Summary
Non-small cell lung carcinoma (NSCLC) is the leading cause of cancer-related death worldwide, with an overall 5-year survival rate of only 10–15% 1. Deregulation of the Ras pathway is a frequent hallmark of NSCLC, often through mutations that directly activate Kras 2. p53 is also frequently inactivated in NSCLC and, since oncogenic Ras can be a potent trigger of p53 3, it seems likely that oncogenic Ras signalling plays a major and persistent part in driving the selection against p53. Hence, pharmacological restoration of p53 is an appealing therapeutic strategy for treating this disease 4. Here, we model the likely therapeutic impact of p53 restoration in a spontaneously evolving mouse model of NSCLC initiated by sporadic oncogenic activation of endogenous Kras 5. Surprisingly, p53 restoration failed to induce significant regression of established tumours although it did result in a significant decrease in the relative proportion of tumours classed as high grade. This is due to selective activation of p53 only in the more aggressive tumour cells within each tumour. Such selective activation of p53 correlates with marked up regulation in Ras signal intensity and induction of the oncogenic signalling sensor p19ARF 6. Our data indicate that p53-mediated tumour suppression is triggered only when oncogenic Ras signal flux exceeds a critical threshold. Importantly, the failure of low-level oncogenic Kras to engage p53 reveals inherent limits in the capacity of p53 to restrain early tumour evolution and to the efficacy of therapeutic p53 restoration to eradicate cancers.
Inactivation of the p53 tumour suppressor pathway is a common feature of human cancers, fostering the attractive notion of restoring p53 function in established tumours as an effective and tumour-specific therapeutic strategy 4. Indeed, p53 restoration was recently shown to trigger dramatic tumour regression in vivo 7–9. While encouraging, these studies utilized tumour models (either transgene 7,9 or radiation-induced 8) driven by preternaturally high levels of oncogenes. Because high-level oncogene activity potently engages p53 via the p19ARF tumour suppressor 6,7,10, p53 restoration has a dramatic impact in these models. Unlike high oncogenic activity, however, low-level expression of dominant oncogenes appears insufficient to engage intrinsic tumour suppression, even though it still suffices to drive tumourigenesis 11,12. This raises the spectre that many epithelial malignancies, initiated as they are by low-level oncogenic signals such as those arising from mutational activation of ras genes in situ, may be insensitive to p53 restoration - at least during certain phases of their evolution. To investigate this possibility we assessed the ability of p53 restoration to trigger tumour regression in the well-characterized Lox-Stop-Lox-KrasG12D (KR) murine tumour model of NSCLC 5 wherein tumourigenesis is driven by sporadic, low-level activation of mutant Kras. This model closely recapitulates its human disease counterpart 13.
After inhalation of adenovirus-Cre, KR mice develop multiple, independently evolving lung tumours, permitting contemporaneous analysis of different disease stages within each animal. KR mice were crossed into the p53KI/KI switchable mouse model in which both alleles of the endogenous p53 gene are replaced by the conditional variant p53ERTAM 14. p53KI/KI mice can be reversibly toggled in vivo between p53 wild-type (wt) and p53 null states by administration or withdrawal of Tamoxifen (Tam). Importantly, once functionally restored in Tam-treated p53KI/KI mice, p53-mediated tumour suppression is triggered only if p53-activating signals are present 7,10.
KrasG12D was sporadically activated in KR;p53KI/+ and KR;p53KI/KI lungs and tumours allowed to develop for 16 weeks. In both genotypes, KrasG12D activation induced a spectrum of lung tumour grades including hyperplasias, adenomas and adenocarcinomas. Like KR;p53-deficient animals 15 (Supplementary Figure 1), KR;p53KI/KI mice exhibit accelerated tumour progression and increased incidence of high-grade tumours relative to their KR;p53KI/+ counterparts. These data affirm that p53 restrains Kras-driven NSCLC yet indicate that, even when combined, KrasG12D activation and p53 inactivation are insufficient to generate malignant tumours without additional, aleatory mutations.
To ascertain its therapeutic impact, p53 function was restored for one week in KR;p53KI/KI lung tumours (Figure 1A). Surprisingly, given the dramatic tumour regression induced by p53 restoration in other models 7–9, p53 restoration had no macroscopically evident impact on these tumours (Figure 1B). Close inspection, however, indicated that p53 restoration did elicit a modest decrease in proliferating tumour cells (Figure 1C; 13.99% Ki67 positive cells per Tam-treated tumours versus 20.97% in controls) and an increase in apoptosis (Supplemental Figure 2 and Figure 1D; 45% of p53-restored tumours contain apoptotic cells versus 13.5% of control tumours). Nevertheless, the distribution of apoptotic cells in tumours following p53 restoration was irregular and clustered (Figure 1E). This high variability in the response to sustained p53 restoration was confirmed by microCT imaging of individual tumours over 7 days. While all control tumours grew during treatment, individual Tam-treated tumours exhibited all possible responses – some grew, others were unchanged, and many shrank (Figure 2A and Supplemental Figure 3). Such variability in tumour response to Tam might reflect heterogeneities among tumor cells in the efficiency of p53 restoration, in the presence of p53-activating signals, or in the engagement of downstream effectors following p53 restoration. To determine which, we first ascertained the efficiency with which Tam restored p53 function in tumours. Mice were treated for 7 days with Tam or vehicle and then exposed to a single dose of γ-radiation (IR) 2 hrs after the last treatment to activate p53 directly. p53 activity was then monitored in individual tumours by assaying induction of the prototypical p53-responsive gene, CDKN1A (p21cip1) 16,17. All tumours showed substantial CDKN1A induction (Figure 2B), indicating that systemic Tam pervasively restores p53 function in all tumours. Hence, the heterogeneity of the therapeutic response to Tam is not a consequence of either variability in Tam-dependent p53 restoration or in the capacity of p53, once activated, to induce CDKN1A. By contrast, when p53 function was restored in the absence of concomitant DNA damage, CDKN1A was induced in only a minority of tumours (Figure 2B). Hence, the variability in response to p53 restoration is because only a minority of tumours harbour endogenous p53-activating signals. Interestingly, whereas we see abundant apoptosis in aggressive tumour cells following p53 restoration, Feldser et al. in an accompanying paper do not 18, even though their mouse lung tumour model driven by spontaneous, sporadic KRas activation is ostensibly similar to ours. The reasons for this are unclear. However, the models differ in several ways. First, the mechanism of KRas activation is different, and may target distinct cell lineages with innately different sensitivities to p53-induced apoptosis. Second, they use Cre-lox recombination to restore p53 function, which is innately less synchronous than in our p53ERTAM model and may make it difficult to see a transient wave of cell death. Cre-lox recombination may also introduce additional genotoxic stresses that further modify p53 output. In the end, however, whether apoptosis or senescence is the principal output of p53 restoration in aggressive tumour cells may not be so important since that both p53-induced apoptosis 7 and senescence 9 are effective at eliciting tumour clearance.
