Abstract
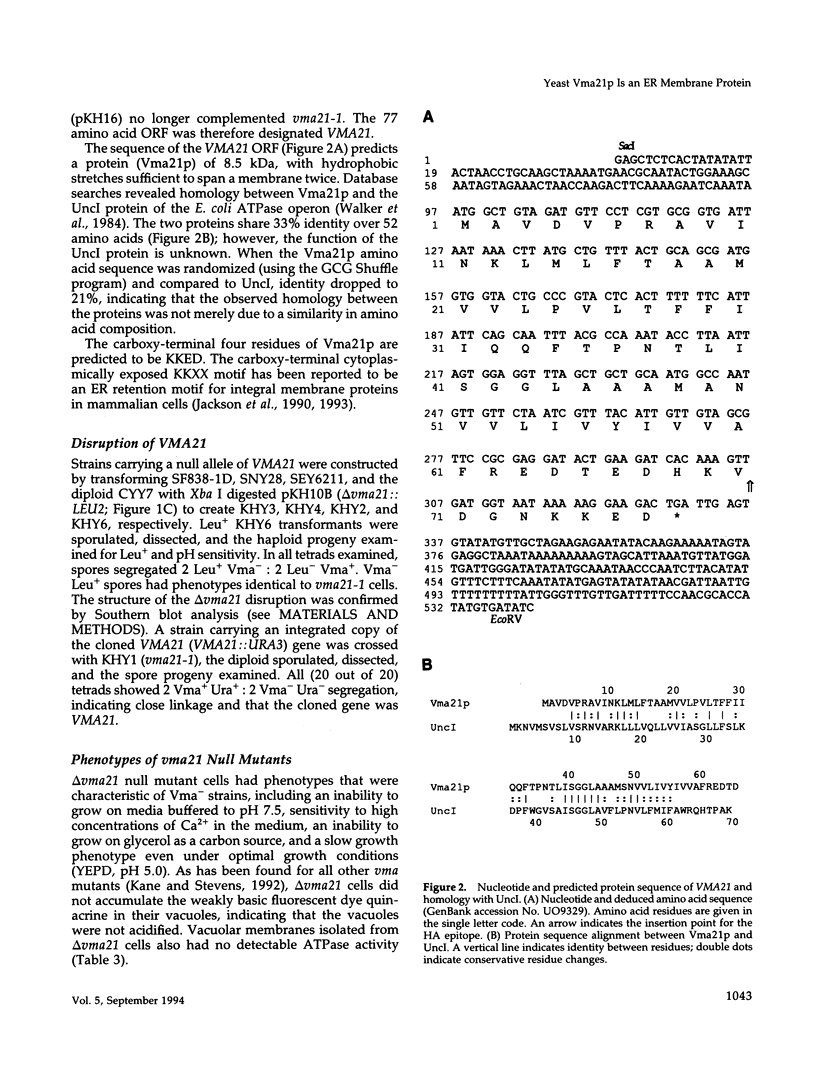
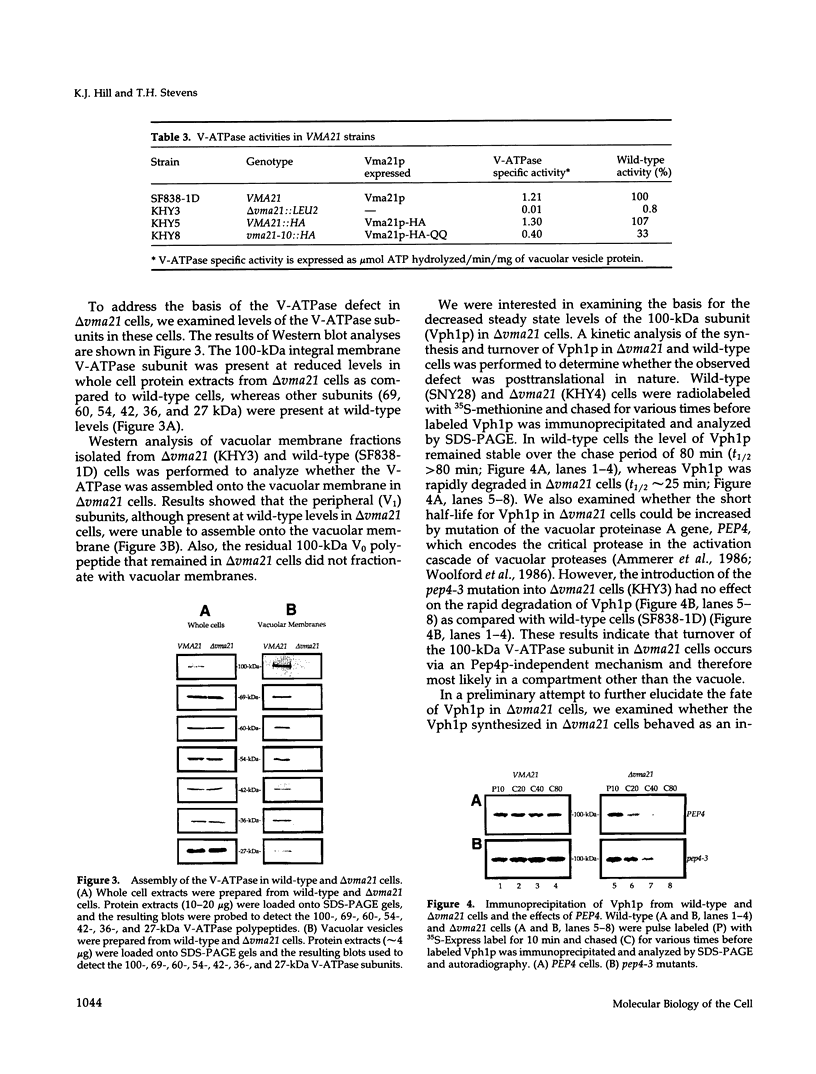
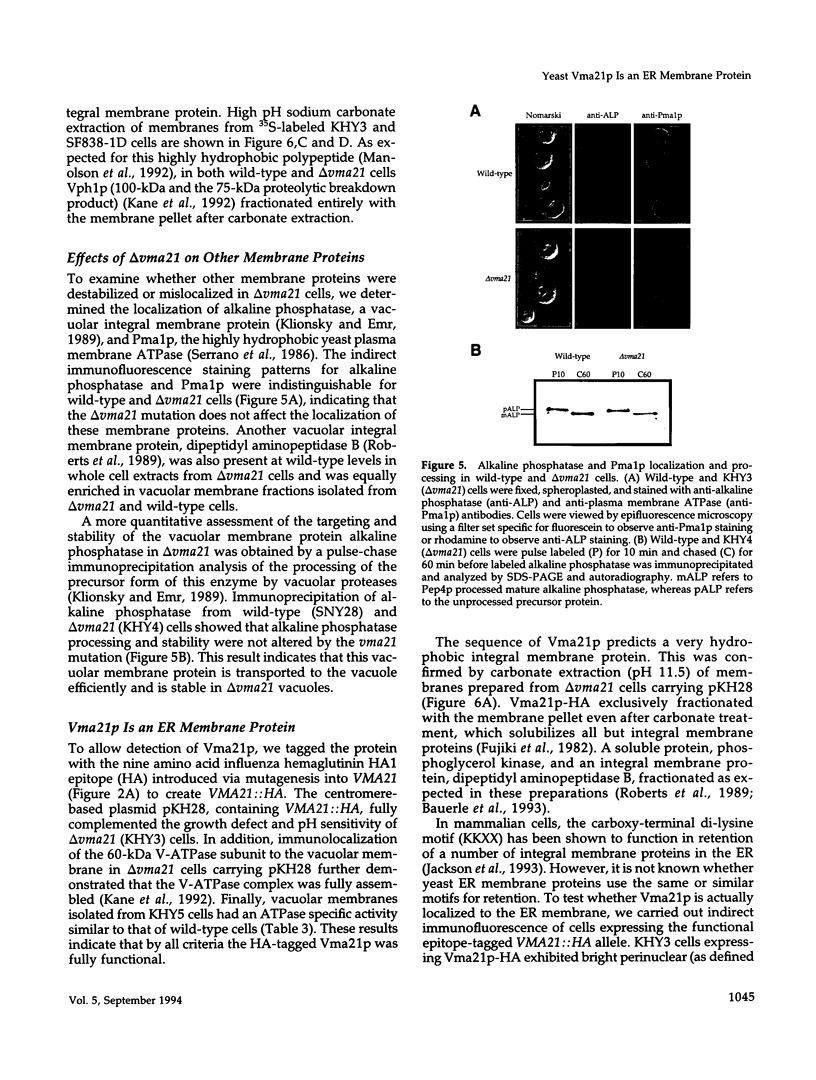
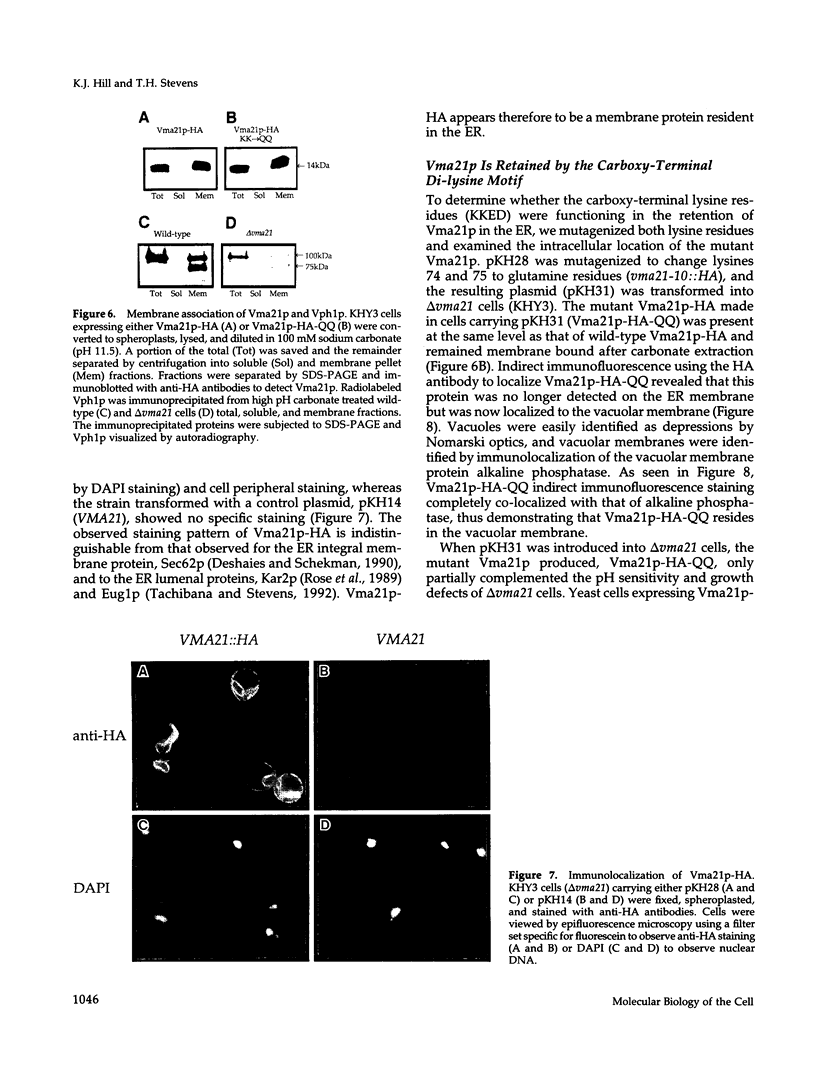
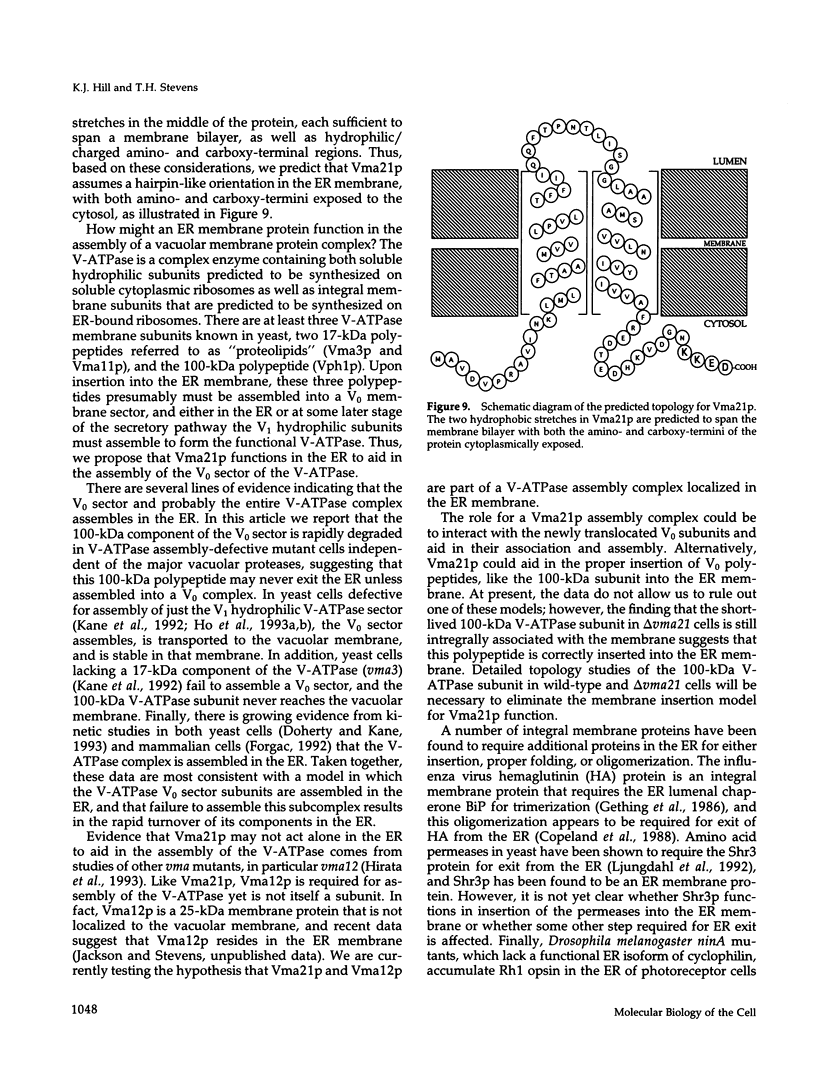
The yeast vacuolar proton-translocating ATPase (V-ATPase) is a multisubunit complex comprised of peripheral membrane subunits involved in ATP hydrolysis and integral membrane subunits involved in proton pumping. The yeast vma21 mutant was isolated from a screen to identify mutants defective in V-ATPase function. vma21 mutants fail to assemble the V-ATPase complex onto the vacuolar membrane: peripheral subunits accumulate in the cytosol and the 100-kDa integral membrane subunit is rapidly degraded. The product of the VMA21 gene (Vma21p) is an 8.5-kDa integral membrane protein that is not a subunit of the purified V-ATPase complex but instead resides in the endoplasmic reticulum. Vma21p contains a dilysine motif at the carboxy terminus, and mutation of these lysine residues abolishes retention in the endoplasmic reticulum and results in delivery of Vma21p to the vacuole, the default compartment for yeast membrane proteins. Our findings suggest that Vma21p is required for assembly of the integral membrane sector of the V-ATPase in the endoplasmic reticulum and that the unassembled 100-kDa integral membrane subunit present in delta vma21 cells is rapidly degraded by nonvacuolar proteases.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Ammerer G., Hunter C. P., Rothman J. H., Saari G. C., Valls L. A., Stevens T. H. PEP4 gene of Saccharomyces cerevisiae encodes proteinase A, a vacuolar enzyme required for processing of vacuolar precursors. Mol Cell Biol. 1986 Jul;6(7):2490–2499. doi: 10.1128/mcb.6.7.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauerle C., Ho M. N., Lindorfer M. A., Stevens T. H. The Saccharomyces cerevisiae VMA6 gene encodes the 36-kDa subunit of the vacuolar H(+)-ATPase membrane sector. J Biol Chem. 1993 Jun 15;268(17):12749–12757. [PubMed] [Google Scholar]
