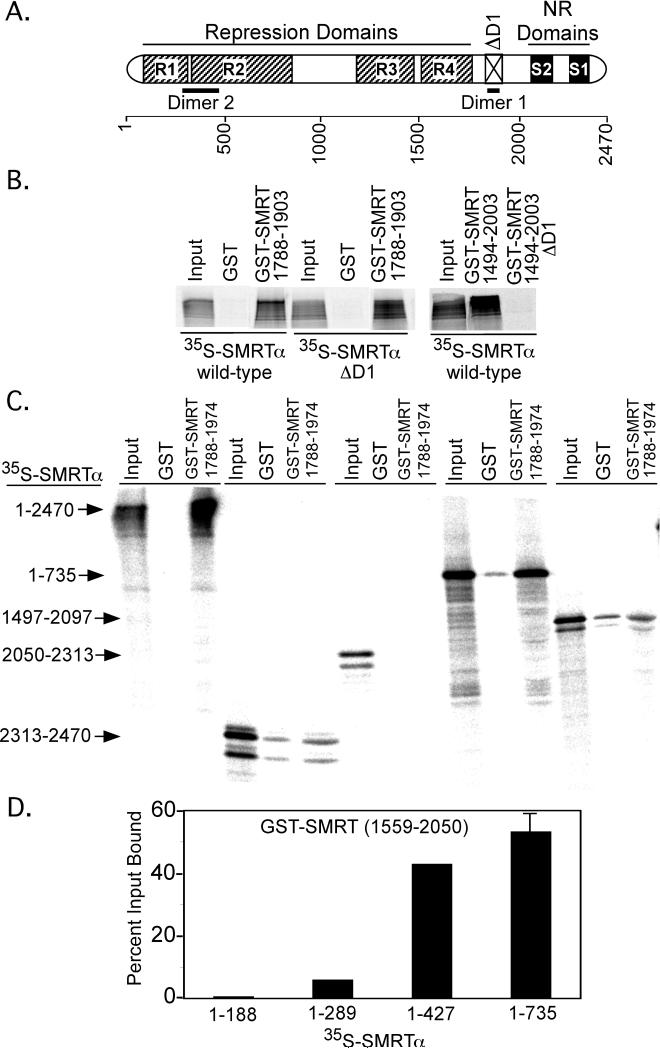
The ability of SMRT to self-associate could potentially be mediated by either homophilic or heterophilic surfaces. A homophilic interaction was ruled out by the observation that a deletion of the Dimer 1 interaction sequence in SMRT (denoted ΔD1 in Figure 3A) from within the 35S-full-length SMRTα had no effect on its ability to bind to the GST-SMRTα (1788-1903) construct (Figure 3B, center 3 lanes), whereas deletion of the same domain from the GST-SMRTα (1788-1903 construct), as expected from Figure 2, eliminated interaction with full-length 35S-SMRTα (Figure 3B, right 3 lanes). Further analysis of a series of 35S-labeled subdomains of SMRTα demonstrated that the Dimer 1 domain [e.g. a GST-SMRTα (1788-1904) construct] interacted most strongly with amino acids 1-735 within the N-terminal domain of SMRTα (Figure 3C, and confirmed by coIP from CV1 cells, data not shown). In contrast, 35S-labeled C-terminal portions of SMRTα representing amino acids 1497-2097, 2050-2313, or 2313-2470 displayed significantly weaker, or no binding, to the GST-SMRTα (1788-1974) construct (Figure 3C). Still further deletion analysis of the SMRTα N-terminal domain demonstrated that SMRTα N-terminal amino acids 290-427 were required for interaction with the GST-SMRTα C-terminal domain (Figure 3D). These findings indicate that the SMRT self-association can take place through an anti-parallel mode of interaction, utilizing a series of contacts between regions centered on amino acids 290-427 and 1788-1903 (denoted “Dimer 1” and “Dimer 2” in Figure 3).
Figure 3. SMRT self-association is mediated in a head-to-tail fashion.
A. A schematic of SMRTα is presented; the N-terminal Dimer 2 site is indicated, as is an internal deletion (1838-1950) of the Dimer 1 site (ΔD1). B. SMRTα self-associates through a heterophilic interaction of the Dimer 1 sequence with a different surface in full-length SMRTα. Dimer 1 deletions (ΔD1) in the GST-SMRT C-terminal domain abolished its interaction with full-length 35S-SMRTα (right panel), whereas the ΔD1 mutation in the full-length 35S-SMRTα did not affect its interaction with the GST-SMRTα C-terminal domain (left panel). GST-pulldown assays were performed as in Figure 2. C. The Dimer 1 domain in the C-terminal third of SMRTα interacts primarily with a Dimer 2 domain within the N-terminal third of SMRTα. 35S-radiolabeled fragments of SMRTα, indicated on the left, were incubated with the GST-only, or with GST-SMRTα (1788-1974), as indicated above the panel. 35S-proteins remaining bound after washing were analyzed by SDS-PAGE and phosphorimager analysis as in Figure 2. D. Dimer 2 domain function maps to/requires codons 289 to 427. The GST-pulldown in panel C was repeated using narrower subdomains of 35S-SMRTα, and the phosphorimager results were quantified.
