Abstract
Bartonella infections from wild mice and voles (Apodemus flavicollis, Mi. oeconomus, Microtus arvalis and Myodes glareolus) were sampled from a forest and old-field habitats of eastern Poland; a complex network of Bartonella isolates, referrable to B. taylorii, B. grahamii, B. birtlesii and B. doshiae, was identified by the sequencing of a gltA fragment, comparable to previous studies of Bartonella diversity in rodents. Nested clade analysis showed that isolates could be assigned to zero- and one-step clades which correlated with host identity and were probably the result of clonal expansion; however, sequencing of other housekeeping genes (rpoB, ribC, ftsZ, groEl) and the 16S RNA gene revealed a more complex situation with clear evidence of numerous recombinant events in which one or both Bartonella parents could be identified. Recombination within gltA was found to have generated two distinct variant clades, one a hybrid between B. taylorii and B. doshiae, the other between B. taylorii and B. grahamii. These recombinant events characterised the differences between the two-step and higher clades within the total nested cladogram, involved all four species of Bartonella identified in this work and appear to have played a dominant role in the evolution of Bartonella diversity. It is clear, therefore, that housekeeping gene phylogenies are not robust indicators of Bartonella diversity, especially when only a single gene (gltA or 16S RNA) is used. Bartonella clades infecting Microtus were most frequently involved in recombination and were most frequently tip clades within the cladogram. The role of Microtus in influencing the frequency of Bartonella recombination remains unknown.
Introduction
Bartonella is a parasite of mammalian erythrocytes and endothelial cells transmitted by blood feeding arthropod ectoparasites [5]. This genus has a well-established alpha taxonomy consisting of 27 species and three sub-species, and several taxa cause significant human disease: B. bacilliformis, responsible for Oroya fever; B. quintana, causing trench fever and B. henselae, causing cat-scratch disease are the most important, but others are recorded as adventitious zoonotic infections, giving the genus the reputation as an emerging human pathogen [6, 7]. Since La Scola et al. [28] suggested that species could be described on the basis of DNA sequences from housekeeping genes, of which RNA polymerase (rpoB) and citrate synthase (gltA) were considered most useful, there has been an explosion of research into the taxonomy of the genus, and GenBank (http://www.ncbi.nlm.nih.gov) now contains over 1,000 sequences of gltA from the genus, mostly from rodents and insectivores, although isolates from hosts as diverse as marsupials and marine mammals have been sequenced.
Surveys of gltA diversity have revealed an impressive range of Bartonella isolates from rodents and their arthropod ectoparasites, which are generally treated as clonal and recombinantly isolated (e.g. [21, 23]). Indeed, the paradigm of isolation from recombination and inferred clonality underpins almost all recent work on Bartonella in a clinical or veterinary setting [8, 19, 30]. Great diversity can occur even across short geographical distances; thus, Welc-Falęciak et al. [44] recorded 16 genotypes of B. taylorii and B. grahamii from a small area of NE Poland, treating all as isolated non-recombinant strains. This level of diversity suggests either implausibly rapid mutational evolution of gltA or a very long independent evolutionary history for clades. The alternative hypothesis is that diversity is a result of recombination. To test this, we have systematically searched for evidence of recombination within a population of Bartonella isolates from rodents in NE Poland, part of the same sample area as Welc-Falęciak et al. [44], utilising a nested clade [41] approach to describe the most parsimonious structure for Bartonella clades within this population, which could then be used to test specific hypotheses about the prevalence of recombination within and between clades.
Methods
Mice (Apodemus flavicollis) and voles (Microtus arvalis, Mi. oeconomus and Myodes glareolus) were live-trapped at monthly intervals between June 2007 and May 2009 in regenerating old-field habitats derived from abandoned agricultural land (post 1990) and in a managed forest at Urwitałt in the Mazury Lake District, NE Poland. These habitats were within one of the locations utilised by Welc-Falęciak et al. [44]. Within this location, the most distant trapping sites (between forest and old fields) were no more than 5 km apart. Detailed trapping protocols have been described previously [35] and conformed with permission granted by the Polish Ethical Committee (permission number 737/2007). The four sampled rodent species represent the greater part of the small mammal fauna in this area. Micromys minutus, Apodemus agrarius, Sicista betulina, Sorex araneus and S. minutus also occur infrequently but were not sampled. At each capture, 50 μl of blood was taken from a tail vein directly into 200 μl of 0.001 M EDTA and used for PCR amplification. The gltA gene locus was amplified as previously described by Norman et al. [33], using primers specific to a fragment corresponding to amino acids 260 to 370 of the Escherichia coli sequence [32]. For selected isolates, additionally fragments of a further five genes were amplified: RNA polymerase beta-subunit gene (rpoB), cell division protein gene (ftsZ), riboflavin synthase gene (ribC), 60 kDa heat-shock protein gene (groEl) and 16S RNA coding gene. The 333 bp rpoB gene fragment was amplified using primers described previously by Renesto et al. [37], modified on the basis of alignment of sequences from GenBank (accession numbers, AF165991, AF165993, AF165995, AB426700, AB426701), with the forward primer, 5′-GCACGATTYGCATCATCATTTTCC-3′ and reverse primer, 5′-CGCATTATGGTCGTATTTGTCC-3′. The PCR of the groEl gene fragment was conducted using the forward primer of Zeaiter et al. [45], 5′-GGAAAAAGTGGGCAATGAAG-3′ and reverse primer designed in this study (on the basis of sequences deposited in GenBank, AF304017, AB426677, AF014833), 5′-TCCTTTAACGGTCAACGCATT-3′, and the obtained product was 752 bp. Primers to 420 bp fragment of ribC gene (forward, 5′-TYGGTTGTGTKGAAGATGT-3′; reverse, 5′-AATAATMAGAACATCAAAAA-3′), 515 bp fragment of ftsZ gene (forward, 5′-CATATGGTTTTCATTACTGCYGGTATGG-3′; reverse, 5′-TTCTTCGCGAATACGATTAGCAGCTTC-3′) and 369 bp fragment of 16S RNA gene (forward, 5′-TCAGAACGAACGCTGGCGGC-3′; reverse, 5′- CGTCATTATCTTCACCGG-3′) were designed in this study on the basis of sequences from GenBank (accession numbers: ribC-AY116635, AY116627, AB426690, AB426689, ftsZ-AF467754, AF467756, AB426641, AB426642, AB426645, AB426647, AB426649, AB426651, NC012846, 16S RNA-Z31349, Z31350, Z31351). All of the genes, after first denaturation in 94 °C for 2 min, were amplified with 40 cycles of 94 °C for 45 s, 47 °C (groEl), 54 °C (16R RNA), 52 °C (rpoB and ribC) or 61 °C (ftsZ) for 45 s, 72 °C for 45 s and followed by a single 7-min extension step in 72 °C. All PCR products were sequenced in both directions. Chromatogram quality was inspected visually, and only sequences derived from a single PCR product (i.e. no ambiguous peaks on the chromatogram) were analysed further. Potential mixed infections were therefore excluded from analysis.