- Beltrán C., Kopecky J., Pan Y. C., Nelson H., Nelson N. Cloning and mutational analysis of the gene encoding subunit C of yeast vacuolar H(+)-ATPase. J Biol Chem. 1992 Jan 15;267(2):774–779. [PubMed] [Google Scholar]
- Boeke J. D., LaCroute F., Fink G. R. A positive selection for mutants lacking orotidine-5'-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197(2):345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- Colley N. J., Baker E. K., Stamnes M. A., Zuker C. S. The cyclophilin homolog ninaA is required in the secretory pathway. Cell. 1991 Oct 18;67(2):255–263. doi: 10.1016/0092-8674(91)90177-z. [DOI] [PubMed] [Google Scholar]
- Cooper A. A., Chen Y. J., Lindorfer M. A., Stevens T. H. Protein splicing of the yeast TFP1 intervening protein sequence: a model for self-excision. EMBO J. 1993 Jun;12(6):2575–2583. doi: 10.1002/j.1460-2075.1993.tb05913.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper A., Bussey H. Yeast Kex1p is a Golgi-associated membrane protein: deletions in a cytoplasmic targeting domain result in mislocalization to the vacuolar membrane. J Cell Biol. 1992 Dec;119(6):1459–1468. doi: 10.1083/jcb.119.6.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland C. S., Zimmer K. P., Wagner K. R., Healey G. A., Mellman I., Helenius A. Folding, trimerization, and transport are sequential events in the biogenesis of influenza virus hemagglutinin. Cell. 1988 Apr 22;53(2):197–209. doi: 10.1016/0092-8674(88)90381-9. [DOI] [PubMed] [Google Scholar]
- Deshaies R. J., Schekman R. Structural and functional dissection of Sec62p, a membrane-bound component of the yeast endoplasmic reticulum protein import machinery. Mol Cell Biol. 1990 Nov;10(11):6024–6035. doi: 10.1128/mcb.10.11.6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty R. D., Kane P. M. Partial assembly of the yeast vacuolar H(+)-ATPase in mutants lacking one subunit of the enzyme. J Biol Chem. 1993 Aug 5;268(22):16845–16851. [PubMed] [Google Scholar]