3.3. SMRT self-association is disrupted by Erk2-mediated phosphorylation
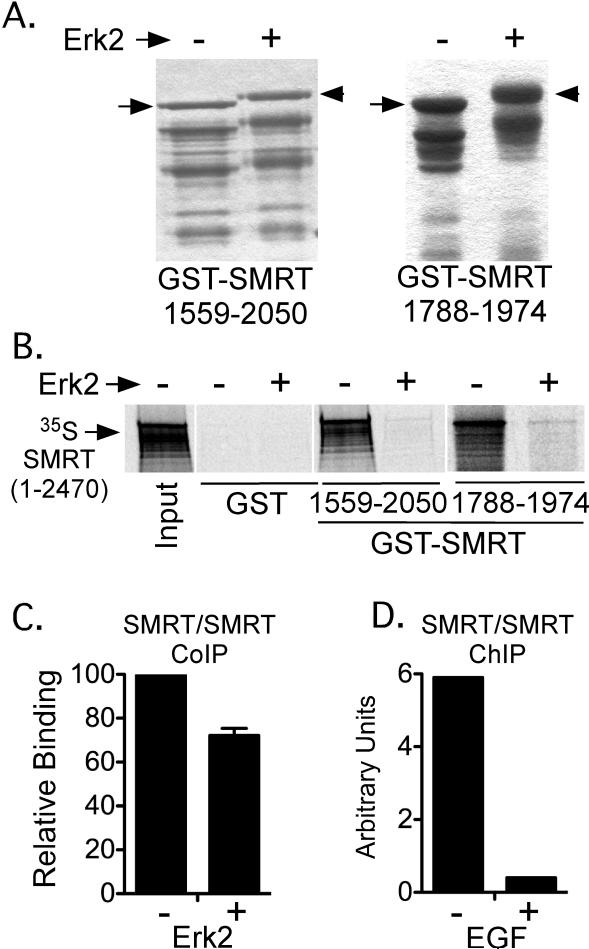
As noted in the Introduction, the interaction between SMRTα and many of its nuclear receptor partners is regulated by MAP kinase cascade signaling. In prior studies we demonstrated that Erk2-mediated phosphorylation of amino acids 1559-2050 in SMRTα plays a role in this regulation by mediating release of the corepressor from TRs and from RARs [Jonas et al., 2007]. Our current finding identifies the same general region as mediating SMRT self-association, leading us to examine if Erk2 might have an impact not only on the nuclear receptor interaction, but also on the broader organization of the SMRT corepressor complex itself. We first examined the effect of Erk2 in our GST-pulldown assays in vitro. Treatment of a purified GST-SMRTα (1559-2050) construct with Erk2 in vitro led to its extensive phosphorylation, which was detected both by use of [γ-] 32P-ATP (data not shown) and by the decreased mobility of the phosphorylated protein on SDS-PAGE (Figure 4A, left panel). Notably the Erk2-phosphorylated GST-SMRTα (1559-2050) construct was severely inhibited in its ability to interact with full-length SMRTα under these conditions (Figure 4B). The same result was observed utilizing the smaller SMRTα (1788-1974) construct (Figure 4A and 4B, right panels). Omitting ATP from the incubation abolished the effect of Erk2, consistent with phosphorylation mediating the inhibition of SMRT dimerization, rather than some other feature of the Erk2 preparation (data not shown). Erk2 expression, or EGF-treatment of CV-1 cells (a strong inducer of MAP kinase phosphorylation of SMRT) also inhibited SMRT self-association in vivo as assayed by either co-immunoprecipitation or by chromatin immunoprecipitation methods (Figure 4C and 4D). We conclude that Erk2 phosphorylation of the SMRT(1788-1974) dimerization surface strongly interferes with its ability to associate with full-length SMRTα.
Figure 4. The ability of SMRTα to self-associate is inhibited by Erk 2 phosphorylation.
A. Erk2 phosphorylates SMRTα in vitro. Purified GST-SMRTα (1559-2050) or GST-SMRT (1788-1974) proteins were treated -/+ with Erk2, were resolved by SDS-PAGE, and were visualized with Coomassie Blue. The change in mobility of the mock-treated (arrow) versus Erk2-treated (arrowhead) proteins is highlighted. Lower molecular weight protein bands represent degradation products. B. Erk2 strongly inhibits SMRTα self-interaction in vitro. The GST-SMRTα constructs, treated -/+ Erk2 as in panel A, were tested for the ability to bind 35S-full-length SMRTα as in Figure 2. C. Erk2 inhibits SMRTα self-association in a co-immunoprecipitation assay. An HA-SMRTα:Myc-SMRTα coimmunoprecipitation assay was performed as in Figure 1, except including an empty expression vector (-), or an expression vector for actived Erk2 (+). The amount of HA-tagged SMRTα co-immunoprecipitating with the Myc-tagged SMRT was quantified. D. EGF inhibits SMRTα self-association on chromatin. A chromosome immunoprecipitation assay was performed as in Figure 2E either in the absence (-) or presence (+) of EGF (an inducer of Erk signaling cascade signaling).