Analysis
Phylogenetic analysis and alignments were carried out using the Mega 4.1 software [39], but examination of resulting cladograms revealed poor resolution of clades at a subspecific level. A network of zero-step and one-step clades was therefore established using the methodology of Nested Clade Analysis and statistical parsimony [41–43] to describe the most likely mutational relationships between Bartonella isolates collected during this work. The cladogram and the limit for statistical parsimony were calculated using TCS [10]. Four isolates, EU014267, EU014269, EU014274 and EU014275, collected from the same location by Welc-Falęciak et al. [44] in 2005, were also included because they represented otherwise missing internal steps within the clade network. Log-linear models were implemented using SPSS v 14.00 to establish significant departures from randomness in the host range of isolates within each nested Bartonella clade [41]. To identify recombination within the gltA gene, the sequenced fragment was first divided into three 100 bp segments and phylogenies generated using the minimum evolution algorithm in Mega 4.1. Discrepancies between these phylogenies were then used to identify potential recombinant gltA sequences, which were analysed further to confirm or reject recombination using the RDP-2 software package [18]. To identify potential recombination events between disparate parts of the genome, isolates from the range of Bartonella gltA clades were sequenced at the other genes described. All distinct genotypes of each gene were treated as distinct alleles and coded as such. Using an MLST approach [14, 29], distinct alleles were then plotted on to the cladogram and evidence was sought of disjunctions between the overall distribution of housekeeping genes and of connections between disparate clades, which could be taken as evidence of a recombinant event. The congruence of the gene phylogenies to each other and to the gltA phylogeny was tested by generating consensus (100 bootstraps) maximum likelihood phylogenies using PhyML ([20], performed on the Montpellier bioinformatics platform and the University of Oslo Bioportal), after first establishing optimal DNA evolution models for each gene using jModelTest [36]. The congruence between trees generated in this way for each gene and trees constrained by the assumption that the gltA phylogeny reflected the evolutionary history of the Bartonella isolates was tested using maximum likelihood ratio tests.
The unique sequences were deposited in GenBank under accession numbers GU338880-GU338885 (16S), GU338887-GU338901 (ftsZ), GU338903-GU338915 and GU338917-GU338924 (ribC), GU338925-GU338936 and GU338938-GU338941 (rpoB), GU338942-GU338976 (gltA), and GU559862-GU559871 and GU559873 (groEl).
Results
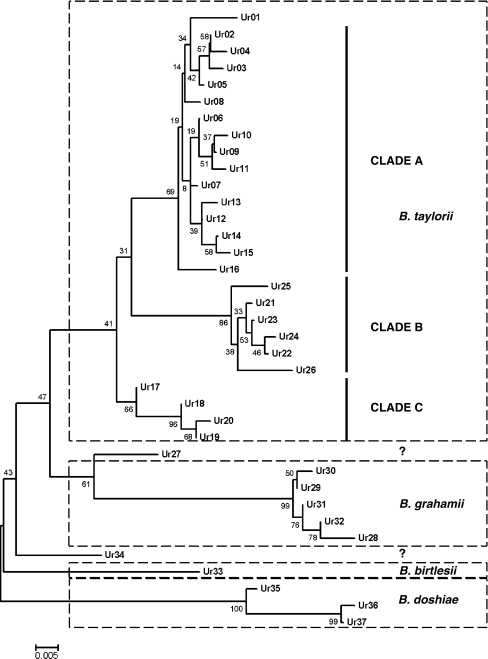
From 1,457 rodents sampled over 2 years (2007–2009), 2,582 blood samples were obtained, of which 579 (22.4%) were positive for Bartonella. Of these, 147 isolates were characterized more fully, sequencing of gltA revealing a rich, complex diversity of strains. Phylogenetic analysis (minimum evolution algorithm; 1,000 bootstrap replicates, Mega 4.1) resulted in 26 (102 isolates) poorly resolved variants (Ur01-Ur26), which clustered in a clade that included the B. taylorii type strain and other B. taylorii isolates from rodents and a further five variants (Ur28-Ur32, 39 isolates), which grouped with type isolates of B. grahamii. A single variant (Ur33, one isolate) could be referred to B. birtlesii while three variants (Ur35-Ur37, three isolates) showed closest similarity to B. doshiae. One isolate (Ur27) clustered persistently with B. grahamii in the cladistic analyses although it showed greatest percent similarity at gltA to B. taylorii while another (Ur34) could not be linked with any previously described species (Fig. 1). Because of the poor resolution within the B. taylorii isolates, nested clade analysis was used to establish hypotheses of mutational relationships between clades and was therefore used to infer the clonal structure of Bartonella isolates. For the 292 bp fragment of gltA, the probability that an individual step in the cladogram is parsimonious is 0.9985 (calculated using TCS). We have therefore treated isolates which cannot be linked to their nearest neighbour by less than three missing mutational steps (P = 0.9899) or to the central clade of the cladogram (as identified by the TCS) by five or less mutational steps (P = 0.9748) as belonging to separate clades. In fact, there is only one example of a clade at this threshold, the case of Ur01 within the B. taylorii clade A. In all other cases, the difference between clades is of the order of a minimum of seven to ten changes at the gltA locus, and assignation to these higher level clades was unambiguous.
Figure 1.