- Forgac M. Structure and properties of the coated vesicle (H+)-ATPase. J Bioenerg Biomembr. 1992 Aug;24(4):341–350. doi: 10.1007/BF00762527. [DOI] [PubMed] [Google Scholar]
- Foury F. The 31-kDa polypeptide is an essential subunit of the vacuolar ATPase in Saccharomyces cerevisiae. J Biol Chem. 1990 Oct 25;265(30):18554–18560. [PubMed] [Google Scholar]
- Fujiki Y., Hubbard A. L., Fowler S., Lazarow P. B. Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J Cell Biol. 1982 Apr;93(1):97–102. doi: 10.1083/jcb.93.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gething M. J., McCammon K., Sambrook J. Expression of wild-type and mutant forms of influenza hemagglutinin: the role of folding in intracellular transport. Cell. 1986 Sep 12;46(6):939–950. doi: 10.1016/0092-8674(86)90076-0. [DOI] [PubMed] [Google Scholar]
- Hirata R., Ohsumk Y., Nakano A., Kawasaki H., Suzuki K., Anraku Y. Molecular structure of a gene, VMA1, encoding the catalytic subunit of H(+)-translocating adenosine triphosphatase from vacuolar membranes of Saccharomyces cerevisiae. J Biol Chem. 1990 Apr 25;265(12):6726–6733. [PubMed] [Google Scholar]
- Hirata R., Umemoto N., Ho M. N., Ohya Y., Stevens T. H., Anraku Y. VMA12 is essential for assembly of the vacuolar H(+)-ATPase subunits onto the vacuolar membrane in Saccharomyces cerevisiae. J Biol Chem. 1993 Jan 15;268(2):961–967. [PubMed] [Google Scholar]
- Ho M. N., Hill K. J., Lindorfer M. A., Stevens T. H. Isolation of vacuolar membrane H(+)-ATPase-deficient yeast mutants; the VMA5 and VMA4 genes are essential for assembly and activity of the vacuolar H(+)-ATPase. J Biol Chem. 1993 Jan 5;268(1):221–227. [PubMed] [Google Scholar]
- Ho M. N., Hirata R., Umemoto N., Ohya Y., Takatsuki A., Stevens T. H., Anraku Y. VMA13 encodes a 54-kDa vacuolar H(+)-ATPase subunit required for activity but not assembly of the enzyme complex in Saccharomyces cerevisiae. J Biol Chem. 1993 Aug 25;268(24):18286–18292. [PubMed] [Google Scholar]