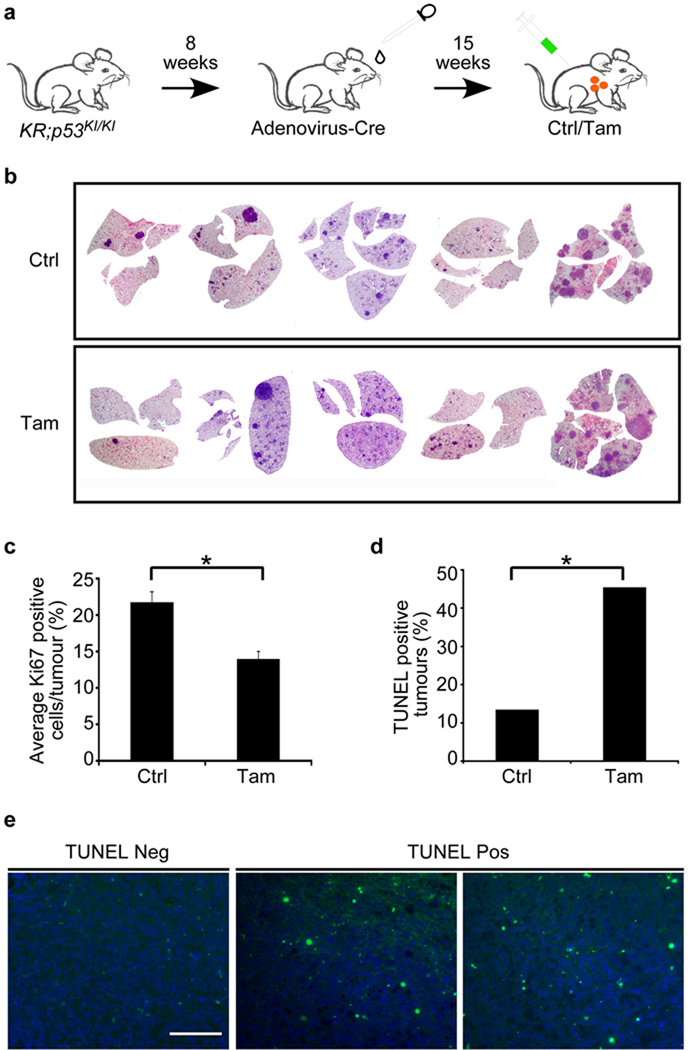
Figure 1. Heterogeneous therapeutic impact of p53 restoration in KrasG12D driven lung tumours.
a. Schematic representation of the experimental treatment regime. KrasG12D was activated in the lung epithelium of 8 week old KR;p53KI/KI mice by adenoviral-Cre nasal inhalation and the resulting tumours treated with Tam or vehicle (Ctrl) 15 weeks after adenoviral infection.
b. Haematoxylin and Eosin staining of lung sections from KR;p53KI/KI mice showing tumour load after 7 daily control (Ctrl) or Tam treatments.
c. Quantification of Ki67 positive cells per lung tumour from 7 day Tam/Ctrl-treated KR;p53KI/KI mice. Error bars indicate standard error of mean (Ctrl: s.e.m=1.20 n=55; Tam: s.e.m=1.31 n=37). * P=0.0003, Student’s t-test.
d. Percent of apoptotic (TUNEL-positive) tumours (scored as a minimum of 1 positive cell per tumour section) in 7 day Ctrl and Tam treated KR;p53KI/KI lungs (n=37 Ctrl; n=22 Tam treated tumours). * P=0.0064, Pearson Chi square.
e. KR;p53KI/KI lung tumours from KR;p53KI/KI treated for 6 hrs with Tam, showing either no discernible TUNEL staining (Neg) or significant levels of TUNEL staining (Pos). Scale bar=100 µm.