Phylogeny of a 292 bp fragment of citrate synthase (gltA) generated using minimum evolution algorithm (MEGA 4.1) with 1,000 bootstrap replicates. Species identities based on nearest match to type sequences from GenBank, clade identities based on second-step clades from the nested clade analysis. Two clades (Ur27 and Ur34) could not be assigned to a species with confidence
The gltA Network
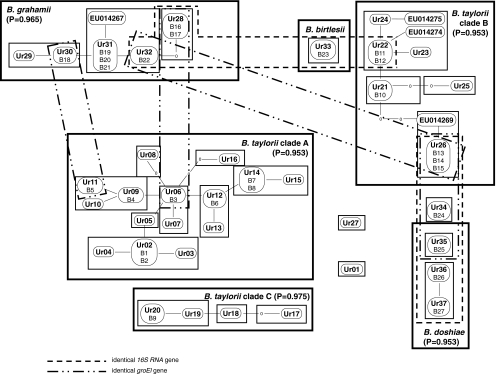
Using NCA, the 39 B. grahamii isolates grouped into a network of five zero-step clades differing by single mutations within gltA (Fig. 2) and a sixth clade (Ur28) separated from its nearest neighbour by two mutations. The probability that this overall 6-step network is parsimonious is 0.9650 (TCS), and the links between the B. grahamii gltA clades were therefore accepted at the 95% level.
Figure 2.
Cladogram of the Bartonella isolates collected in this work, and relevant haplotypes from Welc-Falęciak et al. [44] based on a 292 bp fragment of citrate synthase showing known (perfect match) recombinant events within the cladogram. “Ur01-Ur37” represents unique variants recovered from Bartonella isolates corresponding to zero-step clades in the terminology of Templeton et al. [41]. Boxes represent one-step clades. ‘Missing’ haplotypes (i.e. predicted but not collected) are indicated by 0. Larger clades corresponding to ‘species’ and other higher clades surrounded by heavy boxes. Distances between clades of more than five base changes not marked because of likelihood that these will be recombinant events. Clades sharing groEl and 16S RNA haplotypes between ‘species’ are marked
The 102 isolates of B. taylorii, on the other hand, exhibited a much more complex pattern breaking up into three clusters of clades (B. taylorii clades A, B and C) which were only distantly related to each other and two other clades (Ur01 and Ur27) which could not be parsimoniously linked to any higher grouping. The distances between the major clades are considerable; clade B is 13 nucleotides different to the type A clade while the type C clade differs by seven nucleotides from type A. Ur01 differs by five nucleotides from the central clade of the type A clade and is therefore close to the parsimony limit for this clade. We treat it as independent of the type A clade, the most conservative interpretation of its position.
The B. taylorii clade A consists of fifteen zero-step clades (Fig. 2). None of these clades are more than the three steps from the centre of the network, and most are directly connected by a single step to another clade. Two clades, Ur08 and Ur16, are connected to their nearest neighbour by one and two missing transitions, respectively, but these still remain within the parsimonious limit (P = 0.9530), and the links between these clades were accepted at the 95% level.
The B. taylorii clade B represents the most complex of the B. taylorii clades (Fig. 2). It consists of a core of six zero-step clades (Ur21, Ur22, Ur23, Ur24, EU014274 and EU014275), which are all joined by no more than two mutational steps into a network with Ur21 at its centre. This part of the B. taylorii clade B is clearly parsimonious and is accepted at the 95% level. There is less certainty over the placement of the remaining parts of the network as both Ur25 and a clade containing Ur26 and EU014269 connect into Ur21 by two missing mutational steps. Strictly, according to the criteria established using the TCS, this is a parsimonious network with Ur26 connected to the central clade (Ur22) by five steps, including only two missing steps (P = 0.9530); however, in view of the central position of Ur26 in a number of recombinational events within the network, this may not be the correct position for this clade. The B. taylorii clade C represents a simple and parsimonious network of four zero-step clades.
The B. doshiae clade consists of two zero-step clades which are parsimoniously joined (Ur37 and Ur36) and a third clade (Ur35) which appears to be B. doshiae based on overall sequence similarity, but which differs from Ur36 and Ur37 by, respectively, seven and eight base substitutions.
Recombination Within gltA and the Origin of Novel Bartonella Clades
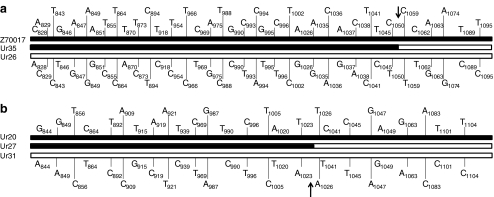
Within this network, four clades remained problematical; Ur01, which appears to be a B. taylorii isolate, Ur35 which appears to represent a form of B. doshiae, and Ur27 and Ur34 which could not be identified with confidence based on their gltA sequences. Recombination within gltA appeared to be involved in the generation of two of these four clades. Ur27 appears, by sequence analysis, to lie between B. taylorii and B. grahamii. On closer examination, it became clear that the first part of the sequenced 292 bp gltA fragment was identical with the gltA sequence for Ur20, part of the B. taylorii clade C. The final part of the gltA fragment, however, was identical at every variable base with the equivalent part of the sequence for Ur31, a clade which forms part of B. grahamii. The recombinant break point was between bases 1024 and 1025 of the gene (numbering according to [32]). Statistical support for this event was high and provided by four algorithms [P = 0.0033, GENECONV; P = 0.00027, MaxChi and Chimaera; P = 0.00000399, 3Seq] within the RDP-2 package and was obvious on visual inspection (Fig. 3).
Figure 3.
Examples of intra-gene recombination events into the gltA gene with polymorphic sites and break points marked on. a Ur35 (B. doshiae) as a recombinant of Ur26 (B. taylorii clade B) and B. doshiae type sequence (Z70017). b Ur27 (species not defined) as a recombinant of Ur20 (B. taylorii clade B) and Ur31 (B. grahamii)
Recombination also appears to have been involved in the generation of the B. doshiae clade Ur35, which seems to be a recombinant clade between B. doshiae (the type sequence, which has not been recovered at Urwitałt) and Ur26 (Fig. 3). In this case, of the 41 variable sites between the three sequences, all are consistent with the parental sequences with a break point between bases 1051 and 1058 of the gene. This event has high statistical support (P = 0.00032, GENECONV; P = 0.021, MaxChi and Chimaera; 0.000007, 3Seq) and is clear on visual inspection (Fig. 3).
