Abstract
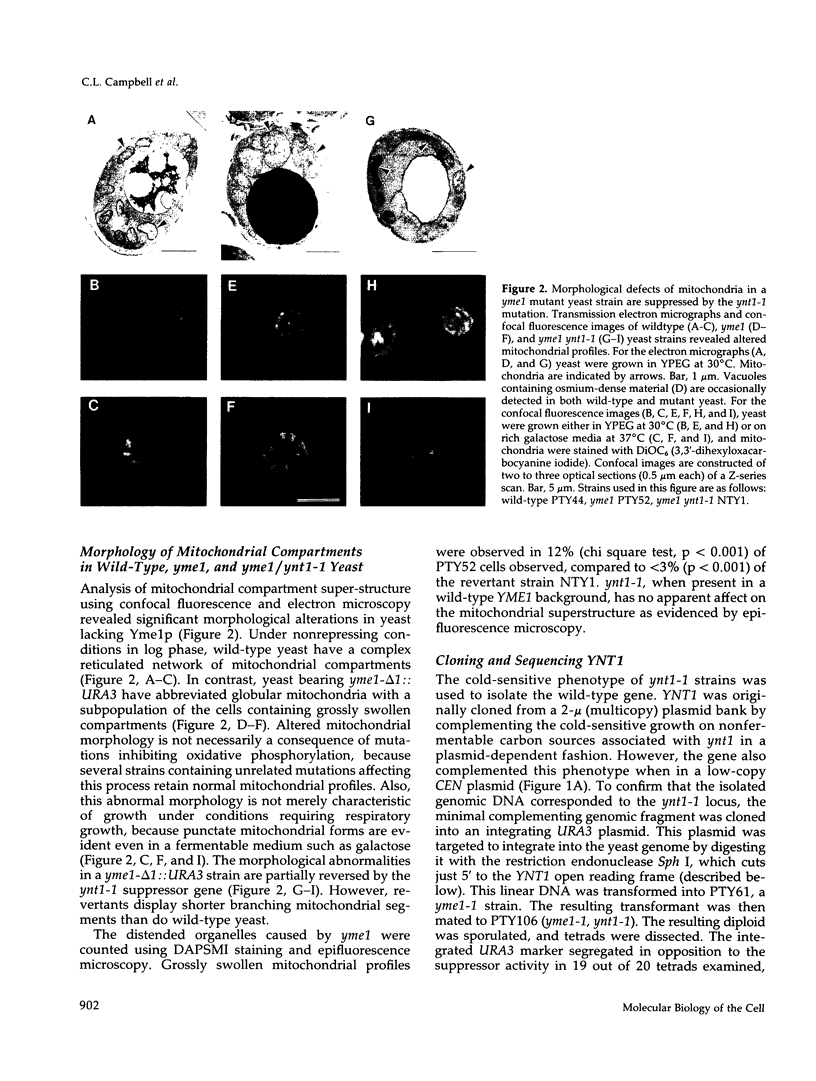
The absence of functional Yme1p, a putative ATP and zinc-dependent protease localized to mitochondria of yeast, results in abnormal mitochondrial function and morphology. Yeast lacking Yme1p lose DNA from mitochondria at an accelerated rate, fail to grow on nonfermentable carbon sources at 37 degrees C, and have severely deficient growth if mitochondrial DNA suffers large deletions or is completely lost. In place of the normal reticulated mitochondrial network, strains lacking Yme1p have punctate mitochondria with some grossly swollen compartments. The growth phenotypes and morphological alterations evident in these mutant yeast can be compensated by a mutation in YNT1, an essential gene in yeast. The sequence of the YNT1 gene product indicates that it is one of a number of related regulatory subunits of the 26S protease. This proteolytic activity is necessary for progression through the cell cycle and has been implicated in the regulation of transcription. Ynt1p is more distantly related to Yme1p.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Begg K. J., Tomoyasu T., Donachie W. D., Khattar M., Niki H., Yamanaka K., Hiraga S., Ogura T. Escherichia coli mutant Y16 is a double mutant carrying thermosensitive ftsH and ftsI mutations. J Bacteriol. 1992 Apr;174(7):2416–2417. doi: 10.1128/jb.174.7.2416-2417.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers B., Goetsch L. Preparation of yeast cells for thin-section electron microscopy. Methods Enzymol. 1991;194:602–608. doi: 10.1016/0076-6879(91)94044-d. [DOI] [PubMed] [Google Scholar]
- Chin D. T., Goff S. A., Webster T., Smith T., Goldberg A. L. Sequence of the lon gene in Escherichia coli. A heat-shock gene which encodes the ATP-dependent protease La. J Biol Chem. 1988 Aug 25;263(24):11718–11728. [PubMed] [Google Scholar]
- Diaz R., Mayorga L. S., Weidman P. J., Rothman J. E., Stahl P. D. Vesicle fusion following receptor-mediated endocytosis requires a protein active in Golgi transport. Nature. 1989 Jun 1;339(6223):398–400. doi: 10.1038/339398a0. [DOI] [PubMed] [Google Scholar]
- Dubiel W., Ferrell K., Pratt G., Rechsteiner M. Subunit 4 of the 26 S protease is a member of a novel eukaryotic ATPase family. J Biol Chem. 1992 Nov 15;267(32):22699–22702. [PubMed] [Google Scholar]
- Fröhlich K. U. Sequence Similarity Presenter: a tool for the graphic display of similarities of long sequences for use in presentations. Comput Appl Biosci. 1994 Apr;10(2):179–183. doi: 10.1093/bioinformatics/10.2.179. [DOI] [PubMed] [Google Scholar]
- Ghislain M., Udvardy A., Mann C. S. cerevisiae 26S protease mutants arrest cell division in G2/metaphase. Nature. 1993 Nov 25;366(6453):358–362. doi: 10.1038/366358a0. [DOI] [PubMed] [Google Scholar]
- Gordon C., McGurk G., Dillon P., Rosen C., Hastie N. D. Defective mitosis due to a mutation in the gene for a fission yeast 26S protease subunit. Nature. 1993 Nov 25;366(6453):355–357. doi: 10.1038/366355a0. [DOI] [PubMed] [Google Scholar]
- Henikoff S., Henikoff J. G. Automated assembly of protein blocks for database searching. Nucleic Acids Res. 1991 Dec 11;19(23):6565–6572. doi: 10.1093/nar/19.23.6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A., Ciechanover A. The ubiquitin system for protein degradation. Annu Rev Biochem. 1992;61:761–807. doi: 10.1146/annurev.bi.61.070192.003553. [DOI] [PubMed] [Google Scholar]