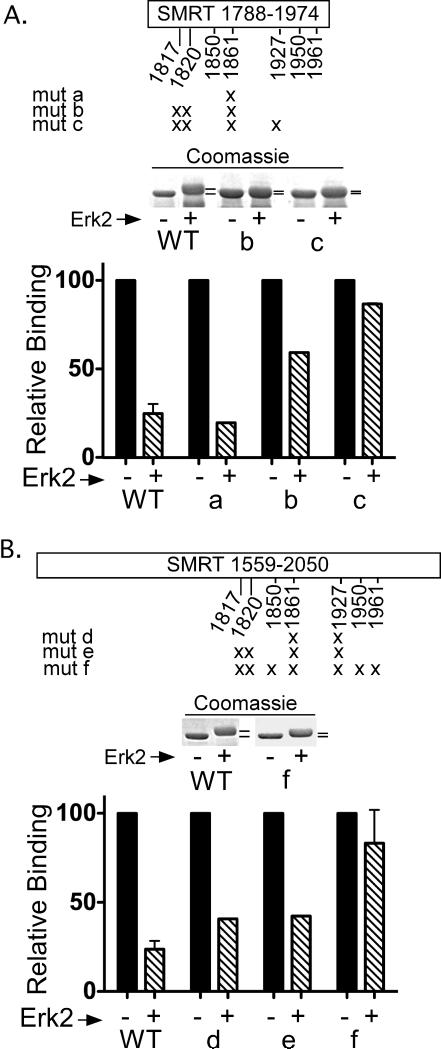
There are 7 Erk2 consensus phosphorylation sites (S/T-P) within the SMRTα (1788-1974) domain [Obenauer et al., 2003] (Figure 5A). We created a series of alanine substitutions at these sites, incubated the resulting GST-SMRTα (1788-1974) mutants with or without Erk2 in vitro, and tested them for the ability to bind to the 35S-radiolabeled full-length SMRTα protein (Figure 5A). Various single or pairwise combinations of these alanine mutants neither fully eliminated Erk2 phosphorylation of the SMRT (1788-1974) construct, nor prevented Erk 2-mediated inhibition of the SMRT/SMRT interaction (e.g. Figure 5A and data not shown). These results indicated that multiple serines or threonines within the (1788-1974) domain were targets for Erk2 phosphorylation and that mutation of multiple sites might be required to detectably affect the Erk2 response. Consistent with this suggestion, a combined mutation of 3 out of the 7 Erk2 consensus phosphorylation sites into alanines both detectably decreased the overall extent of Erk2 phosphorylation of the GST-SMRTα (1788-1974) construct and partially blocked the ability of Erk2 to disrupt the SMRT/SMRT interaction (Figure 5A, mut b); conversely, introduction of aspartic acids as phospho-mimics at these positions impaired SMRT self-association even in the absence of Erk2 signaling (data not shown). Additional mutation of 4 out of the 7 Erk2 consensus phosphorylation sites to alanines virtually eliminated the ability of Erk2 to interfere with the SMRT/SMRT interaction (Figure 5A, mut c).
Figure 5. A cluster of Erk2 phosphorylation sites regulate SMRTα self-association.
A. A schematic of the SMRTα (1788-1974) domain is presented; 7 Erk consensus phosphorylation sites (S/T-P) are highlighted. Mutant constructs bearing alanine substitutions at these positions (“X”) are indicated. The ability of the wild-type or mutant GST-SMRTα (1788-1974) construct to be shifted in mobility by incubation with Erk2 (middle panel) and to bind full-length 35S-SMRTα in a GST-pulldown (bottom panel) were determined as in Figure 4. B. The analysis of Erk2 phosphylation sites was extended to the larger SMRTα (1559-2050) domain, using the same methodology as in panel A.
Intriguingly, when the equivalent four alanine mutations were introduced into a longer GST-SMRTα (1559-2050) construct, they impaired, but did not fully abolish the inhibitory effects of Erk2 on the ability of this expanded construct to recruit full-length SMRT in the GST pulldown (Figure 5B, mut e). SMRTα (1559-2050) has 19 putative Erk2 sites and substitution of seven of these sites by alanines reduced the overall level of Erk2-induced phosphorylation and once again abrogated the ability of Erk2 to inhibit the SMRT/SMRT interaction (Figure 5B, mut f). This result indicates that complete dimerization domain of SMRT includes sequences that extend beyond the minimum (1788-1974) region, and that multiple Erk2 sites, both within and adjacent to the minimum dimerization domain, are involved in inhibition of SMRT homodimerization by Erk2 in vitro.
3.4. Erk2 phosphorylation results in a global reorganization of the SMRT corepressor complex and inhibits SMRT-mediated repression
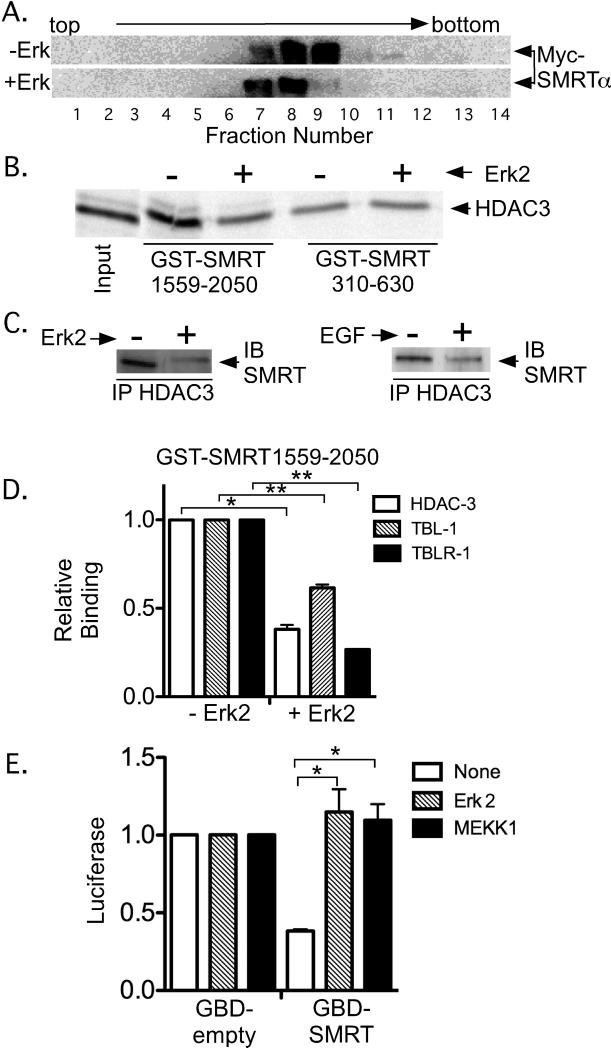
As previously noted, SMRT exists in cells as large protein complexes composed of multiple protein subunits [Guenther et al., 2000, Li et al., 2000]. To test the effect of Erk2 on this complex, we transfected CV1 cells with SMRTα plus or minus a constitutively-active form of Erk2. We subsequently lysed these cells, fractionated the lysates by ultracentrifugation in 5-25% glycerol gradient, and analyzed each fraction for SMRTα by SDS-PAGE immunoblotting (Figure 6A). Consistent with prior studies, in the absence of Erk2 the bulk of the SMRTα sedimented as large complexes greater than 1 million daltons in apparent molecular weight (Figure 6A). Interestingly, introduction of the active Erk2 resulted in a reduction in the apparent size of SMRT corepressor complex, suggesting that Erk2 changes the overall organization and/or composition of the SMRT complex in cells (Figure 6A).