Recombination Across the Genome
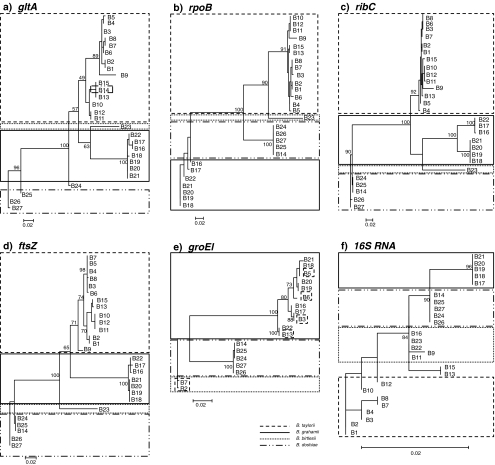
Twenty-seven isolates (B1-B27), chosen to give the greatest possible phylogenetic coverage of obtained gltA phylogeny, were sequenced at the other gene loci. The genes that were used allowed analysis of recombinant events spanning the genome. In general, the pattern of alleles of rpoB, ribC and ftsZ mirrored the pattern of gltA clades and supported the assignation of clades based on gltA (Fig. 4), but although the consensus phylogenies of ribC and rpoB were similar, the difference between them and that of gltA was nevertheless significant (0.01 < P < 0.05, maximum likelihood ratio test). The ftsZ gene phylogeny was less congruent with the gltA gene phylogeny (P > 0.05, maximum likelihood ratio test).
Figure 4.
Comparison of phylogenies obtained with the metabolic genes analysed in this work: gltA, ftsZ, ribC, rpoB, groEl and gene coding 16S RNA. Phylogenies generated using maximum likelihood (PhyML) with 100 bootstrap replicates. Species identities boxed, based on nearest match to type sequences from GenBank. Note the lack of support for B. taylorii with groEl
The pattern of alleles of these other genes established conclusively that Ur35 and Ur34 were indeed related to Ur36 and Ur37 within B. doshiae. The most notable differences in the distribution of these housekeeping gene alleles are listed in Table 1. The most important concerned Ur26. Three isolates of this clade were sequenced, all with identical gltA; however, while two isolates resembled B. taylorii based on other housekeeping genes and supported placement within the B. taylorii clade B, the third isolate was identical at all other genes with Ur34 and Ur35 and was B. doshiae-like. This appears therefore to be a case of a recombinant exchange of gltA from B. taylorii into a B. doshiae clade. The next most distinctive example of recombination is the presence of an alternative ribC allele (differing by 32 or 33 base substitutions) in clades Ur28 and Ur32, suggesting a recombinant event dividing B. grahamii into two groups. The other important difference is the presence of an alternative ftsZ allele (differing by eight substitutions) within Ur02, suggesting a recombinant event into this part of the B. taylorii clade A.
Table 1.
Evidence of recombination within clades
| B. taylorii clade A | B. taylorii clade B | B. grahamii | B. doshiae | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B3 | B4 | B5 | B6 | B7 | B8 | B1 | B2 | B11 | B12 | B13 | B15 | B10 | B19 | B18 | B20 | B21 | B22 | B16 | B17 | B26 | B27 | B25 | B24 | |
| gltA | – | 1 | 2 | 1 | 2 | 2 | 2 | 2 | – | 0 | 5 | 5 | 1 | – | 1 | 0 | 0 | 1 | 3 | 3 | – | 1 | 7 | 30 |
| rpoB | – | 3 | 4 | 4 | 1 | 1 | 3 | 3 | – | 0 | 4 | 4 | 0 | – | 0 | 0 | 0 | 3 | 4 | 6 | – | 0 | 0 | 0 |
| ribC | – | 6 | 3 | 0 | 1 | 0 | 3 | 3 | – | 1 | 5 | 5 | 1 | – | 0 | 0 | 0 | 32 | 33 | 32 | – | 2 | 5 | 5 |
| ftsZ | – | 0 | 0 | 1 | 0 | 1 | 8 | 8 | – | 0 | 5 | 4 | 0 | – | 0 | 0 | 0 | 3 | 4 | 5 | – | 0 | 5 | 5 |
| groEl | – | ND | 10 | 11 | 68 | ND | ND | 70 | ND | ND | ND | ND | ND | – | 3 | 0 | 6 | 16 | 9 | 9 | – | 0 | 1 | 1 |
| 16S RNA | 0 | ND | ND | 1 | 1 | 1 | – | 2 | 2 | 2 | 5 | – | 0 | 0 | 0 | 4 | 4 | 0 | – | 0 | 0 | 0 | ||
Number of pairwise differences between an isolate considered typical of each clade (B3, B11, B19 and B26) and other isolates from that clade. Numbers in italics denote changes of up to five bases which are assumed due to mutational drift. Numbers in bold denote larger changes which suggest recombination
ND not done
The other two genes sequenced, groEl and the 16S RNA gene, show much greater evidence of recombination across the network of Bartonella clades, and for both loci, the consensus phylogenies were very different from that of gltA (P > 0.001, maximum likelihood ratio test). Although the 16S RNA gene of all isolates was sequenced successfully, there were several cases of groEl not amplifying successfully, complicating interpretation for this gene. Nevertheless, there were clear examples of groEl exchanged between clades (Table 2). Three examples were identified where B. taylorii isolates shared different groEl alleles with B. grahamii isolates. In addition, there were two examples of gltA clades (Ur28, Ur31) in which different isolates had radically different groEl alleles. An intriguing observation was that two B. taylorii clade A clades had groEl alleles most similar to that of the B. birtlesii type sequence (AM690315). It is not clear whether these represent two distinct recombinant events involving as yet uncollected B. birtlesii isolates from the Urwitałt environment or evidence of an older recombinant event into B. taylorii which has subsequently undergone mutational drift and disruption by other recombinations from B. grahamii. A similarly complex pattern of recombination was noted for the 16S RNA gene. Eleven 16S RNA alleles were collected from the sequenced isolates, some of which were widely distributed. One allele, found in the B. birtlesii isolate, was also identified in the B. grahamii clades Ur32 and Ur28 and in Ur22 (B. taylorii clade B). These links between clades are illustrated in Fig. 2.
Table 2.
