Abstract
Focal adhesion (FA) formation is induced by extracellular matrix-stimulated integrin clustering and activation of receptors for diffusible factors. Leupaxin (LPXN) is a member of the paxillin family of FA proteins expressed in many cancer cell lines. We found activation of gastrin-releasing peptide receptor (GRPr) by bombesin (BN) stimulated LPXN translocation from cytoplasm to FAs. Using mutagenesis, we identified LIM3 as the primary FA targeting domain for LPXN and showed BN-induced LPXN tyrosine phosphorylation on residues 22, 62 and 72. A LIM3 point mutant of LPXN failed to target to FAs and had no BN-stimulated tyrosine phosphorylation. Conversely, a non-phosphorylatable mutant (Y22/62/72F) translocated to FAs after BN addition. Stimulation of FA formation using vinblastine also induced LPXN translocation and tyrosine phosphorylation. Therefore, dynamic LPXN tyrosine phosphorylation requires translocation to FAs. LPXN and paxillin had opposite roles in adhesion to collagen I (CNI) in MDA-MB-231 breast cancer cells. LPXN siRNA stimulated whereas paxillin siRNA inhibited cell adhesion. Knockdown of both LPXN and paxillin behaved similarly to paxillin knockdown alone, suggesting LPXN's function in adhesion might depend on paxillin. Additionally, LPXN regulated cell spreading on CNI but not on fibronectin whereas paxillin knockdown suppressed spreading on both substrates. These results demonstrate that although LPXN and paxillin's FA targeting and tyrosine phosphorylation are similar, each protein has distinct functions.
Key words: focal adhesion, tyrosine phosphorylation, bombesin, adhesion, spreading
Introduction
Proliferation, morphogenesis and motility of tumor cells and immune cells involve interactions with the local environment which is comprised of neighboring cells and the extracelluar matrix (ECM). Binding of diffusible factors (“growth factors”) with cognate receptors initiates many local (autocrine and paracrine) cell-cell interactions. In addition, cells directly respond to and interact with each other and the ECM via adhesion receptors. Signaling cascades generated in response to either diffusible factors or adhesion receptor ligands cooperate via cross-talk to regulate enzyme activities and gene expression essential for functional characteristics of tumors and the immune response.
There are several adhesion receptor families including integrins, cadherins, selectins and immunoglobulin superfamily members.1 Integrins are heterodimeric receptors composed of α and β subunits whose combinations determine their specificity for different ECM proteins.2,3 The binding of integrins to the ECM induces integrin clustering and recruitment of various intracellular proteins such as vinculin, talin, α-actinin, focal adhesion kinase (FAK) and paxillin to form macromolecular complexes.4–6 These integrin-based adhesion complexes are heterogeneous and dynamic structures that differ in morphology, composition and function. Several types of integrin-based adhesions have been described including “classic” actin stress fiber-linked focal adhesions (FAs), dot-like focal complexes (FXs) and elongated fibronectin-bound fibrillar adhesions (FBs).7,8 The assembly and turnover of these adhesion complexes is largely regulated by the Rho GTPase family and tyrosine phosphorylation of FA proteins. Rac activity induces the formation of FXs, whereas activation of Rho leads to the growth of FAs and enhances FBs formation.9–11 Recently, tyrosine phosphorylation of paxillin was implicated as a major switch to control different adhesion phenotypes.12
Besides direct ECM-mediated integrin clustering, activation of receptor tyrosine kinases and G protein-coupled receptors (GPCRs) can also stimulate FA formation.13,14 Bombesin (BN) is an example of a diffusible ligand known to stimulate membrane ruffling, stress fiber and FA formation.13,15,16 BN-like peptides signal through a family of GPCRs, including gastrin-releasing peptide receptor (GRPr).17 The agonist-occupied GRPr activates Gαq to induce phospholipase C-β-mediated hydrolysis of phosphatidylinositides that leads to Ca2+ mobilization and activation of PKC.17 Evidence also suggests that GRPr couples to Gα12/13 to regulate Rho and Rac activity.18 BN-like peptides are chemoattractants for a variety of cells such as macrophages, leukocytes and small cell lung carcinoma cells.19–21
Leupaxin (LPXN) is a member of the paxillin family of adapter proteins initially characterized as a paxillin homologue preferentially expressed in hematopoietic cells.22 Like paxillin, LPXN contains two types of protein-protein interaction domains: repeated leucine-aspartate (LD) motifs at the N-terminus, followed by LIM (Lin-11 Isl-1 Mec-3) domains at the C-terminus. LPXN has also been shown to be tyrosine phosphorylated in lymphoblastoid cells and osteoclasts.22,23 However, very little is known about the regulation of LPXN localization and phosphorylation. In contrast to paxillin, LPXN function is just beginning to be explored and little is known about the distinct roles of each paxillin family member. In osteoclasts, LPXN is involved in bone resorption and localizes to the podosomal/sealing zone complex that is analogous to FAs in other cells.23 LPXN has also been shown to stimulate PC3 prostate cancer cell migration.24 Since BN stimulates FA formation and tyrosine phosphorylation of FAK and paxillin,13,16,25 activation of GRPr by BN could a useful experimental paradigm to study the regulation of LPXN localization and tyrosine phosphorylation. Examining the functions of LPXN and paxillin in cells endogenously expressing both family members (such as MDA-MB-231 breast cancer cells) would be a way to begin to identify unique roles for each family member.