- Koning A. J., Lum P. Y., Williams J. M., Wright R. DiOC6 staining reveals organelle structure and dynamics in living yeast cells. Cell Motil Cytoskeleton. 1993;25(2):111–128. doi: 10.1002/cm.970250202. [DOI] [PubMed] [Google Scholar]
- Magnani M., Serafini G., Antonelli A., Malatesta M., Gazzanelli G. Evidence for a particulate location of ubiquitin conjugates and ubiquitin-conjugating enzymes in rabbit brain. J Biol Chem. 1991 Nov 5;266(31):21018–21024. [PubMed] [Google Scholar]
- McConnell S. J., Stewart L. C., Talin A., Yaffe M. P. Temperature-sensitive yeast mutants defective in mitochondrial inheritance. J Cell Biol. 1990 Sep;111(3):967–976. doi: 10.1083/jcb.111.3.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell S. J., Yaffe M. P. Nuclear and mitochondrial inheritance in yeast depends on novel cytoplasmic structures defined by the MDM1 protein. J Cell Biol. 1992 Jul;118(2):385–395. doi: 10.1083/jcb.118.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami Y., Matsufuji S., Kameji T., Hayashi S., Igarashi K., Tamura T., Tanaka K., Ichihara A. Ornithine decarboxylase is degraded by the 26S proteasome without ubiquitination. Nature. 1992 Dec 10;360(6404):597–599. doi: 10.1038/360597a0. [DOI] [PubMed] [Google Scholar]
- Ohana B., Moore P. A., Ruben S. M., Southgate C. D., Green M. R., Rosen C. A. The type 1 human immunodeficiency virus Tat binding protein is a transcriptional activator belonging to an additional family of evolutionarily conserved genes. Proc Natl Acad Sci U S A. 1993 Jan 1;90(1):138–142. doi: 10.1073/pnas.90.1.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechsteiner M., Hoffman L., Dubiel W. The multicatalytic and 26 S proteases. J Biol Chem. 1993 Mar 25;268(9):6065–6068. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnall R., Mannhaupt G., Stucka R., Tauer R., Ehnle S., Schwarzlose C., Vetter I., Feldmann H. Identification of a set of yeast genes coding for a novel family of putative ATPases with high similarity to constituents of the 26S protease complex. Yeast. 1994 Sep;10(9):1141–1155. doi: 10.1002/yea.320100903. [DOI] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989 May;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsness P. E., Fox T. D. Nuclear mutations in Saccharomyces cerevisiae that affect the escape of DNA from mitochondria to the nucleus. Genetics. 1993 May;134(1):21–28. doi: 10.1093/genetics/134.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsness P. E., White K. H., Fox T. D. Inactivation of YME1, a member of the ftsH-SEC18-PAS1-CDC48 family of putative ATPase-encoding genes, causes increased escape of DNA from mitochondria in Saccharomyces cerevisiae. Mol Cell Biol. 1993 Sep;13(9):5418–5426. doi: 10.1128/mcb.13.9.5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomoyasu T., Yuki T., Morimura S., Mori H., Yamanaka K., Niki H., Hiraga S., Ogura T. The Escherichia coli FtsH protein is a prokaryotic member of a protein family of putative ATPases involved in membrane functions, cell cycle control, and gene expression. J Bacteriol. 1993 Mar;175(5):1344–1351. doi: 10.1128/jb.175.5.1344-1351.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiebel F. F., Kunau W. H. The Pas2 protein essential for peroxisome biogenesis is related to ubiquitin-conjugating enzymes. Nature. 1992 Sep 3;359(6390):73–76. doi: 10.1038/359073a0. [DOI] [PubMed] [Google Scholar]
- Zhaung Z. P., McCauley R. Ubiquitin is involved in the in vitro insertion of monoamine oxidase B into mitochondrial outer membranes. J Biol Chem. 1989 Sep 5;264(25):14594–14596. [PubMed] [Google Scholar]