Figure 6. Erk2 alters the composition of the SMRT corepressor complex.
A. Erk 2 treatment shifts the SMRT complex to a slower sedimenting species. CV-1 cells were transfected with pSG5-Myc-SMRTα, the actived Erk2 construct, or an equivalent empty vector, and the lysates were resolved by velocity ultracentrifugation. Fractions collected from the gradient were resolved by SDS-PAGE and were visualized by immunoblotting with anti-Myc antibodies. B. Erk2 treatment inhibits interaction between GST-SMRTα (1559-2050) and HDAC3 in vitro. Immobilized GST-SMRTα (310-630) or GST-SMRTα (1559-2050) were treated -/+ with Erk2 as in Figure 4, incubated with 35S-radiolabeled HDAC3, and 35S-proteins remaining bound after washing were eluted and visualized by SDS-PAGE/phosphorimager analysis. C. Erk 2 or EGF treatment inhibits the interaction between SMRT and HDAC3 in CV-1 cells. Left panel: HDAC3 and SMRTα were expressed, plus or minus an activated Erk2 construct, as in panel A. The cell lysates were immunoprecipitated with anti-HDAC3 sera, resolved by SDS-PAGE, and any coprecipitated SMRT was visualized with anti-SMRT antibodies (arrows). Right panel: The same experiment was repeated but the cells were treated -/+ 100 ng/ml EGF rather than using an Erk2 construct. D. Erk2 treatment also inhibits the interaction of GST-SMRTα (1559-2050) with TBL-1 and TBLR-1. The experiment in panel B was repeated using radiolabeled HDAC3, TBL-1, or TBLR-1 and the results quantified (binding in the absence of Erk2 is defined as 1). The mean and standard error are presented (n=4). * and ** indicate a P<0.05 or <0.01 respectively. E. Introduction of Erk2 or MEK1 inhibits SMRT-mediated transcriptional repression. A GAL4-DBD construct (GBD) or a GAL4-DBD-SMRTα construct (GBD-SMRTα) was introduced into CV-1 cells together with a GAL 17-mer luciferase reporter (Jonas et al., 2007). The transfections also included empty vector, an expression vector for activated MEK-1, or an expression vector for activated Erk2. After 48 hrs the cells were lysed and luciferase activity was calculated; reporter gene activity for the empty GDB construct is defined as 1. The mean and standard error is presented (n=4); * indicates a P<0.05.
To better define the nature of these Erk2-induced changes to the SMRT complex, we analyzed the composition of the complex, plus or minus Erk2, in more detail. HDAC3 is a known physical and functional component of the untreated SMRT/NCoR corepressor complex and has been shown to bind to two distinct regions of SMRT and NCoR [Codina et al., 2005, Guenther et al., 2001, Li et al., 2000]. The N-terminal HDAC3 docking site (also denoted DAD for deacetylase activation domain), maps to amino acids 412-480 in SMRTα and not only binds HDAC3, but also is a strong, allosteric activator of its deacetylase activity [Guenther et al., 2001]. The C-terminal HDAC3 docking surface of SMRTα, mapping to amino acids 1559-2050, physically recruits HDAC3, but is not known to be necessary for its enzymatic activity [Codina et al., 2005]. Intriguingly, although phosphorylation of the SMRT DAD domain (SMRT 310-630) by Erk2 had no effect on its interaction with HDAC3, the same Erk2 treatment of the SMRT (1559-2050) fragment inhibited the latter's interaction with HDAC3 (Figure 6B). An overall reduction in the amount of HDAC3 coimmunoprecipitated from cells with full-length SMRTα was also noted in response to introduction of Erk2 or its upstream activator, EGF, in cultured cells (Figure 6C). Extending the in vitro analysis to additional known components of the SMRT corepressor complex, we observed that phosphorylation of SMRT (1559-2050) by Erk2 not only inhibited the HDAC3 interaction, but also that of TBL-1 and TBLR-1 (Figure 6D). We conclude that MAPK signaling changes the architecture of the SMRT complex in multiple ways.
To determine if these rearrangements altered corepressor activity, we assayed the ability of a GAL4DBD-SMRTα fusion to repress a Gal17-mer promoter-luciferase reporter construct. As previously observed [Wong and Privalsky, 1998], the GAL4DBD-SMRTα construct repressed expression of this reporter relative to the non-recombinant GAL4DBD control (Figure 6E, compare “GBD-SMRTα” to “GBD”). Co-expression of either a constitutive Erk2, or MEK1 (an activator of endogenous Erks) reversed this repression (Figure 6E). We conclude that Erk2 not only releases the SMRT corepressor complex from many of its nuclear receptor partners (Jonas et al. 2007) but also results in a rearrangement of the corepressor complex itself and an attenuation in its ability to repress transcription.