In this study, we demonstrate that activation of GRPr by BN stimulates LPXN translocation to FAs and tyrosine phosphorylation on Y22, 62 and 72. FA targeting is required and sufficient for dynamic tyrosine phosphorylation of LPXN. Like paxillin, LIM3 is the primary focal adhesion targeting domain for LPXN. Using siRNA-mediated knockdown in MDA-MB-231 cells, we found that although LPXN and paxillin share a similar mechanism to localize to FAs, they have distinct roles in cell adhesion and spreading on different ECM substrates.
Results
Cancer cell lines express various levels of paxillin family members.
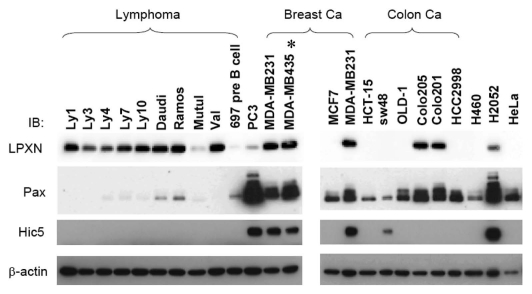
Much less is known about LPXN than the other two family members, paxillin and Hic-5. To begin to understand LPXN functions in cells and compare them with other paxillin family members, we examined LPXN expression by western blotting in a panel of different cancer cell lines. As shown in Figure 1, although LPXN was reported to be preferentially expressed in hematopoietic cells,22 in addition to lymphoma cell lines, we found LPXN expressed in cell lines derived from mesotheliomas, colon, breast and prostate cancer. LPXN expression in several solid tumor cell lines including MDA-MB-231, MDA-MB-435, Colo205 and Colo201 was comparable to that in lymphoid tumor lines, suggesting that LPXN might have an important function in many cell types.
Figure 1.
There is wide variation of LPXN, paxillin and Hic-5 expression in human tumor cell lines. 20 µg of cell lysate from different human cancer cell lines were analyzed by anti-LPXN (LPXN), anti-paxillin (Pax) or anti-Hic5 (Hic5) western blotting. Equal loading was confirmed by anti-β-actin immunoblotting. Tumor types that are not specified in the panel: PC3 (prostate carcinoma), H460 (lung carcinoma), H2052 (mesothelioma) and HeLa (cervical carcinoma). *MDA-MB-435 was shown to be of melanoma origin recently.54,55
BN stimulates LPXN translocation to focal adhesions in BALB/c 3T3 fibroblasts and PC3 cancer cells.