- Hoffman C. S., Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57(2-3):267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- Jackson M. R., Nilsson T., Peterson P. A. Identification of a consensus motif for retention of transmembrane proteins in the endoplasmic reticulum. EMBO J. 1990 Oct;9(10):3153–3162. doi: 10.1002/j.1460-2075.1990.tb07513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson M. R., Nilsson T., Peterson P. A. Retrieval of transmembrane proteins to the endoplasmic reticulum. J Cell Biol. 1993 Apr;121(2):317–333. doi: 10.1083/jcb.121.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane P. M., Kuehn M. C., Howald-Stevenson I., Stevens T. H. Assembly and targeting of peripheral and integral membrane subunits of the yeast vacuolar H(+)-ATPase. J Biol Chem. 1992 Jan 5;267(1):447–454. [PubMed] [Google Scholar]
- Kane P. M., Stevens T. H. Subunit composition, biosynthesis, and assembly of the yeast vacuolar proton-translocating ATPase. J Bioenerg Biomembr. 1992 Aug;24(4):383–393. doi: 10.1007/BF00762531. [DOI] [PubMed] [Google Scholar]
- Kane P. M., Yamashiro C. T., Stevens T. H. Biochemical characterization of the yeast vacuolar H(+)-ATPase. J Biol Chem. 1989 Nov 15;264(32):19236–19244. [PubMed] [Google Scholar]
- Klionsky D. J., Emr S. D. Membrane protein sorting: biosynthesis, transport and processing of yeast vacuolar alkaline phosphatase. EMBO J. 1989 Aug;8(8):2241–2250. doi: 10.1002/j.1460-2075.1989.tb08348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Ljungdahl P. O., Gimeno C. J., Styles C. A., Fink G. R. SHR3: a novel component of the secretory pathway specifically required for localization of amino acid permeases in yeast. Cell. 1992 Oct 30;71(3):463–478. doi: 10.1016/0092-8674(92)90515-e. [DOI] [PubMed] [Google Scholar]