4. DISCUSSION
4.1. SMRT proteins self-associate
The SMRT and NCoR corepressors are each approximately 250 kDa in molecular weight, but exist in cells as much larger complexes that contain an assortment of additional effector and architectural proteins [Guenther et al., 2000, Guenther and Lazar, 2003, Li et al., 2000, Wen et al., 2000, Yoon et al., 2003, Zhang et al., 2002]. These auxiliary proteins include chromatin modifying and remodeling enzymes (such as HDAC3 and members of the SWI/SNF family), ubiquitin ligases (such as TBL1/TBLR1), and a variety of additional polypeptides that appear to play scaffolding or stabilizing roles (such as GPS2). Notably the stoichiometry of these different subunits in the overall complex is not fully understood, and, in fact, there is evidence for the existence of multiple corepressor complexes in cells containing distinct subunit compositions. We therefore sought to better understand the overall organization of the corepressor complexes recruited by SMRT, and the regulatory phenomenon that might impact on this organization.
We report here that SMRT has the ability to self-associate into protein dimers, with the strongest interaction being an anti-parallel one between regions encompassing amino acids 290-427 and 1788-1903. We can detect this head-to-tail interaction by a wide variety of means, including GST-pulldown, co-immunoprecipitation from cells, and ChIP analysis. This SMRT homodimerization helps account for the relatively large size of the corepressor complexes found in cells, which exceed the sums of the molecular weights predicted from a monomeric assembly of the most commonly attributed corepressor subunits. Intriguingly, many auxiliary proteins, including HDAC3, TBL1 and TBLR1, have been reported to interact with two distinct regions of SMRT, with one interaction surface localized in the C-terminal regions of SMRT, and another localized closer to the N-terminus (e.g. [Codina et al., 2005, Guenther et al., 2000, Li et al., 2000, Yoon et al., 2003]. Given our observation that SMRT homodimerizes, it is possible that what appears to be two distinct interaction surfaces for each of these auxiliary proteins are, in reality, two portions of a composite docking site formed by the two antiparallel molecules of SMRT. SMRT self-association may therefore serve to stabilize the overall structure of the corepressor complex by furnishing two contact surfaces for each of these auxiliary proteins. Conversely, several auxiliary corepressor proteins have been shown to bind to identical or overlapping sites within SMRT, raising the question of how two of these different corepressor subunits could be simultaneously recruited into the same corepressor complex [Jepsen and Rosenfeld, 2002, Lazar, 2003, Lee et al., 2001, Moehren et al., 2004, Ordentlich et al., 2001, Perissi et al., Privalsky, 2004, Stanya and Kao, 2009]; this paradox is resolved if there are two (or more) molecules of SMRT per complex, thereby providing two copies of each docking surface for the recruitment of two different coregulatory proteins.
NCoR is a close paralog of SMRT, and the two corepressor proteins play overlapping, but non-identical roles in physiology and development [Jepsen et al., 2000, Jepsen and Rosenfeld, 2002, Jepsen et al., 2007, Nofsinger et al., 2008]. In addition to its ability to self-dimerize, we also observed an ability of the SMRTα (1788-1974) domain to “heterodimerize” with full-length NCoRω. This leads to the suggestion that SMRT and NCoR may be able to co-assemble into shared corepressor complexes that presumably would display a combinatorial mixture of the properties of both paralogs. Intriguingly, in our hands, this interaction was non-reciprocal: the region of NCoRω equivalent to SMRTα (1788-1974) failed to interact with either full-length SMRTα or full-length NCoRω (NV and MLP, unpublished observations). This may be a technical limitation of our assay, or may represent a fundamental divergence in the contacts by which NCoR and SMRT assemble into larger corepressor complexes.
Other transcriptional corepressors are also known to form dimers or higher order oligomers. For example, the Groucho family of corepressors play multiple roles in metazoan development and are known to homotetramerize; Groucho oligomerization has been reported to be required for repression[Chen et al., 1998, Song et al., 2004]. Similarly, CtBP-interacting protein, a corepressor that binds to many proteins involved in cell cycle control, is known to form homodimers [Dubin et al., 2004].
4.2. SMRT self-association is regulated by Erk2 phosphorylation
We have previously reported that the MAP kinase cascade is an important regulator of SMRT corepressor function, and that different tiers of this cascade can induce corepressor release from its nuclear receptor partners and redistribution of SMRT from the nucleus into the cytoplasm [Hong and Privalsky, 2000, Jonas and Privalsky, 2004, Jonas et al., 2007]. In the current study, we have shown that components of the same MAP kinase cascade also regulate the ability of SMRT to self-associate, and can alter the overall composition and organization of the corepressor complex itself. Most significantly, Erk2 phosphorylation of a clustered group of serine and thronines with the SMRT dimerization domain results in abrogation of SMRT self-association in vitro. Similar effects of Erk2 on SMRT self-dimerization were observed in vivo using co-immunoprecitation and ChIP based assays. Using ultracentrifugation analysis, we also observed a parallel Erk2 induced decrease in the size of native SMRT complexes in cells that indicates the change in SMRT self-association has a broader impact on the overall structure of the corepressor complex. Multiple Erk2 phosphorylation sites are involved in mediating this inhibition of self-association; although more modest effects were observed with less comprehensive mutagenesis, it was necessary to mutate 4 out of 7 in the core SMRTα (1788-1903) region, or 7 out of the 11 possible Erk2 consensus sites in the extended SMRTα (1559-2050) dimerization domain, to strongly inhibit ERK 2 inhibition. This inhibitory effects of Erk2 on SMRT self-association therefore represents a newly recognized mechanism by which the composition of the SMRT holo-corepressor complex itself is regulated by cell signaling pathways.