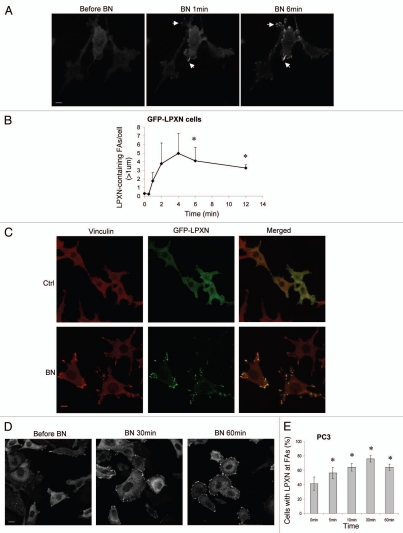
LPXN shares an overall modest sequence identity to paxillin (37%) but has much higher sequence homology in the corresponding LD motifs and LIM domains (60–80%). We sought to identify any conserved and distinct properties between LPXN and paxillin. LPXN was reported to localize to focal adhesions (FAs) in PC3 prostate cancer cells in steady state.32 However, the mechanism by which LPXN targets to FAs is not known. Bombesin (BN) stimulates robust FA formation in fibroblasts (Fig. 2C, left).16 Therefore, we utilized activation of GRPr by BN as a way of inducing FA formation in order to study the dynamic regulation of LPXN focal adhesion targeting. We generated BALB/c 3T3 fibroblasts that stably express both myc-tagged-GRPr (myc-GRPr) and GFP-tagged LPXN (GFP-LPXN cells). Activation of myc-GRPr by BN rapidly changed GFP-LPXN localization from the cytoplasm to FAs (Fig. 2A–C) confirmed by anti-vinculin immunofluorescence staining. In contrast, GFP alone did not localize to FAs, indicating that BN-stimulated LPXN focal adhesion targeting is not due to GFP (data not shown). BN stimulation of GFP-LPXN cells is an excellent model system to study the dynamic regulation of LPXN localization since activation of GRPr by BN stimulated dramatic GFP-LPXN translocation from the cytoplasm to FAs. GFP-LPXN FA translocation peaked approximately 4–5 min after BN addition (Fig. 2B) and GFP-LPXN remained at FAs throughout the time of observation (30 min; data not shown). To determine if endogenous LPXN can target to FAs dynamically, we studied LPXN localization in PC3 cells that express endogenous GRPr and LPXN.24,33 BN induced LPXN translocation to FAs in PC3 cells (Fig. 2D and E). However, the amplitude of BN-stimulated translocation was smaller and the time course slower than that in transfected BALB/c 3T3 fibroblasts. In PC3 cells, peak LPXN translocation occurred at 30–60 min.
Figure 2.
BN stimulates LPXN translocation to focal adhesions in BALB/c fibroblasts and PC3 prostate cancer cells. (A and B) BALB/c fibroblasts stably expressing GRPr and GFP-LPXN (GFP-LPXN cells) were plated in glass-bottomed dishes and imaged in real time by confocal microscopy before and after addition of BN to a final concentration of 100 nM. (A) Representative images from a movie with GFP-LPXN cells. Arrows indicate some of the LPXN-containing focal adhesions (FAs). (B) GFP-LPXN containing FAs were counted if greater than 1 µm in size and GFP-LPXN translocation was expressed as LPXN-containing FAs/cell at various times after BN addition. Results shown are the mean ± s.e.m. of four live cell movies. (C) Serum-starved GFP-LPXN cells were treated with DMEM (Ctrl) or 100 nM BN for 10 min, fixed and then stained with anti-vinculin antibody (red) to visualize FAs. Co-localization of GFP-LPXN and vinculin (yellow) was evaluated by merging the two images (merged). (D and E) PC3 cells received no treatment (before BN, 0 min) or were treated with 100 nM BN for various times as indicated. Cells were then fixed and stained with anti-LPXN antibody. Approximately 50–100 cells were scored blindly for each time point. Data shown are representative images (D) and quantification by determining percentage of cells with LPXN at FAs and is expressed as the mean ± s.e.m. of three experiments (E). *p < 0.05 compared with time 0 (before BN). Bars, 10 µm (A); 13 µm (C); 15 µm (D).
LIM3 is essential for LPXN focal adhesion targeting.
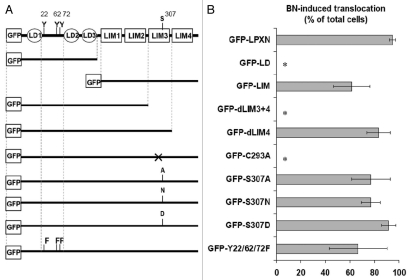
Localization of paxillin to FAs is primarily mediated through LIM3 domain.34 To determine which domain(s) of LPXN are responsible for its focal adhesion targeting, myc-GRPr expressing BALB/c 3T3 fibroblasts (myc-GRPr cells) were stably transfected with GFP-tagged deletion mutants of LPXN (Fig. 3A). We initially examined an amino-terminal construct containing only the LD domains (GFP-LD) and a carboxyl-terminal construct containing only the four LIM domains (GFP-LIM). GFP-LD failed to localize to FAs after BN treatment. In contrast, GFP-LIM translocated to FAs after addition of BN, indicating that the main focal adhesion targeting domain lies in the LIM domains of LPXN (Fig. 3A and B). Unlike full length LPXN, GFP- LIM localized to nucleus in addition to cytosol and FAs (Sup. Fig. S1), suggesting a nuclear export signal (NES) is present in the LD domains of LPXN.