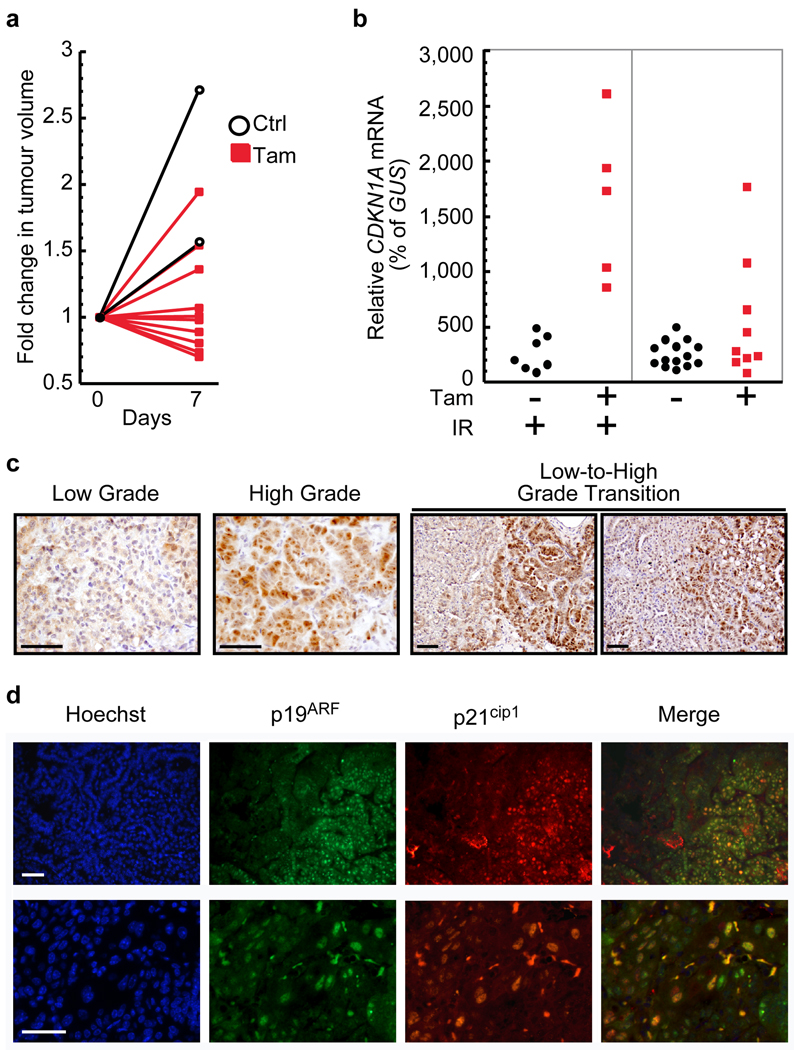
Figure 2. Heterogeneous p53 activation and p19ARF up-regulation in KR;p53KI/KI tumours.
a. MicroCT-derived plots depicting changes in tumour volume during a 7-day treatment. 10 independent tumours are shown before (day 0) and after (day 7) daily Tam (red lines, filled symbols) or sham (black lines, open symbols) treatments.
b. Taqman analysis of CDKN1A expression in individual laser-captured lung tumours from KR;p53KI/KI mice treated for 7 days with vehicle (black circles) or Tam (red squares). Tumours were harvested 24 hrs after the final Ctrl/Tam treatment. Where indicated (IR +, left panel) mice were exposed to a single dose of γ-radiation 2 hrs after the last Tam/Ctrl treatment. Each circle/square represents a single tumour.
c. IHC data comparing levels of p19ARF expression in low and high-grade tumours as well as in transitional lesions exhibiting both low and high-grade features. Scale bars=50 µm.
d. Co-immunostaining for p19ARF and p21cip1 in KR;p53KI/KI lung tumours from mice treated for 6 hrs with Tam. Representative fields shown, one at low (upper panel) and one at high magnification (lower panel). Scale bar=50 µm.
Although p53 may be activated by a wide-range of stress signals, recent in vivo studies implicate induction of p19ARF by oncogenic signalling as the critical p53-activating trigger in established tumours 7,10. Since oncogenic Ras can be a potent inducer of p19ARF 19, we assayed for p19ARF expression in KR;p53KI/KI lung tumours. Immunohistochemical analysis (IHC) of KR;p53KI/KI lungs revealed p19ARF expression to be highly heterogeneous – generally limited to specific regions of certain tumours. Stratification of lung tumours into low and high-grade, the latter comprising mostly adenocarcinomas (Supplemental Figure 4) 20, revealed that p19ARF was confined mostly to high-grade tumours. High p19ARF cells were only rarely observed in low-grade tumours and, when present, were restricted to small, sporadic foci. Close examination of transitional tumours comprising clearly defined high and low-grade regions showed p19ARF to be highly expressed only in high-grade/carcinoma areas (Figure 2C).
Since p19ARF is a potent activator of p53, we next ascertained whether the high-grade regions expressing elevated p19ARF coincide with those that spontaneously activate p53 following restoration. p53 function was acutely restored in KR;p53KI/KI mice and tumours analyzed for expression of p19ARF and p21cip1. Upon p53 restoration, tumour areas positive for p19ARF overlapped extensively with those positive for p21cip1 (Figure 2D): ~70% of p19ARF–positive cells from Tam-treated mice stained positive for p21cip1 compared with 2% of control. That p19ARF plays a causal role in engaging p53-mediated tumour suppression in high-grade tumours was corroborated by the rapid cessation of cell proliferation specific to p19ARF-positive regions following p53 restoration (Figure 3A – Tam, two upper rows). By contrast, proliferation remained high in p19ARF-negative tumours after p53-restoration (Figure 3A –Tam, two lower rows). Of note, no γ-H2AX staining DNA damage foci were detected in KR;p53KI/KI lung tumours, although they were readily evident in tumours from γ-irradiated mice (Supplemental Figure 5). The remarkable overlap between p53 activation and p19ARF expression strongly implicates p19ARF, and not DNA damage, as the endogenous signal responsible for triggering p53 in high-grade lung tumours.