4.3. Erk2 phosphorylation also inhibits interaction of SMRTα with HDAC3, TBL1 and TBLR1 and interferes with SMRT-mediated repression
The ability of SMRT and NCoR to mediate transcriptional repression does not appear to be innate to the SMRT or NCoR polypeptides per se, but rather these corepressor proteins serve as platforms that recruit the actual effectors of transcriptional repression, such as HDAC3 [Alenghat et al., 2008, Fischle et al., 2002, Guenther et al., 2001, Ishizuka and Lazar, 2003, Yang et al., 2002]. Additional proteins recruited to the SMRT/NCoR platform appear to contribute to the architecture of the overall complex, such as GPS2, or participate in release of the corepressor assembly on gene activation, such as the ubiquitin ligases TBL1 and TBLR1 [Guenther et al., 2000, Perissi et al., 2004, Zhang et al., 2002]. As noted above, HDAC3 interacts with two distinct domains of SMRT, the N-terminal DAD domain and the C-terminal (1559-2050) domain. Interestingly, Erk2 phosphorylation of SMRT destabilized the HDAC3 interaction with the latter, but not the former. The role of the HDAC3 interaction with the SMRT(1559-2050) domain is poorly understood; conceivably the inhibition of the SMRT(1559-2050) contact may reduce the specific activity of the HDAC activity per receptor complex, may alter the ability of HDAC3 to access specific substrates by changing the topology of this enzyme in the corepressor complex, or may affect an as-yet unknown HDAC3/SMRT function. Notably the binding of TBL1 and TBLR2 to SMRTα were also inhibited by Erk2, as was the ability of SMRT to repress in a GAL4DBD-tethered reporter gene assay.
Taken as a whole, our experiments demonstrate that multiple aspects of SMRT corepressor complex structure and function are under the control of MAP kinase cascades operating in cells. As previously reported, the MEKK1 and MEK1 tiers of these cascades regulate the subcellular localization of SMRT, whereas Erk2 regulates the physical interaction of the SMRT protein with many of its nuclear receptor partners. As now shown here, Erk2 is also a key modifier of the architecture of the corepressor complex itself, and regulates the homodimerization of the SMRT platform, the specific auxiliary proteins recruited by that platform, and the ability of that platform to mediate repression in cells. The distinct SMRT complexes that form plus or minus kinase, may serve to target different transcription factor partners, or may mediate distinct forms of transcriptional regulation when tethered to different target genes.
Supplementary Material
Varlakhanova et al. Appendix Figure 1 Supplemental information for reviewers
ACKNOWLEDGEMENTS
The authors thank Liming Liu for superb technical assistance. This work was supported by Public Health Service/National Institute of Diabetes and Digestive and Kidney Diseases award RO1DK53528.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Alenghat T, Meyers K, Mullican SE, Leitner K, Adeniji-Adele A, Avila J, Bucan M, Ahima RS, Kaestner KH, Lazar MA. Nuclear receptor corepressor and histone deacetylase 3 govern circadian metabolic physiology. Nature. 2008;456:997–1000. doi: 10.1038/nature07541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Nguyen PH, Courey AJ. A role for Groucho tetramerization in transcriptional repression. Mol. Cell. Biol. 1998;18:7259–7268. doi: 10.1128/mcb.18.12.7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JD, Evans RM. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature. 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- Codina A, Love JD, Li Y, Lazar MA, Neuhaus D, Schwabe JW. Structural insights into the interaction and activation of histone deacetylase 3 by nuclear receptor corepressors. Proc. Natl. Acad. Sci. U.S.A. 2005;102:6009–6014. doi: 10.1073/pnas.0500299102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downes M, Ordentlich P, Kao HY, Alvarez JG, Evans RM. Identification of a nuclear domain with deacetylase activity. Proc. Natl. Acad. Sci. U.S.A. 2000;97:10330–10335. doi: 10.1073/pnas.97.19.10330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubin MJ, Stokes PH, Sum EY, Williams RS, Valova VA, Robinson PJ, Lindeman GJ, Glover JN, Visvader JE, Matthews JM. Dimerization of CtIP, a BRCA1- and CtBP-interacting protein, is mediated by an N-terminal coiled-coil motif. J. Biol. Chem. 2004;279:26932–26938. doi: 10.1074/jbc.M313974200. [DOI] [PubMed] [Google Scholar]
- Eisold M, Asim M, Eskelinen H, Linke T, Baniahmad A. Inhibition of MAPK-signaling pathway promotes the interaction of the corepressor SMRT with the human androgen receptor and mediates repression of prostate cancer cell growth in the presence of antiandrogens. J. Mol. Endocrinol. 2009;42:429–435. doi: 10.1677/JME-08-0084. [DOI] [PubMed] [Google Scholar]
- Emrick MA, Hoofnagle AN, Miller AS, Ten Eyck LF, Ahn NG. Constitutive activation of extracellular signal-regulated kinase 2 by synergistic point mutations. J. Biol. Chem. 2001;276:46469–46479. doi: 10.1074/jbc.M107708200. [DOI] [PubMed] [Google Scholar]
- Fischle W, Dequiedt F, Hendzel MJ, Guenther MG, Lazar MA, Voelter W, Verdin E. Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol. Cell. 2002;9:45–57. doi: 10.1016/s1097-2765(01)00429-4. [DOI] [PubMed] [Google Scholar]