Figure 3.
LIM3 is essential for LPXN focal adhesion targeting. BALB/c fibroblasts stably expressing GRPr and GFP-tagged LPXN constructs were imaged in real time by confocal microscopy before and after addition of BN. (A) Schematic illustration of GFP-tagged LPXN constructs. (B) Quantification of BN-induced translocation of LPXN constructs is expressed as “BN-induced translocation”, defined as percentage of cells demonstrating a BN-induced increase in LPXN-containing FA number or size. Results shown are the mean ± s.e.m. of at least three experiments. *p < 0.05 compared with GFP-LPXN.
To further determine which LIM domain(s) of LPXN are responsible for focal adhesion targeting, truncation mutants that delete only LIM4 or LIM3 + 4 were fused to GFP and stably transfected into myc-GRPr cells (Fig. 3A). LPXN with LIM4 deleted (GFP-dLIM4) translocated efficiently to FAs. However, further removal of LIM3 (GFP-dLIM3 + 4) completely abolished focal adhesion targeting, implicating LIM3 as the primary domain regulating LPXN focal adhesion localization (Fig. 3A and B). Finally, we generated GFP-C293A, a full length LPXN construct with a point mutation at a Cys residue in LIM3 domain predicted to be essential for LIM3 structure (Fig. 3A and B; Sup. Fig. S1).35 Vinculin staining confirmed the presence of FAs in GFP-C293A expressing cells (Sup. Fig. S2), but GFP-C293A showed no focal adhesion translocation after BN treatment, confirming that LIM3 is the principle focal adhesion targeting domain for LPXN as was reported by Tanaka et al. while this manuscript was in preparation using a LIM3 deletion mutant of mouse LPXN.36
Serine phosphorylation of the paxillin LIM domains was implicated in temporal control of its focal adhesion targeting.37 Non-phosphorylatable (LIM3S481A) and phospho-mimic (LIM3S481D) mutants of paxillin retarded and increased focal adhesion targeting respectively upon adhesion to fibronectin.37 We mutated the equivalent serine residue in LPXN LIM3 domain to determine if serine phosphorylation can regulate LPXN focal adhesion localization. Non-phosphorylatable (GFP-S307A/N) or phospho-mimic (GFP-S307D) mutants translocated after BN treatment (Fig. 3A and B; Sup. Fig. S3). In addition, the translocation time courses of these serine mutants were similar to wild type (data not shown), suggesting that phosphorylation of Ser307 in LIM3 is not important for LPXN focal adhesion localization.
BN stimulates LPXN Tyr phosphorylation on Y22, 62 and 72 in BALB/c 3T3 fibroblasts.
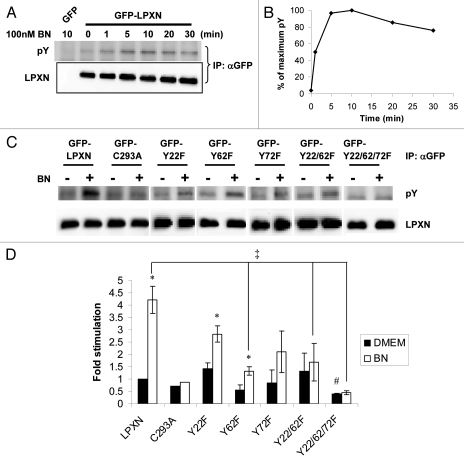
Tyrosine phosphorylation of paxillin is regulated by integrin engagement with the ECM or by different classes of growth factor receptors including GRPr.25,38,39 Tyrosine phosphorylation of paxillin is involved in regulating assembly and forms of adhesions.12 LPXN is tyrosine phosphorylated in JY8 lymphoblastoid cells and osteoclasts.22,23 However, it is not clear whether LPXN undergoes dynamic phosphorylation. To determine if BN stimulates LPXN tyrosine phosphorylation, sub-confluent GFP-LPXN cells were serum starved overnight, then treated with 100 nM BN for various times. Phosphorylation of GFP-LPXN was detected by immunoprecipitating GFP-LPXN with anti-GFP antibody, followed by anti-phosphotyrosine western blotting. BN induced rapid tyrosine phosphorylation of LPXN. BN-stimulated phosphorylation reached a maximum within 5–10 min after BN addition and sustained at or near the maximum level for up to 30 min (Fig. 4A and B).