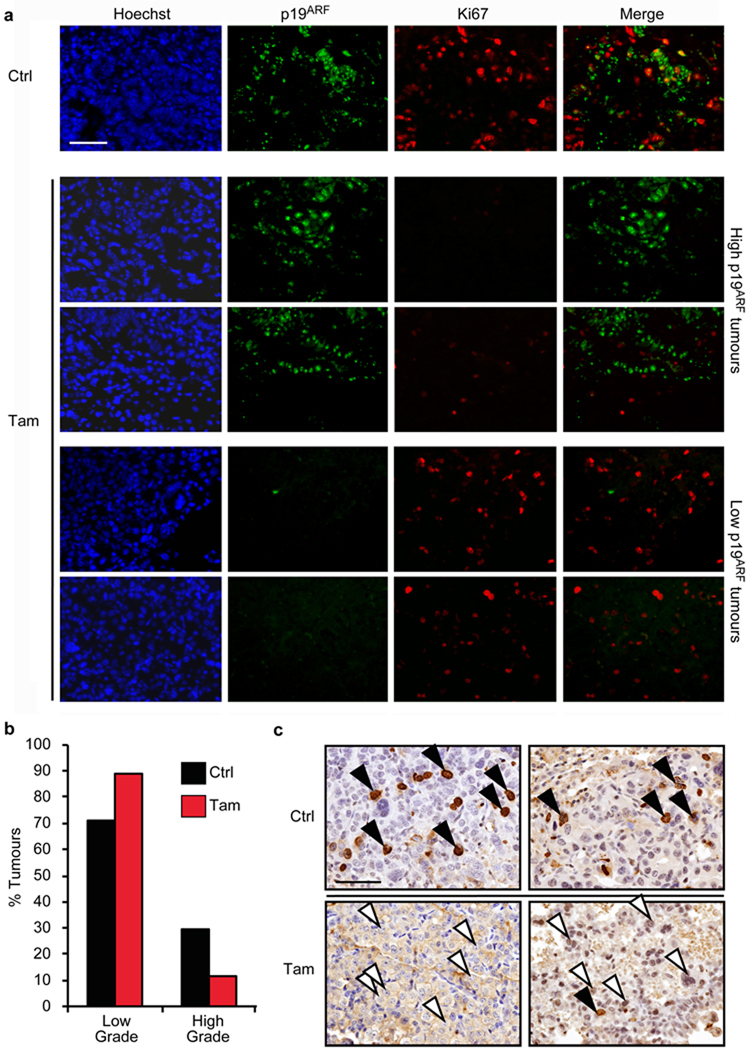
Figure 3. p53 restoration targets high-grade, but not low-grade, lung tumour cells.
a. Co-immunostaining for p19ARF and the proliferation marker Ki67 in lung tumours from KR;p53KI/KI mice treated for 24 hrs with vehicle (Ctrl, upper row) or Tam (four lower rows). Row 2 and 3 illustrate the profound anti-proliferative impact (low Ki67) of p53 restoration in tumours with high p19ARF levels. By contrast, the lower two rows show lack of growth inhibition following p53 restoration in tumours lacking detectable p19ARF. Scale bar = 50 µm.
b. Quantification of low versus high-grade tumour frequencies in lungs of KR;p53KI/KI mice treated for 7 days with either vehicle (Ctrl) or Tam (n=143 Ctrl; n=163 Tam). P=0.0001, Pearson Chi square.
c. Representative images show IHC for BrdU in high-grade tumours from 7-day treated Ctrl (Ctrl, upper panel) or Tam mice (lower panel). BrdU was administered 2 hrs before harvesting. Arrows highlight high-grade cells in each tumour (filled, BrdU positive and open, BrdU negative). Scale bars=50 µm.
Although germ-line p53 deficiency significantly accelerates lung tumour progression and malignancy in KR mice 15, our data indicate that p53 tumour suppression acts only at later stages of tumour evolution. Since p53 is specifically activated in the most aggressive tumour cells, its restoration in a mixed tumour population should lead to a shift downwards in assigned tumour grade. Indeed, 7 days of p53 restoration in KR;p53KI/KI mice harbouring a mixture of low and high-grade tumours elicited a downward shift in the frequency of high-grade tumours (from 29% to 11%) and a pro rata increase in the proportion of low-grade tumours (from 71% to 89%) (Figure 3B and Supplemental Figure 6). The percentage of BrdU-positive high-grade cells was also dramatically reduced following treatment (Figure 3C).
Our data show that the p19ARF/p53 pathway is only engaged in high-grade KR;p53KI/KI cells, even though all tumour cells harbour oncogenic KrasG12D. Hence, oncogenic activity of Kras is not alone sufficient to induce p19ARF and engage p53-mediated tumour suppression. Interestingly, recent in vivo studies indicate that intrinsic tumour suppression is only engaged when oncogenic signals are preternaturally elevated 11,12. Such observations echo in vitro data showing that expression of oncogenic KrasG12D from its endogenous promoter induces proliferation and immortalization whereas KrasG12D over-expression engages p53-dependent replicative senescence 21,22. Since marked up-regulation of the MAPK-pathway is a characteristic feature of advanced lung tumours in both mice 15 and NSCLC in humans 23, we asked whether induction of p19ARF in high-grade tumours is a consequence of elevated flux through the Ras signalling network. Indeed, immunostaining showed a remarkably tight spatial concordance of tumour cells exhibiting elevated ERK phosphorylation (p-ERK), a signature of downstream Ras signalling, and those with high p19ARF (Figures 4A and Supplemental Figure 7) – the cell-by-cell overlap between up-regulation of p19ARF and p-ERK was 91.2% (n=1312; STDEV: 3.77). Hence, increased flux through oncogenic KrasG12D is the likely mechanism for both malignant progression and concomitant activation of (and eventual counter-selection against) the p19ARF/p53 tumour suppressor pathway.