- Glass CK, Rosenfeld MG. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- Guan KL, Dixon JE. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal. Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- Guenther MG, Barak O, Lazar MA. The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol. Cell. Biol. 2001;21:6091–6101. doi: 10.1128/MCB.21.18.6091-6101.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Lane WS, Fischle W, Verdin E, Lazar MA, Shiekhattar R. A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40-repeat protein linked to deafness. Genes Dev. 2000;14:1048–1057. [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Lazar MA. Biochemical isolation and analysis of a nuclear receptor corepressor complex. Methods Enzymol. 2003;364:246–257. doi: 10.1016/s0076-6879(03)64014-0. [DOI] [PubMed] [Google Scholar]
- Heinzel T, Lavinsky RM, Mullen TM, Söderstrom M, Laherty CD, Torchia J, Yang WM, Brard G, Ngo SD, Davie JR, Seto E, Eisenman RN, Rose DW, Glass CK, Rosenfeld MG. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature. 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- Hong SH, Privalsky ML. The SMRT corepressor is regulated by a MEK-1 kinase pathway: inhibition of corepressor function is associated with SMRT phosphorylation and nuclear export. Mol. Cell. Biol. 2000;20:6612–6625. doi: 10.1128/mcb.20.17.6612-6625.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SH, Wong CW, Privalsky ML. Signaling by tyrosine kinases negatively regulates the interaction between transcription factors and SMRT (silencing mediator of retinoic acid and thyroid hormone receptor) corepressor. Mol. Endocrinol. 1998;12:1161–1171. doi: 10.1210/mend.12.8.0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SH, Yang Z, Privalsky ML. Arsenic trioxide is a potent inhibitor of the interaction of SMRT corepressor with its transcription factor partners, including the PML-RARα oncoprotein found in human acute promyelocytic leukemia. Mol. Cell. Biol. 2001 doi: 10.1128/MCB.21.21.7172-7182.2001. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hörlein AJ, Näär AM, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamei Y, Söderström M, Glass CK, Al. E. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature. 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- Huang EY, Zhang JS, Miska EA, Guenther MG, Kouzarides T, Lazar MA. Nuclear receptor corepressors partner with class II histone deacetylases in a Sin3-independent repression pathway. Genes Dev. 2000;14:45–54. [PMC free article] [PubMed] [Google Scholar]
- Ishizuka T, Lazar MA. The N-CoR/histone deacetylase 3 complex is required for repression by thyroid hormone receptor. Mol. Cell. Biol. 2003;23:5122–5131. doi: 10.1128/MCB.23.15.5122-5131.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen FC, Girardi AJ, Gilden RV, Koprowski H. Infection of human and simian tissue cultures with Rous Sarcoma Virus. Proc. Natl. Acad. Sci. USA. 1964;52:53–59. doi: 10.1073/pnas.52.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jepsen K, Hermanson O, Onami TM, Gleiberman AS, Lunyak V, Mcevilly RJ, Kurokawa R, Kumar V, Liu F, Seto E, Hedrick SM, Mandel G, Glass CK, Rose DW, Rosenfeld MG. Combinatorial roles of the nuclear receptor corepressor in transcription and development. Cell. 2000;102:753–763. doi: 10.1016/s0092-8674(00)00064-7. [DOI] [PubMed] [Google Scholar]
- Jepsen K, Rosenfeld MG. Biological roles and mechanistic actions of co-repressor complexes. J. Cell. Sci. 2002;115:689–698. doi: 10.1242/jcs.115.4.689. [DOI] [PubMed] [Google Scholar]
- Jepsen K, Solum D, Zhou T, Mcevilly RJ, Kim HJ, Glass CK, Hermanson O, Rosenfeld MG. SMRT-mediated repression of an H3K27 demethylase in progression from neural stem cell to neuron. Nature. 2007;450:415–419. doi: 10.1038/nature06270. [DOI] [PubMed] [Google Scholar]
- Jonas BA, Privalsky ML. SMRT and N-CoR corepressors are regulated by distinct kinase signaling pathways. J. Biol. Chem. 2004;279:54676–54686. doi: 10.1074/jbc.M410128200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas BA, Varlakhanova N, Hayakawa F, Goodson M, Privalsky ML. Response of SMRT (silencing mediator of retinoic acid and thyroid hormone receptor) and N-CoR (nuclear receptor corepressor) corepressors to mitogen-activated protein kinase kinase kinase cascades is determined by alternative mRNA splicing. Mol. Endocrinol. 2007;21:1924–1939. doi: 10.1210/me.2007-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PL, Sachs LM, Rouse N, Wade PA, Shi YB. Multiple N-CoR complexes contain distinct histone deacetylases. J. Biol. Chem. 2001;276:8807–8811. doi: 10.1074/jbc.C000879200. [DOI] [PubMed] [Google Scholar]
- Kao HY, Downes M, Ordentlich P, Evans RM. Isolation of a novel histone deacetylase reveals that class I and class II deacetylases promote SMRT-mediated repression. Genes Dev. 2000;14:55–66. [PMC free article] [PubMed] [Google Scholar]