Figure 4.
BN stimulates LPXN Tyr phosphorylation on Y22, 62 and 72 at focal adhesions in fibroblasts. (A and B) Time course of BN-induced LPXN tyrosine phosphorylation. Serum-starved BALB/c fibroblasts stably expressing GRPr and GFP-LPXN or GFP received no treatment (0 min) or were treated with 100 nM BN for various times as indicated. GFP or GFP-LPXN was immunoprecipitated by anti-GFP antibody, followed by anti-phosphotyrosine (pY) western blotting. Equal loading was confirmed by anti-LPXN western blotting. Results shown are representative blots (A) and the average quantification of two experiments (B). (C and D) Serum-starved BALB/c fibroblasts stably expressing GRPr- and GFP-tagged wide-type LPXN or LPXN point mutants were treated with DMEM (−) or 100 nM BN (+) for 10 min. Cell lysates were processed as in (A and B). For each construct, tyrosine phosphorylation was normalized to expression and quantified relative to basal GFP-LPXN tyrosine phosphorylation (defined as 1). Results shown are representative blots (C) and the mean ± s.e.m. of at least three experiments (D). *p < 0.05 for difference within each cell line between BN-stimulated and unstimulated (basal) tyrosine phosphorylation. ‡ and #p < 0.05 for difference in BN-stimulated or basal tyrosine phosphorylation between a GFP-LPXN mutant and GFP-LPXN respectively.
LPXN Y22 was identified as a phosphorylation site by mass spectrometry in pervanadate pretreated Jurkat T cells.40–42 Y72 was reported to be phosphorylated by Lyn kinase in transfected 293T cells and in A20 B cells upon B cell antigen receptor stimulation.43 There are three candidate Tyr phosphorylation sites in the LD domains: Y22, Y62 and Y72 (Fig. 3A). To identify sites of BN-stimulated tyrosine phosphorylation, we mutated one, two or all candidate phosphorylation sites in the LD domains and stably transfected them into myc-GRPr cells. As shown in Figure 4C and D, Y62 mutants (GFP-Y62F, GFP-Y22/62F and GFP-Y22/62/72F) showed reduced BN-induced tyrosine phosphorylation relative to GFP-LPXN (Fig. 4D, ‡), suggesting Y62 is a primary phosphorylation site in response to BN. However, GFP-Y62F still had significant BN-stimulated tyrosine phosphorylation above basal (Fig. 4D, *) whereas GFP-Y22/62F and GFP-Y22/62/72F did not, suggesting that Y22 was also phosphorylated after BN addition. Although in each experiment with GFP-Y72F, BN-stimulated tyrosine phosphorylation was greater than basal and lower than BN-stimulated GFP-LPXN tyrosine phosphorylation, there was large interexperiment variability. This resulted in non-significant differences in BN-stimulated tyrosine phosphorylation between GFP-Y72F and GFP-LPXN (p = 0.067) and non-significant difference between basal and BN-stimulated tyrosine phosphorylation of GFP-Y72 (p = 0.24) unless a paired analysis was performed (in which case p = 0.024). However, the triple Tyr mutant, GFP-Y22/62/72F, demonstrated a statistically lower BN-stimulated tyrosine phosphorylation compared with GFP-LPXN (Fig. 4D, ‡), and no BN-induced tyrosine phosphorylation above DMEM control (Fig. 4D). GFP-Y22/62/72F was the only mutant to have statistically lower basal phosphorylation compared with GFP-LPXN (Fig. 4D, #p = 0.00004). This suggests Y72 is also phosphorylated. Taken together, these data indicate that Y22, Y62 and Y72 were all phosphorylated after BN addition. There is a LPXN-specific band detected at baseline with the anti-phosphotyrosine antibody in all constructs, even in GFP-Y22/62/72F. To determine if this band was due to phospho-LPXN or reactivity of the antibody with non-phosphorylated LPXN, we treated immunoprecipitated samples with tyrosine phosphatase. However, we were unable to prevent GFP-LPXN from degrading after tyrosine phosphatase treatment (data not shown). Therefore, we can not exclude the possibility that there is basal Tyr phosphorylation of LPXN before BN addition on unidentified residues in the LIM domains.
Targeting of LPXN to focal adhesions is required and sufficient for its dynamic Tyr phosphorylation.