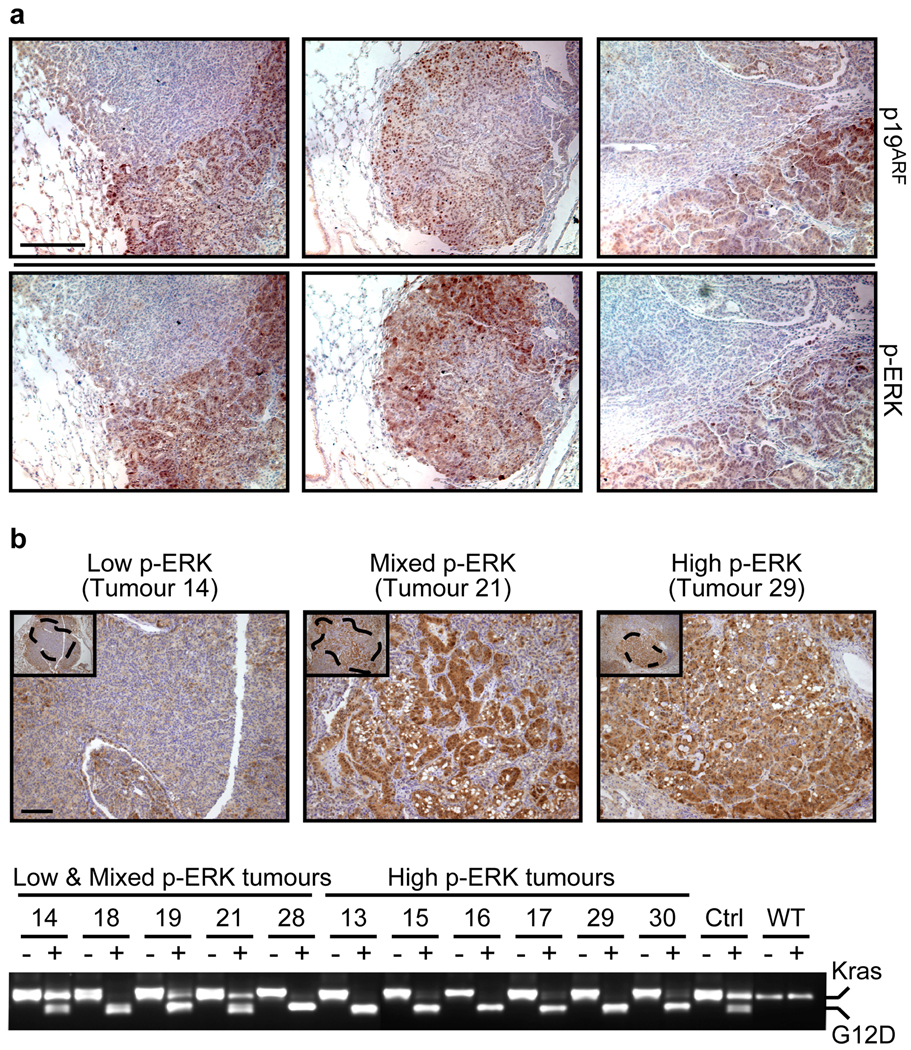
Figure 4. High-grade lung tumours exhibit increased Kras signalling.
a. IHC for p19ARF and p-ERK in consecutive sections of three independent low-to-high-grade transition tumours from KR;p53KI/KI mice. Scale bar = 200 µm.
b. Kras allele analysis was performed on genomic DNA from KR;p53KI/KI lung tumours following laser capture microdissection. p-ERK IHC was used to define areas of low, mixed or high p-ERK (upper panel, Scale bar=50µm) and consecutive slides used for LCM of defined regions (see dotted areas). DNA was isolated from LCM material and the Kras genomic region amplified by PCR and digested with HindIII (lower panel). For each tumour, the undigested (−) and digested (+) PCR fragments were run alongside and the wt (Kras, higher band) and mutant alleles (G12D, lower band) are indicated. Control lung tissue from heterozygous (KrasG12D/+: Ctrl) and wild-type (WT) mice was also analyzed.
Many potential mechanisms might underlie the dramatic up-regulation of p-ERK we observe in high-grade lung tumours, including changes in Kras copy number (known to occur in human NSCLC), secondary inactivation of the wt Kras allele, inactivation of Kras negative feedback mechanisms and incidental activation of cooperating oncogenes 24–27. Initial analysis of whole low versus high-grade tumours suggested downregulation of Sprouty 2 or loss of the wt Kras allele as possible mechanisms for Kras signal up-regulation in high p-ERK tumours (Supplemental Figure 8). Since elevated Ras signalling is a property peculiar to high-grade tumour regions, we used p-ERK staining to demarcate high, low and mixed p-ERK areas of individual tumours (Figure 4B, upper panel). These tumour regions were individually laser microdissected and their genomic DNA extracted and assessed for the relative copy representation of wt versus mutant Kras alleles. We saw variable levels of wt Kras retention in the low/mixed p-ERK tumour tissues, ranging from 100% in the low p-ERK tumour 14 through to partial or total loss in the mixed grade tumours (e.g. 21 and 18). Remarkably, the wt Kras allele was lost in all high p-ERK tumours (Figure 4B, lower panel) and the mutant Kras allele often duplicated (Supplemental Figure 9). Overall, across all tumour samples Kras allelic imbalance, a known mechanism by which Ras signal strength is elevated 27, correlated tightly with high p-ERK.