- Manolson M. F., Proteau D., Preston R. A., Stenbit A., Roberts B. T., Hoyt M. A., Preuss D., Mulholland J., Botstein D., Jones E. W. The VPH1 gene encodes a 95-kDa integral membrane polypeptide required for in vivo assembly and activity of the yeast vacuolar H(+)-ATPase. J Biol Chem. 1992 Jul 15;267(20):14294–14303. [PubMed] [Google Scholar]
- Mellman I., Fuchs R., Helenius A. Acidification of the endocytic and exocytic pathways. Annu Rev Biochem. 1986;55:663–700. doi: 10.1146/annurev.bi.55.070186.003311. [DOI] [PubMed] [Google Scholar]
- Nelson H., Mandiyan S., Nelson N. A conserved gene encoding the 57-kDa subunit of the yeast vacuolar H+-ATPase. J Biol Chem. 1989 Jan 25;264(3):1775–1778. [PubMed] [Google Scholar]
- Nelson H., Nelson N. The progenitor of ATP synthases was closely related to the current vacuolar H+-ATPase. FEBS Lett. 1989 Apr 10;247(1):147–153. doi: 10.1016/0014-5793(89)81259-1. [DOI] [PubMed] [Google Scholar]
- Raymond C. K., Howald-Stevenson I., Vater C. A., Stevens T. H. Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol Biol Cell. 1992 Dec;3(12):1389–1402. doi: 10.1091/mbc.3.12.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts C. J., Nothwehr S. F., Stevens T. H. Membrane protein sorting in the yeast secretory pathway: evidence that the vacuole may be the default compartment. J Cell Biol. 1992 Oct;119(1):69–83. doi: 10.1083/jcb.119.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts C. J., Pohlig G., Rothman J. H., Stevens T. H. Structure, biosynthesis, and localization of dipeptidyl aminopeptidase B, an integral membrane glycoprotein of the yeast vacuole. J Cell Biol. 1989 Apr;108(4):1363–1373. doi: 10.1083/jcb.108.4.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts C. J., Raymond C. K., Yamashiro C. T., Stevens T. H. Methods for studying the yeast vacuole. Methods Enzymol. 1991;194:644–661. doi: 10.1016/0076-6879(91)94047-g. [DOI] [PubMed] [Google Scholar]