Examples of whole-gene exchange between distant clades of Bartonella within the recombinant network
| Isolates (zero-step clade, bacteria species) | Genes with identical sequence |
|---|---|
| B17 (Ur28, B. grahamii) and B18–B21 (Ur30, Ur32; B. grahamii) | 16S RNA |
| B16 (Ur28, B. grahamii); B22 (Ur32, B. grahamii); B23 (Ur23, B. birtlesii) and B11 (Ur22, B. taylorii clade B) | 16S RNA |
| B13, B15 (Ur26, B. taylorii clade B) and B14 (Ur26, B. doshiae) | gltA |
| B17 (Ur28, B. grahamii) and B3 (Ur06, B. taylorii clade A) | groEl |
| B18 (Ur30, B. grahamii) and B5 (Ur11, B. taylorii clade A) | groEl |
| B22 (Ur32, B. grahamii) and B13 (Ur26, B. taylorii clade B) | groEl |
Only examples where genes were identical in sequence in both partners are included
The Ecology of Bartonella Clades
The first-step gltA clades showed a degree of both host and habitat specificity (Table 3). Log-linear contingency analysis revealed a significant association between clade and host identity (first-step clade x host species, χ 2 = 266.63, df = 69, P < 0.001). A significant association was also noted with habitat, but it is not clear whether this is due rather to the habitat specificity of the rodent species. At second-step level, significance was noted between clade identity and host family (second-step clade x host family, χ 2 = 139.51, df = 13, P < 0.001).
Table 3.
Identity of zero-step, one-step and two-step clades of Bartonella collected during this work and their relationship with infected host
| Second-step clade | First-step clade | Zero-step clade | Internal clade | Number of isolates | |||
|---|---|---|---|---|---|---|---|
| My. glareolus | A. flavicollis | Mi. arvalis | Mi. oeconomus | ||||
| 1 | 1 | Ur01 | NA | 1 | |||
| 2 | 2 | Ur02 | Y | 3 (B1, B2) | |||
| Ur03 | N | 1 | |||||
| Ur04 | N | 1 | |||||
| 3 | Ur05 | Y | 3 | ||||
| 3 | 4 | Ur06 | Y | 14 | 2 (B3) | ||
| Ur07 | N | 2 | |||||
| 5 | Ur08 | N | 1 | ||||
| 6 | Ur09 | Y | 13 (B4) | ||||
| Ur10 | N | 1 | |||||
| Ur11 | N | 1 (B5) | |||||
| 4 | 7 | Ur12 | Y | 2 | 1 (B6) | ||
| Ur13 | N | 1 | |||||
| 8 | Ur14 | Y | 23 (B7, B8) | ||||
| Ur15 | N | 2 | |||||
| 5 | 9 | Ur16 | N | 1 | |||
| 6 | 10 | Ur17 | N | 1 | |||
| 11 | Ur18 | Y | 1 | ||||
| 12 | Ur29 | Y | 4 | ||||
| Ur20 | N | 1 (B9) | |||||
| 7 | 13 | Ur21 | Y | 5 (B10) | |||
| 14 | Ur22 | Y | 3 (B11, B12) | ||||
| Ur23 | N | 1 | |||||
| Ur24 | Y | 1 | |||||
| 15 | Ur25 | N | 1 | ||||
| 8 | 16 | Ur26 | N | 10 (B13, B15) | 1 (B14) | ||
| 9 | 17 | Ur27 | NA | 1 | |||
| 10 | 18 | Ur28 | N | 4 (B16, B17) | |||
| 19 | Ur29 | N | 1 | ||||
| Ur30 | Y | 1 | 2 (B18) | ||||
| 20 | Ur31 | Y | 16 (B19, B20) | 10 (B21) | 1 | ||
| Ur32 | Y | 4 (B22) | |||||
| 11 | 21 | Ur33 | NA | 1 (B23) | |||
| 12 | 22 | Ur34 | NA | 1 (B24) | |||
| 13 | 23 | Ur35 | NA | 1 (B25) | |||
| 14 | 24 | Ur36 | NA | 1 (B26) | |||
| Ur37 | NA | 1 (B27) | |||||
Information about the placement of the clade. Codes in parentheses indicate isolates characterised in more detail
Y internal clade, N tip clade, NA does not apply
The frequency of occurrence of the gltA clades was consistent with predictions of the nested clade analysis, although clade prevalence was not included in the original algorithm. More isolates were collected from internal clades (Table 3) than from the tip zero-step clades, and this distribution also showed a relationship with host identity, with tip clades more commonly encountered in Microtus than the stem clades, which occurred in Myodes and Apodemus (χ 2 = 42.50, df = 6, P < 0.001). This was reflected in the molecular diversity of isolates collected from each host. In the case of Microtus spp., an average of only 2.5 isolates per clade were identified (13 distinct gltA variants from 33 sequenced isolates). In My. glareolus on the other hand, seven isolates per clade (seven distinct gltA variants from 53 isolates) were collected. Bartonella from A. flavicollis showed intermediate diversity, with 3.1 isolates per clade (20 distinct gltA variants from 62 isolates).
Discussion
It is customary to treat Bartonella isolates as clonal [21] and to assume that meaningful phylogenies can be generated from single gene alignments [23, 25] despite convincing genomic evidence that the genus undergoes genetic re-organisation and recombination [2, 38]. The present work makes it clear that these bacteria frequently recombine, to the extent that it proved impossible to use an algorithm such as eBurst [13] to demonstrate clonal multiplication. Instead, nested clade analysis [41] allowed us to visualise the diversity of gltA alleles in Bartonella and then to test specific hypotheses about their distribution. Nested clade analysis in phylogeography [40] has been criticised [31], but we have used the method as originally envisaged [41] to partition character states (host specificity or sequence variation in other genes) between the network of clades established using one gene (gltA). The network of first-step clades (joined by single mutational steps) are probably clonal; three such clones were identified as ‘B. grahamii’, and 16 as ‘B. taylorii’, with two clones of ‘B. doshiae’ and a single clone of ‘B. birtlesii’ within the overall Bartonella population from rodents within this small area of forest and old fields. There was compelling evidence for recombination between all four Bartonella taxa identified, hitherto regarded as valid species, and recombination within the gltA locus had generated two clades which were otherwise difficult to assign to species.