Long-lived organisms must solve the problem of suppressing cancer without compromising the facility of normal cells to proliferate. This requires an accurate means of distinguishing between normal and oncogenic signals. However, emerging evidence hints at a “flaw” in how our tumour suppressor pathways have evolved – rather than responding to the aberrant signal persistence that is actually responsible for oncogenesis, mammalian intrinsic tumour suppressor pathways have instead evolved to respond to the unusual elevation in signal intensity that often (but not invariably) accompanies oncogenic activation 11. Paradoxically, therefore, low-level oncogenic activities may be more efficient at initiating tumourigenesis than high-level oncogenic signals because they “fall beneath the radar” of tumour surveillance 28: high-level oncogenic signals, which appear necessary to drive progression to malignancy, are tolerable only once p53 function has been quelled.
At first glance, our data showing limited therapeutic impact of restoring p53 in established lung tumours appear at odds with previous studies 7–9. However, such studies utilized advanced, relatively homogenous tumours driven by high levels of oncogenic signalling that had already engaged the ARF pathway – hence the dramatic impact of re-instating p53. By contrast, the spontaneously evolving lung tumours that afflict KR mice are initiated by sporadic oncogenic activation of endogenous Kras at a level insufficient to engage p53. Our data suggest that it is only relatively late in their evolution, at the point when sporadic elevation of Ras signalling precipitates tumours into aggressive, high-grade lesions, that the p53 pathway is triggered. Such considerations offer a compelling rationale for the long-baffling observation that selection for p53 pathway inactivation arises relatively late in the evolution of many solid human tumours.
The inability of low-level oncogenic signalling to engage p53 also casts a cautionary shadow over the potential efficacy of p53 restoration in treating cancer. Established tumours are typically comprised of heterogeneous clades of neoplastic clones that encompass all phases of oncogenic evolution. Although p53 restoration might cull the most malignant cells, less aggressive tumour cells driven by low-level oncogenic signals would presumably survive to evolve another day. At best, then, p53 restoration as a single therapy would be a means of temporary tumour containment rather than eradication.
METHODS SUMMARY
Tumour induction and treatment
Animals were maintained under UCSF IACUC-approved protocols. KR 5 and p53KI mice 14 progeny were infected with Adenovirus-CRE (5 × 107 pfu/mouse) by nasal inhalation at 8 weeks of age 5. p53 function was restored by intraperitoneal injection of Tamoxifen (1 mg/mouse/daily) 7,10,14. Where appropriate, mice were irradiated (4 Gy) 2 hr after Ctrl/Tam treatment using a Mark 1–68 137Cesium source (0.637 Gy/min). A minimum of 5 mice per cohort was used for each experiment.
Immunohistochemistry and immunoflourescence
Primary antibodies used were p19ARF (gift from CJ. Sherr and MF Roussel 29); p21 (BD Pharmigen #556430); Ki67 (SP6 Neomarkers); P-ERK (Cell Signaling Technologies #4376) and phospho-histone H2AX (Upstate #05-636). They were detected with HRP-/Alexa-conjugated secondary antibodies. An Apoptag™ kit (Chemicon) was used for TUNEL.
LCM, expression and copy number analysis
For CDKN1A Taqman analysis 8, LCM isolation of frozen samples 30 was followed by RNA preparation (Arcturus PicoPure RNA Isolation kit, Arcturus Engineering) and cDNA production (iScript cDNA Synthesis kit, Bio-Rad). For copy number analysis, LCM (Zeiss P.A.L.M) collection of paraffin samples was followed by DNA isolation (QIAamp® DNA Micro Kit #56304) and Taqman (probes: β-Actin: Mm00607939_s1; Kras: Mm03053281_s1, Applied Biosystems) or PCR (primers: KrasHind3_F GCCATTAGCTGCTACAAAACAGTA and KrasHind3_R CCTCTATCGTAGGGTCGTACTCAT). Following PCR the KrasG12D and Kraswt alleles were distinguished by the presence of a KrasG12D-specific HindIII site in the amplified fragment (WT = 400 bp; KrasG12D = 300 +100 bp).
MicroCT X-Ray Tomography
Pre- (day 0) and post-therapy (day 7) MicroCT data was acquired using a FLEX™ X-O™ system (Gamma Medica-Ideas, Northridge, CA). Only clearly discrete tumours were measured.
Immunoblot Analysis
Whole-cell lysates from dissected tumour halves were immunoblotted with anti-Spry 2 (Abcam ab50317), Dusp6 (Santa Cruz sc-28902) or β-actin (Sigma A5441) antibodies.
METHODS
Mice, adenoviral infection and treatments
Animals were maintained in SPF conditions under UCSF IACUC-approved protocols. KP 5 and p53KI mice 14 were crossed and KP and KP;p53KI/KI animals were infected by nasal inhalation with Adenovirus-CRE (5 × 107 pfu/mouse) at 8 weeks of age, as described 5. p53 function was restored by treating mice with Tamoxifen (1 mg/mouse/daily) delivered by intraperitonial injection, as described 7,10,14. Where appropriate, mice were irradiated (4 Gy) 2 hr after Ctrl/Tam treatment using a Mark 1–68 137Cesium source (0.637 Gy/min). A minimum of 5 mice per cohort were used for each experiment.