- Rose M. D., Misra L. M., Vogel J. P. KAR2, a karyogamy gene, is the yeast homolog of the mammalian BiP/GRP78 gene. Cell. 1989 Jun 30;57(7):1211–1221. doi: 10.1016/0092-8674(89)90058-5. [DOI] [PubMed] [Google Scholar]
- Rose M. D., Novick P., Thomas J. H., Botstein D., Fink G. R. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60(2-3):237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano R., Kielland-Brandt M. C., Fink G. R. Yeast plasma membrane ATPase is essential for growth and has homology with (Na+ + K+), K+- and Ca2+-ATPases. Nature. 1986 Feb 20;319(6055):689–693. doi: 10.1038/319689a0. [DOI] [PubMed] [Google Scholar]
- Shih C. K., Wagner R., Feinstein S., Kanik-Ennulat C., Neff N. A dominant trifluoperazine resistance gene from Saccharomyces cerevisiae has homology with F0F1 ATP synthase and confers calcium-sensitive growth. Mol Cell Biol. 1988 Aug;8(8):3094–3103. doi: 10.1128/mcb.8.8.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989 May;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana C., Stevens T. H. The yeast EUG1 gene encodes an endoplasmic reticulum protein that is functionally related to protein disulfide isomerase. Mol Cell Biol. 1992 Oct;12(10):4601–4611. doi: 10.1128/mcb.12.10.4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida E., Ohsumi Y., Anraku Y. Purification and properties of H+-translocating, Mg2+-adenosine triphosphatase from vacuolar membranes of Saccharomyces cerevisiae. J Biol Chem. 1985 Jan 25;260(2):1090–1095. [PubMed] [Google Scholar]
- Umemoto N., Ohya Y., Anraku Y. VMA11, a novel gene that encodes a putative proteolipid, is indispensable for expression of yeast vacuolar membrane H(+)-ATPase activity. J Biol Chem. 1991 Dec 25;266(36):24526–24532. [PubMed] [Google Scholar]
- Vaitukaitis J. L. Production of antisera with small doses of immunogen: multiple intradermal injections. Methods Enzymol. 1981;73(Pt B):46–52. doi: 10.1016/0076-6879(81)73055-6. [DOI] [PubMed] [Google Scholar]