Bartonella has an established binomial taxonomy, which is assumed to reflect underlying biological reality. There has been frequent speculation concerning the extreme diversity of Bartonella (over 1,000 sequences of gltA from this genus are already deposited in GenBank) as an evolutionary and phylogenetic problem [12, 21, 23] while at the same time the contribution of recombination to genome evolution in the genus is clear [2, 38]. Despite frequent cautions against the use of single-gene phylogenies in microbial taxonomy, the reliance on binomial taxonomy and single gene sequences (most frequently gltA or 16S RNA) for diagnosis has led to important judgements in zoonotic medicine and wildlife disease. Thus the reputation of rodent-infecting B. grahamii and B. vinsonii arupensis as emerging human pathogens (e.g. [15, 24]) rests upon the isolation of single clades from humans diagnosed by gene sequences which are prone to recombination. Similarly, B. henselae (cat and human parasite transmitted by fleas) has been recorded from marine mammals, transmitted by Cyamus [16]; the use of a wider range of loci in this work might have clarified the exact strains capable of such an ecologically diverse zoonosis. The present work makes it clear that this approach is both unreliable and potentially highly misleading, and the use of these sequences, especially the 16S RNA marker, for diagnostic purposes should be undertaken only with extreme caution.
Although genetic re-organisation is clear at a genomic level [2], the stability of housekeeping genes is assumed, between which pathogenicity islands are free to evolve [38]. Saenz et al. [38] suggest that recombinant duplication of pathogenicity-related loci led to the evolution of the largest, most complex Bartonella genomes, with up to 69 virulence-related genes (including plasmid encoded copies, see [2]). However, this phylogeny [38] was based only on type isolates; while B. grahamii is a relatively homogenous taxon [3], we have no such confidence in respect of B. taylorii or B. doshiae. We have shown that the latter intergrades with B. taylorii, and there is more evidence of interchange between B. grahamii, B. doshiae and B. birtlesii and the different clades of B. taylorii than there is of interchange between B. taylorii clades themselves. This network of links within B. taylorii presents a problem for wider Bartonella taxonomy. As currently constituted, phylogenies based on gltA (see Fig. 1) which include the three B. taylorii clades identified in this work span a huge range of isolates from rodents, Eurasian insectivores and bats [11]. If the isolates from insectivores and bats are considered not to be B. taylorii, then the current B. taylorii will need to be divided. The scope of B. taylorii as a natural taxon therefore needs experimental review.
Sequencing of B. tribocorum and B. grahamii genomes has shown that groEl and 16S RNA genes lie within a high plasticity region [2, 38], 1624722 and 2084545 bases from the origin of replication. The position of these genes, adjacent to virB/virD and vbh pathogenicity islands, suggests that they could be the most useful genes in predicting pathogenic potential in Bartonella isolates. The situation with gltA is most remarkable; previously the stability of this gene has been assumed, but it is now clear that novel variants of this gene are actively generated by recombination. In this respect, Bartonella appears similar to Wolbachia [1], and recombination within gltA may be quite widespread in this group of bacteria.
The recombinant network of Bartonella isolates extends across four species of rodents in two distinct but related biotypes, old managed forest and recently (20 years) abandoned agricultural land. Nested clade analysis based on gltA had sufficient resolving power to identify clades of B. taylorii and B. grahamii associated with particular host species and with habitat. B. taylorii and B. grahamii as species are normally considered to lack host specificity [3, 4], but in the present work both zero-step clades and, to a large extent, first-step clades nested within the cladogram were host specific. Thus, B. taylorii clade B was recovered only from voles; three of the first-step clades within this clade came from Microtus spp. in the abandoned field system, and two first-step clades came from My. glareolus in the forest. Similarly, within the B. taylorii clade C, all isolates (eight belonging to four one-step clades) came from A. flavicollis. The larger B. taylorii clade A and B. grahamii were more complex, and it was only within these clades that first-step clades infecting more than one host species were noted. The most characteristic was the large B. grahamii clade Ur31; 27 isolates were recorded from three rodent species, but the majority (26 isolates) came from the woodland rodents, My. glareolus and A. flavicollis. The stem zero-step clades (Ur06, Ur31, Ur22) within each clade contained the greatest number of isolates, as would be predicted for the oldest clades from which most others were descended [9]. More interestingly, these were clades involving forest rodents; whereas, isolates infecting Microtus in the abandoned field system tended to belong to tip clades collected from only one or two hosts. As a result, clade diversity was highest in Microtus spp. and lowest in My. glareolus. Almost all of the recombinant connections noted in this work, with the exception of that between B. grahamii Ur30 and B. taylorii Ur11 (exchange of a groEl), involved Microtus as one of the partners. It is worth pointing out that Ur26 in this context is a large clade of 11 isolates from Microtus, all with an identical gltA sequence but composed of both B. doshiae and B. taylorii. This clade took part in many of the long-range recombinant exchanges across the network. This suggests a role for Microtus in the diversification of Bartonella, although this rodent has the strictest habitat specificity [26, 34] and is unlikely to act as a bridge between rodent communities. The low diversity of isolates from My. glareolus supports the role of this host supporting clonal expansion within the forest environment but not favouring genetic diversification or exchange of genetic material with other isolates. The competence of natural Bartonella clades is unknown, but B. grahamii at least carries a diverse complement of plasmids and phages and contains numerous phage sequences intercalated into the genome [2]. It is not clear therefore whether this diversity of transforming factors extends across all the species and isolates examined in this work or is restricted to the ‘B. grahamii sensu stricto’ clades such as Ur29, 30 and 31, which are most similar to the Uppsala isolate sequenced by Berglund et al. [2]. Assuming that all isolates are equally competent, then the importance of Microtus relative to Myodes in fostering recombinant isolates is perhaps due to differences in ectoparasite vector ecology between the two species [17, 35]. This is a topic worthy of further research in understanding the role of recombination in generating novel genotypes of differing pathogenicity.
This study has revealed a huge diversity of Bartonella over very short geographical distances; sequencing just the 300 bp gltA fragment revealed 37 distinct terminal clades among 147 isolates from a small area of forest and abandoned agricultural land. Sequencing of the additional housekeeping loci revealed further diversity, at least within some clades, and possibly all isolates would prove unique if sequenced at sufficient loci. Other studies (e.g. [21, 22, 44]) report similar diversity, which may be a feature of the biology of Bartonella. The agricultural land was abandoned only in 1990, prior to which Microtus must have been relatively rare [27]. We may therefore have witnessed an explosion in Bartonella diversity linked to the expansion of Microtus following ecological succession in the abandoned fields. Whatever the reasons for the considerable recombinant diversity of Bartonella, this is clearly an ideal system in which to study recombination in natural populations of parasitic bacteria, as recombinant events are sufficiently common to be observed in real time but not so frequent as to obscure all signal of clonal lineages. This system provides a model for the study of Bartonella evolution at a fine scale, to identify in natural populations the mechanisms which allow specific evolutionary lineages to become human pathogens.