Immnunohistochemistry and immunoflourescence
IHC stainings were performed on z-fix fixed, 5 µm paraffin embedded tissue sections. Sections were incubated overnight at 4°C with the following primary antibodies: p19ARF (gift from CJ. Sherr and MF Roussel 29); p21 (BD Pharmigen #556430, San Jose, CA) ; Ki67 (SP6, Neomarkers: Fremont, CA); P-ERK (Cell Signaling Technologies #4376, Danvers, MA), phospho-Histone H2AX (Upstate #05-636, Billerica, MA). Antibodies were detected using Vectastain ABC™ detection (Vector Laboratories, Burlingame, CA) or with specific biotinylated secondaries (anti-Rat biotinylated, Vector Laboratories BA-4001 and anti-Rabbit biotinylated, Dako #E0432) followed with stable diamobenzidine treatment (Invitrogen, Carlsbad, CA). Alternatively, Alexa-conjugated mouse, rat or rabbit IgG antibodies were used (Molecular Probes). TUNEL staining was performed using the Apoptag™ flourescein labeled kit (Chemicon) according to the manufacturers directions.
Laser capture microdissection, expression and copy number analysis
For RNA analysis 30 µm sections from fresh frozen lung tissue were fixed, stained and laser capture microdissected, as previously described 30. Total RNA was isolated and DNase I treated using the Arcturus PicoPure RNA Isolation kit (Arcturus Engineering, Mountain View, CA). cDNA was produced utilising iScript cDNA Synthesis kit (Bio-Rad, Hercules, CA). Real time quantitative PCR (Q-PCR) was performed as previously described 8. For copy number analysis 5 µm sections were briefly de-paraffinised and laser capture microdissected using a Zeiss P.A.L.M. LCM microscope. Genomic DNA was isolated using the QIAamp® DNA Micro Kit #56304 and analysed by Taqman or PCR. Copy number Taqman analysis was carried out using the following probes from Applied Biosystems: β-Actin: Mm00607939_s1; Kras: Mm03053281_s1. PCR was performed using the following Kras-specific primers: KrasHind3_F GCCATTAGCTGCTACAAAACAGTA and KrasHind3_R CCTCTATCGTAGGGTCGTACTCAT. Due to the presence of a unique HindIII restriction site in the KrasG12D allele, the mutant and wt alleles can be distinguished based on their HindIII restriction-digestion profile (WT = 400 bp and KrasG12D = 300 +100 bp).
Micro-computed X-ray tomography
Computed tomography (CT) was performed using a micro CT system (FLEX™ X-O™, Gamma Medica-Ideas, Northridge, CA) with an x-ray source with 75 kVp and 0.315 mA. CT data were acquired as 512 projections over 120 seconds of continuous x-ray exposure. Pre-therapy CT data were acquired as the baseline time point and post-therapy CT performed after 7 days of sustained Tamoxifen administration. Only clearly discrete tumours were picked for volume measurements. Volumes of interest were drawn on axial slices, and the total tumour volumes were calculated planimetrically.
Immunoblot analysis
Whole-cell lysates from dissected tumour halves were prepared and immunoblotted with anti-Spry 2 (Abcam ab50317, Cambridge, MA), Dusp6 (Santa Cruz sc-28902) or β-actin (Sigma A5441, St. Louis, MO) antibodies.
Supplementary Material
ACKNOWLEDGEMENTS
We are indebted to T. Jacks for the KR mice, C. Sherr and M. Roussel for the p19ARF antibody, M. Dail and A-T. Maia for advice on Kras copy number analysis and V. Weinberg for guidance on statistical analysis. We also thank D. Tuveson and all the members of the Evan laboratory for their sage comments. This work was supported by grants NCI CA98018, NCI CA100193, AICR 09-0649, the Ellison Medical Foundation and from the Samuel R. Waxman Cancer Research Foundation (all to GIE). M.R.J. is the Enrique Cepero, PhD Fellow of the Damon Runyon Cancer Research Foundation.
Footnotes
AUTHOR CONTRIBUTIONS
C.P.M. designed this study with help from M.R.J. and G.I.E. C.P.M. and M.R.J. performed all experiments with assistance from D.G. and F.M.. C.P.M., M.R.J. and G.I.E. analyzed and interpreted the data. A.K. graded all tumours. D.M.P. and Y.S. performed the MicroCT analysis. F.R. and R.K. helped maintain the mouse colony. C.P.M. and G.I.E. wrote the paper with help from M.R.J. and all authors contributed to editing.