- Walker J. E., Saraste M., Gay N. J. The unc operon. Nucleotide sequence, regulation and structure of ATP-synthase. Biochim Biophys Acta. 1984 Sep 6;768(2):164–200. doi: 10.1016/0304-4173(84)90003-x. [DOI] [PubMed] [Google Scholar]
- Wilcox C. A., Redding K., Wright R., Fuller R. S. Mutation of a tyrosine localization signal in the cytosolic tail of yeast Kex2 protease disrupts Golgi retention and results in default transport to the vacuole. Mol Biol Cell. 1992 Dec;3(12):1353–1371. doi: 10.1091/mbc.3.12.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson I. A., Niman H. L., Houghten R. A., Cherenson A. R., Connolly M. L., Lerner R. A. The structure of an antigenic determinant in a protein. Cell. 1984 Jul;37(3):767–778. doi: 10.1016/0092-8674(84)90412-4. [DOI] [PubMed] [Google Scholar]
- Wolf D. H., Fink G. R. Proteinase C (carboxypeptidase Y) mutant of yeast. J Bacteriol. 1975 Sep;123(3):1150–1156. doi: 10.1128/jb.123.3.1150-1156.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolford C. A., Daniels L. B., Park F. J., Jones E. W., Van Arsdell J. N., Innis M. A. The PEP4 gene encodes an aspartyl protease implicated in the posttranslational regulation of Saccharomyces cerevisiae vacuolar hydrolases. Mol Cell Biol. 1986 Jul;6(7):2500–2510. doi: 10.1128/mcb.6.7.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashiro C. T., Kane P. M., Wolczyk D. F., Preston R. A., Stevens T. H. Role of vacuolar acidification in protein sorting and zymogen activation: a genetic analysis of the yeast vacuolar proton-translocating ATPase. Mol Cell Biol. 1990 Jul;10(7):3737–3749. doi: 10.1128/mcb.10.7.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- te Heesen S., Rauhut R., Aebersold R., Abelson J., Aebi M., Clark M. W. An essential 45 kDa yeast transmembrane protein reacts with anti-nuclear pore antibodies: purification of the protein, immunolocalization and cloning of the gene. Eur J Cell Biol. 1991 Oct;56(1):8–18. [PubMed] [Google Scholar]