Acknowledgements
This study was supported by the Polish Ministry of Science and Higher Education grant no. N N303 093134.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- 1.Baldo L, Bordenstein S, Wernegreen JJ, Werren JH. Widespread recombination throughout Wolbachia genomes. Mol Biol Evol. 2005;23:437–449. doi: 10.1093/molbev/msj049. [DOI] [PubMed] [Google Scholar]
- 2.Berglund EC, Frank AC, Calteau A, Petterson OV, Granberg F, Eriksson AS, Naslund K, Holmberg M, Lindroos H, Andersson SGE. Run off replication of host adaptability genes is associated with gene transfer agents in the genome of mouse infecting Bartonella grahamii. PLoS Genet. 2009;5:e1000546. doi: 10.1371/journal.pgen.1000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berglund EC, Ehrenborg C, Pettersson OV, Granberg F, Näslund K, Holmberg M, Andersson SGE. Genome dynamics of Bartonella grahamii in micro-populations of woodland rodents. BMC Genomics. 2010;11:152. doi: 10.1186/1471-2164-11-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Birtles RJ, Harrison TG, Molyneux DH. Grahamella in small woodland mammals in the UK—isolation, prevalence and host specificity. Ann Trop Med Parasitol. 1994;88:317–327. doi: 10.1080/00034983.1994.11812872. [DOI] [PubMed] [Google Scholar]
- 5.Birtles RJ. Bartonellae as elegant haemotropic parasites. Ann NY Acad Sci. 2005;1063:270–279. doi: 10.1196/annals.1355.044. [DOI] [PubMed] [Google Scholar]
- 6.Boulouis HJ, Chang CC, Henn JB, Hasten RW, Chomel BB. Factors associated with the rapid emergence of zoonotic Bartonella infections. Vet Res. 2005;36:383–410. doi: 10.1051/vetres:2005009. [DOI] [PubMed] [Google Scholar]
- 7.Breitschwerdt EB, Maggi RG, Chomel BB, Lappin MR. Bartonellosis: an emerging infectious disease of zoonotic importance to animals and human beings. Vet Emerg Crit Care (San Antonio) 2009;20:8–30. doi: 10.1111/j.1476-4431.2009.00496.x. [DOI] [PubMed] [Google Scholar]
- 8.Breitschwerdt EB, Maggi RG, Cadenas MB, de Paiva V, Diniz PP. A groundhog, a novel Bartonella sequence and my father’s death. Emerg Infect Dis. 2009;15:2080–2086. doi: 10.3201/eid1512.090206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crandall KA, Templeton AR. Empirical tests of some predictions from coalescent theory with applications to intraspecific phylogeny reconstruction. Genetics. 1993;134:959–969. doi: 10.1093/genetics/134.3.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clement M, Posada D, Crandall KA. TCS: a computer program to estimate gene geneologies. Mol Ecol. 2000;9:1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- 11.Concannon R, Wynn-Owen K, Simpson VR, Birtles RJ. Molecular characterisation of haemoparasites infecting bats (Microchiroptera) from bats. Parasitology. 2005;131:489–496. doi: 10.1017/S0031182005008097. [DOI] [PubMed] [Google Scholar]
- 12.Ehrenborg C, Handley S, Ellis B, Mills J, Holmberg M. Bartonella grahamii infecting rodents display high genetic diversity over short geographic distances. Ann NY Acad Sci. 2003;990:233–235. doi: 10.1111/j.1749-6632.2003.tb07369.x. [DOI] [PubMed] [Google Scholar]
- 13.Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol. 2004;186:1518–1530. doi: 10.1128/JB.186.5.1518-1530.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feil E, Zhou J, Maynard Smith J, Spratt BG. A comparison of the nucleotide sequences of the adk and recA genes of pathogenic and commensal Neisseria species: evidence for extensive inter-species recombination within adk. J Mol Evol. 1996;43:631–640. doi: 10.1007/BF02202111. [DOI] [PubMed] [Google Scholar]
- 15.Fenollar F, Sire S, Raoult D. Bartonella vinsonii subsp. arupensis as an agent of blood culture-negative endocarditis in a human. J Clin Microbiol. 2005;43:945–947. doi: 10.1128/JCM.43.2.945-947.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harms CA, Maggi RG, Breitschwerdt EB, Clemons-Chevis CL, Solangi M, Rotstein DS, Fair PA, Hansen LJ, Hohn AA, Lovewell GN, McLellan WA, Pabst DA, Rowles TK, Schwacke LH, Townsend FI, Wells RS. Bartonella species detection in captive, stranded and free ranging cetaceans. Vet Res. 2008;39:59. doi: 10.1051/vetres:2008036. [DOI] [PubMed] [Google Scholar]
- 17.Harris PD, Paziewska A, Zwolińska L, Siński E. Seasonality of the ectoparasite community of woodland rodents in a Mazurian forest, Poland. Wiad Parazytol. 2009;55:377–388. [PubMed] [Google Scholar]
- 18.Heath L, van der Walt E, Varsani A, Martin DP. Recombinant patterns in aphthoviruses mirror those found in other picornaviruses. J Virol. 2006;80:11827–11832. doi: 10.1128/JVI.01100-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henn JB, Chomel BB, Boulois HJ, Kasten RW, Murray WJ, Bar-Gal GK, King R, Courreau JF, Baneth G. Bartonella rochalimae in raccoons, coyotes and red foxes. Emerg Infect Dis. 2009;15:1984–1986. doi: 10.3201/eid1512.081692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guindon S, Gascuel O. A simple, fast and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 21.Inoue K, Kabeya H, Kosoy MH, Bai Y, Smirnov G, McColl D, Artsob H, Maruyama S. Evolutionary and geographical relationships of Bartonella grahamii isolates from wild rodents by multi-locus sequencing analysis. Microb Ecol. 2009;57:534–541. doi: 10.1007/s00248-009-9488-x. [DOI] [PubMed] [Google Scholar]
- 22.Inoue K, Kabeya H, Shiratori H, Ueda K, Kosoy MY, Chomel BB, Boulouis HJ, Maruyama S. Bartonella japonica sp. nov. and Bartonella silvatica sp. nov., isolated from Apodemus mice in Japan. Int J Syst Evol Microbiol. 2009 doi: 10.1099/ijs.0.011528-0. [DOI] [PubMed] [Google Scholar]