- Lazar MA. Nuclear receptor corepressors. Nucl Recept Signal. 2003;1:e001. doi: 10.1621/nrs.01001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JW, Lee YC, Na SY, Jung DJ, Lee SK. Transcriptional coregulators of the nuclear receptor superfamily: coactivators and corepressors. Cell. Mol. Life Sci. 2001;58:289–297. doi: 10.1007/PL00000856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Lin Q, Wang W, Wade P, Wong J. Specific targeting and constitutive association of histone deacetylase complexes during transcriptional repression. Genes Dev. 2002;16:687–692. doi: 10.1101/gad.962502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JW, Wang J, Wang JX, Nawaz Z, Liu JM, Qin J, Wong JM. Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. EMBO J. 2000;19:4342–4350. doi: 10.1093/emboj/19.16.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moehren U, Eckey M, Baniahmad A. Gene repression by nuclear hormone receptors. Essays Biochem. 2004;40:89–104. doi: 10.1042/bse0400089. [DOI] [PubMed] [Google Scholar]
- Nagy L, Kao HY, Chakravarti D, Lin RJ, Hassig CA, Ayer DE, Schreiber SL, Evans RM. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- Nofsinger RR, Li P, Hong SH, Jonker JW, Barish GD, Ying H, Cheng SY, Leblanc M, Xu W, Pei L, Kang YJ, Nelson M, Downes M, Yu RT, Olefsky JM, Lee CH, Evans RM. SMRT repression of nuclear receptors controls the adipogenic set point and metabolic homeostasis. Proc. Natl. Acad. Sci. U.S.A. 2008;105:20021–20026. doi: 10.1073/pnas.0811012105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obenauer JC, Cantley LC, Yaffe MB. Scansite 2.0: Proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 2003;31:3635–3641. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordentlich P, Downes M, Evans RM. Corepressors and nuclear hormone receptor function. Curr. Top. Microbiol. Immunol. 2001;254:101–116. doi: 10.1007/978-3-662-10595-5_5. [DOI] [PubMed] [Google Scholar]
- Perissi V, Aggarwal A, Glass CK, Rose DW, Rosenfeld MG. A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell. 2004;116:511–526. doi: 10.1016/s0092-8674(04)00133-3. [DOI] [PubMed] [Google Scholar]
- Perissi V, Jepsen K, Glass CK, Rosenfeld MG. Deconstructing repression: evolving models of co-repressor action. Nat. Rev. Genet. 11:109–123. doi: 10.1038/nrg2736. [DOI] [PubMed] [Google Scholar]
- Privalsky ML. Regulation of SMRT and N-CoR corepressor function. Cur. Topics Microbiol. Immunol. 2001;254:117–136. doi: 10.1007/978-3-662-10595-5_6. [DOI] [PubMed] [Google Scholar]
- Privalsky ML. The role of corepressors in transcriptional regulation by nuclear hormone receptors. Ann. Rev. Physiol. 2004;66:315–360. doi: 10.1146/annurev.physiol.66.032802.155556. [DOI] [PubMed] [Google Scholar]
- Sande S, Privalsky ML. Identification of TRACs (T3 receptor-associating cofactors), a family of cofactors that associate with, and modulate the activity of, nuclear hormone receptors. Mol. Endocrinol. 1996;10:813–825. doi: 10.1210/mend.10.7.8813722. [DOI] [PubMed] [Google Scholar]
- Seol W, Mahon MJ, Lee YK, Moore DD. Two receptor interacting domains in the nuclear hormone receptor corepressor RIP13/N-CoR. Mol. Endocrinol. 1996;10:1646–1655. doi: 10.1210/mend.10.12.8961273. [DOI] [PubMed] [Google Scholar]
- Song H, Hasson P, Paroush Z, Courey AJ. Groucho oligomerization is required for repression in vivo. Mol. Cell. Biol. 2004;24:4341–4350. doi: 10.1128/MCB.24.10.4341-4350.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanya KJ, Kao HY. New insights into the functions and regulation of the transcriptional corepressors SMRT and N-CoR. Cell Div. 2009;4:7. doi: 10.1186/1747-1028-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen YD, Perissi V, Staszewski LM, Yang WM, Krones A, Glass CK, Rosenfeld MG, Seto E. The histone deacetylase-3 complex contains nuclear receptor corepressors. Proc. Natl. Acad. Sciences U.S.A. 2000;97:7202–7207. doi: 10.1073/pnas.97.13.7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CW, Privalsky ML. Transcriptional repression by the SMRT-mSin3 corepressor: multiple interactions, multiple mechanisms, and a potential role for TFIIB. Mole. Cell. Biol. 1998;18:5500–5510. doi: 10.1128/mcb.18.9.5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WM, Tsai SC, Wen YD, Fejer G, Seto E. Functional domains of histone deacetylase-3. J. Biol. Chem. 2002;277:9447–9454. doi: 10.1074/jbc.M105993200. [DOI] [PubMed] [Google Scholar]
- Yoon HG, Chan DW, Huang ZQ, Li J, Fondell JD, Qin J, Wong J. Purification and functional characterization of the human N-CoR complex: the roles of HDAC3, TBL1 and TBLR1. Embo J. 2003;22:1336–1346. doi: 10.1093/emboj/cdg120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamir I, Harding HP, Atkins GB, Hörlein A, Glass CK, Rosenfeld MG, Lazar MA. A nuclear hormone receptor corepressor mediates transcriptional silencing by receptors with distinct repression domains. Molecular and Cellular Biology. 1996;16:5458–5465. doi: 10.1128/mcb.16.10.5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Kalkum M, Chait BT, Roeder RG. The N-CoR-HDAC3 nuclear receptor corepressor complex inhibits the JNK pathway through the integral subunit GPS2. Mol Cell. 2002;9:611–623. doi: 10.1016/s1097-2765(02)00468-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Varlakhanova et al. Appendix Figure 1 Supplemental information for reviewers