REFERENCES
- 1.Jemal A, et al. Cancer statistics, 2006. CA Cancer J Clin. 2006;56:106–130. doi: 10.3322/canjclin.56.2.106. [DOI] [PubMed] [Google Scholar]
- 2.Meuwissen R, Berns A. Mouse models for human lung cancer. Genes Dev. 2005;19:643–664. doi: 10.1101/gad.1284505. [DOI] [PubMed] [Google Scholar]
- 3.Serrano M, Lin A, McCurrach M, Beach D, Lowe S. Oncogenic ras provokes premature cell senescence associated with accumulation pf p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 4.Wang W, El-Deiry WS. Restoration of p53 to limit tumor growth. Curr Opin Oncol. 2008;20:90–96. doi: 10.1097/CCO.0b013e3282f31d6f. [DOI] [PubMed] [Google Scholar]
- 5.Jackson EL, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kamijo T, et al. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell. 1997;91:649–659. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- 7.Martins CP, Brown-Swigart L, Evan GI. Modeling the therapeutic efficacy of p53 restoration in tumors. Cell. 2006;127:1323–1334. doi: 10.1016/j.cell.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 8.Ventura A, et al. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661–665. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- 9.Xue W, et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445:656–660. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christophorou MA, Ringshausen I, Finch AJ, Swigart LB, Evan GI. The pathological response to DNA damage does not contribute to p53-mediated tumour suppression. Nature. 2006;443:214–217. doi: 10.1038/nature05077. [DOI] [PubMed] [Google Scholar]
- 11.Murphy DJ, et al. Distinct Thresholds Govern Myc's Biological Output In Vivo. Cancer Cell. 2008;14:447–457. doi: 10.1016/j.ccr.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sarkisian CJ, et al. Dose-dependent oncogene-induced senescence in vivo and its evasion during mammary tumorigenesis. Nat Cell Biol. 2007;9:493–505. doi: 10.1038/ncb1567. [DOI] [PubMed] [Google Scholar]
- 13.Sweet-Cordero A, et al. An oncogenic KRAS2 expression signature identified by cross-species gene-expression analysis. Nat Genet. 2005;37:48–55. doi: 10.1038/ng1490. [DOI] [PubMed] [Google Scholar]
- 14.Christophorou MA, et al. Temporal dissection of p53 function in vitro and in vivo. Nat Genet. 2005;37:718–726. doi: 10.1038/ng1572. [DOI] [PubMed] [Google Scholar]
- 15.Jackson EL, et al. The differential effects of mutant p53 alleles on advanced murine lung cancer. Cancer Res. 2005;65:10280–10288. doi: 10.1158/0008-5472.CAN-05-2193. [DOI] [PubMed] [Google Scholar]
- 16.Dulic V, et al. p53-dependent inhibition of cyclin-dependent kinase activities in human fibroblasts during radiation-induced G1 arrest. Cell. 1994;76:1013–1023. doi: 10.1016/0092-8674(94)90379-4. [DOI] [PubMed] [Google Scholar]
- 17.El Deiry WS, et al. WAF1/CIP1 is induced in p53-mediated G1 arrest and apoptosis. Cancer Res. 1994;54:1169–1174. [PubMed] [Google Scholar]
- 18.Feldser D, et al. Stage-specific sensitivity to p53 restoration in lung cancer. Nature. 2010 doi: 10.1038/nature09535. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palmero I, Pantoja C, Serrano M. p19ARF links the tumour suppressor p53 to Ras. Nature. 1998;395:125–126. doi: 10.1038/25870. [DOI] [PubMed] [Google Scholar]
- 20.Nikitin AY, et al. Classification of proliferative pulmonary lesions of the mouse: recommendations of the mouse models of human cancers consortium. Cancer Res. 2004;64:2307–2316. doi: 10.1158/0008-5472.can-03-3376. [DOI] [PubMed] [Google Scholar]
- 21.Guerra C, et al. Tumor induction by an endogenous K-ras oncogene is highly dependent on cellular context. Cancer Cell. 2003;4:111–120. doi: 10.1016/s1535-6108(03)00191-0. [DOI] [PubMed] [Google Scholar]
- 22.Tuveson DA, et al. Endogenous oncogenic K-ras(G12D) stimulates proliferation and widespread neoplastic and developmental defects. Cancer Cell. 2004;5:375–387. doi: 10.1016/s1535-6108(04)00085-6. [DOI] [PubMed] [Google Scholar]
- 23.Vicent S, et al. ERK1/2 is activated in non-small-cell lung cancer and associated with advanced tumours. Br J Cancer. 2004;90:1047–1052. doi: 10.1038/sj.bjc.6601644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shaw AT, et al. Sprouty-2 regulates oncogenic K-ras in lung development and tumorigenesis. Genes Dev. 2007;21:694–707. doi: 10.1101/gad.1526207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wagner P, et al. In situ evidence of KRAS amplification and association with increased p21 levels in Non-Small Cell Lung Carcinoma. American Journal of Clinical Pathology. 2009;132:500–505. doi: 10.1309/AJCPF10ZUNSOLIFG. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Z, et al. Dual specificity phosphatase 6 (DUSP6) is an ETS-regulated negative feedback mediator of oncogenic ERK signaling in lung cancer cells. Carcinogenesis. 2010;31:577–586. doi: 10.1093/carcin/bgq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Z. Wildtype Kras2 can inhibit lung carcinogenesis in mice et al. Nat Genet. 2001;29:25–33. doi: 10.1038/ng721. [DOI] [PubMed] [Google Scholar]
- 28.Junttila MR, Evan GI. p53, a Jack of all trades but master of none. Nat Rev Cancer. 2009;9:821–829. doi: 10.1038/nrc2728. [DOI] [PubMed] [Google Scholar]
- 29.Bertwistle D, Zindy F, Sherr CJ, Roussel MF. Monoclonal antibodies to the mouse p19(Arf) tumor suppressor protein. Hybrid Hybridomics. 2004;23:293–300. doi: 10.1089/hyb.2004.23.293. [DOI] [PubMed] [Google Scholar]
- 30.Lawlor ER, et al. Reversible Kinetic Analysis of Myc Targets In vivo Provides Novel Insights into Myc-Mediated Tumorigenesis. Cancer Res. 2006;66:4591–4601. doi: 10.1158/0008-5472.CAN-05-3826. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.