- 23.Inoue K, Maruyanra S, Kabeya H, Hagiya K, Izumi Y, Une Y, Yoshikawa Y. Exotic small mammals as potential reservoirs of zoonotic Bartonella spp. Emerg Infect Dis. 2009;15:526–532. doi: 10.3201/eid1504.081223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kerkhoff FT, Bergmans AM, van Der Zee A, Rothova A. Demonstration of Bartonella grahamii DNA in ocular fluids of a patient with neuroretinitis. J Clin Microbiol. 1999;37:4034–4038. doi: 10.1128/jcm.37.12.4034-4038.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kosoy MY, Regnery RL, Tzianabos T, Marston EL, Jones DC, Green D, Maupin GO, Olson JG, Childs JE. Distribution, diversity and host specificity of Bartonella in rodents from the Southeastern United States. Am J Trop Med Hyg. 1997;57:578–588. doi: 10.4269/ajtmh.1997.57.578. [DOI] [PubMed] [Google Scholar]
- 26.Kozakiewicz M, Gortat T, Kozakiewicz A, Barkowska M. Effects of habitat fragmentation on four rodent species in a Polish farm landscape. Landscape Ecol. 1999;14:391–400. doi: 10.1023/A:1008070610187. [DOI] [Google Scholar]
- 27.Kozakiewicz M, Kozakiewicz A. Long-term dynamics and biodiversity changes in the mosaic of agricultural and forest habitats. Ann Zool Fenn. 2008;45:263–269. [Google Scholar]
- 28.La Scola B, Zeaiter Z, Khamis A, Raoult D. Gene-sequence-based criteria for species definition in bacteriology: the Bartonella paradigm. Trends Microbiol. 2003;11:318–321. doi: 10.1016/S0966-842X(03)00143-4. [DOI] [PubMed] [Google Scholar]
- 29.Maiden MCJ, Bygraves JA, Feil E. Multilocus sequence typing: a portable approach to the identification of clades within populations of pathogenic microorganisms. Proc Natl Acad Sci USA. 1998;95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morick D, Osinga N, Gruys E, Harrus S. Identification of a Bartonella species in the harbour seal (Phoca vitulina) and in seal lice (Echinophthirius horridus) Vector Borne Zoonotic Dis. 2009;9:751–753. doi: 10.1089/vbz.2008.0202. [DOI] [PubMed] [Google Scholar]
- 31.Nielsen R, Beaumont MA. Statistical inferences in phylogeography. Mol Ecol. 2009;18:1034–1037. doi: 10.1111/j.1365-294X.2008.04059.x. [DOI] [PubMed] [Google Scholar]
- 32.Nguyen NT, Maurus R, Stokell DJ, Ayed A, Duckworth HW, Brayer GD. Comparative analysis of folding and substrate binding sites between regulated hexameric type II citrate synthases and unregulated dimeric type I enzymes. Biochemistry. 2001;40:13177–13187. doi: 10.1021/bi010408o. [DOI] [PubMed] [Google Scholar]
- 33.Norman AF, Regnery R, Jameson P, Greene C, Krause DC. Differentiation of Bartonella-like isolates at the species level by PCR-restriction fragment length polymorphism in the citrate synthase gene. J Clin Microbiol. 1995;33:1797–1803. doi: 10.1128/jcm.33.7.1797-1803.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Panzacchi M, Linnell JDC, Melis C, Odden M, Odden J, Gorini L, Andersen R. Effect of land use on small mammal abundance and diversity in a forest-farmland mosaic in south-eastern Norway. For Ecol Manage. 2010;259:1536–1545. doi: 10.1016/j.foreco.2010.01.030. [DOI] [Google Scholar]
- 35.Paziewska A, Zwolińska L, Harris PD, Bajer A, Siński E. Utilisation of rodent species by larvae and nymphs of hard ticks (Ixodidae) in two habitats in North East Poland. Exp Appl Acarol. 2010;50:79–91. doi: 10.1007/s10493-009-9269-8. [DOI] [PubMed] [Google Scholar]
- 36.Posada D. jModelTest: phylogenetic model averaging. Mol Biol Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 37.Renesto P, Gouvernet J, Drancourt M, Roux V, Raoult D. Use of rpoB gene analysis for detection and identification of Bartonella species. J Clin Microbiol. 2001;39:430–437. doi: 10.1128/JCM.39.2.430-437.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saenz HL, Engel P, Stoeckli MC, Lanz C, Raddatz G, Vayssier-Taussat M, Birtles R, Schuster SC, Dehio C. Genomic analysis of Bartonella identifies type IV secretion systems as host adaptability factors. Nat Genet. 2007;39:1469–1476. doi: 10.1038/ng.2007.38. [DOI] [PubMed] [Google Scholar]
- 39.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 40.Templeton AR. Nested clade analyses of phylogeographic data: testing hypotheses about gene flow and population history. Mol Ecol. 1998;7:381–397. doi: 10.1046/j.1365-294x.1998.00308.x. [DOI] [PubMed] [Google Scholar]
- 41.Templeton AR, Boerwinkle E, Sing CF. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping. I. Basic theory and an analysis of alcohol dehydrogenase activity in Drosophila. Genetics. 1987;117:343–351. doi: 10.1093/genetics/117.2.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Templeton AR, Crandall KA, Sing CF. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data: III Cladogram estimation. Genetics. 1992;132:619–633. doi: 10.1093/genetics/132.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Templeton AR, Sing CF. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping. IV. Nested analyses with cladogram uncertainty and recombination. Genetics. 1993;134:659–669. doi: 10.1093/genetics/134.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Welc-Falęciak R, Paziewska A, Bajer A, Behnke JM, Siński E. Bartonella spp. infections in rodents from different habitats in the Mazury Lake District, Northeast Poland. Vector Borne Zoonotic Dis. 2008;8:467–474. doi: 10.1089/vbz.2007.0217. [DOI] [PubMed] [Google Scholar]
- 45.Zeaiter Z, Fournier PE, Ogata H, Raoult D. Phylogenetic classification of Bartonella species by comparing groEL sequences. Int J Syst Evol Microbiol. 2002;52:165–171. doi: 10.1099/00207713-52-1-165. [DOI] [PubMed] [Google Scholar]