The time course of BN-induced LPXN tyrosine phosphorylation is very similar to its translocation to FAs, implying that there might be a causal relationship between these two events (compare Figs. 2B with 4B). GFP-C293A, which fails to translocate to FAs had no BN-induced tyrosine phosphorylation (Figs. 3B and 4C and D), strongly suggesting that focal adhesion targeting is required for BN-induced LPXN tyrosine phosphorylation. Conversely, GFP-Y22/62/72F (no BN-stimulated tyrosine phosphorylation, Fig. 4D) still translocated to FAs after BN treatment (Fig. 3B; Sup. Fig. S3), indicating that LPXN focal adhesion targeting does not require tyrosine phosphorylation.
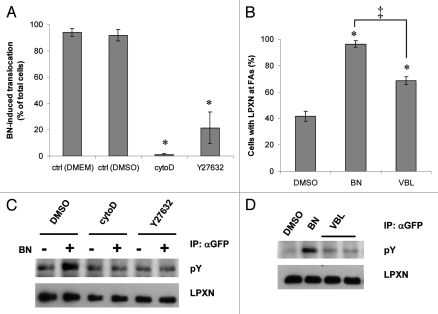
To further test the idea that focal adhesion targeting is necessary for LPXN tyrosine phosphorylation, we utilized pharmacological reagents known to regulate FA dynamics and studied their effects on LPXN focal adhesion targeting and tyrosine phosphorylation. Actin polymerization and Rho activity are required for FA formation. Cytochalasin D (cytoD), an actin polymerization inhibitor, completely eliminated LPXN translocation (Fig. 5A). Inhibition of Rho and Rho-associated kinase (ROCK) activity by exoenzyme C3 and Y27632 also prevented LPXN focal adhesion targeting (Fig. 5A and exoenzyme C3 data not shown). Cytochalasin D and Y27632 also abolished BN-induced LPXN tyrosine phosphorylation (Fig. 5C) but did not reduce the basal LPXN tyrosine phosphorylation. Microtubule disruption induces FA formation in a Rho-dependent manner.44–47 Addition of vinblastine (VBL), a microtubule disrupting agent, stimulated some LPXN translocation but less than BN (Fig. 5B, ‡). VBL also stimulated less LPXN tyrosine phosphorylation than BN (Fig. 5D). The amplitude of LPXN translocation correlated with the amount of tyrosine phosphorylation on LPXN (Fig. 5B and D). Based on these results, we conclude that LPXN translocation reflects focal adhesion formation, which could be induced by BN/GRPr signaling or VBL. Targeting of LPXN to FAs is not only required but also sufficient for its dynamic tyrosine phosphorylation in fibroblasts.
Figure 5.
Focal adhesion targeting is required and sufficient for LPXN tyrosine phosphorylation. (A) GFP-LPXN cells were incubated in vehicle control (ctrl DMEM or ctrl DMSO) or in medium containing 2 µM cytochalasin D (cytoD) or 30 µM Y27632 for 1 h or 30 min (Y27632 only). Cells were then imaged by confocal microscopy before and after addition of BN. BN-stimulated LPXN translocation was quantitated as in Figure 3B (means ± s.e.m. of at least four experiments). (B) GFP-LPXN cells were incubated in medium containing 0.06% DMSO for 1 h, 100 nM BN for 10 min or 1 µM VBL for 1 h, fixed and LPXN translocation was measured as percentage of cells with LPXN at FAs. (C) Serum-starved GFP-LPXN cells were pretreated with vehicle control (DMSO), cytochalasin D or Y27632 as in (A), then stimulated with DMEM (−) or 100 nM BN (+) for 10 min. Lysates were immunoprecipitated with anti-GFP antibody, followed by anti-phosphotyrosine (pY) or anti-LPXN western blotting. (D) Serum-starved GFP-LPXN cells were incubated in medium containing 0.06% DMSO for 1 h, 100 nM BN for 10 min or 1 µM VBL for 1 h. LPXN tyrosine phosphorylation was determined as in (C). *p < 0.05 compared with vehicle control. ‡p < 0.05 for difference in VBL and BN-induced LPXN translocation.
LPXN knockdown enhances but paxillin knockdown inhibits cell adhesion to collagen I.
Our data show that LPXN is similar to paxillin in the regulation of its FA targeting and tyrosine phosphorylation: they utilize the same targeting domain and require the same signals to be tyrosine phosphorylated.25,34 However, LPXN and paxillin may have different functions at FAs. Paxillin is involved in many processes including cell adhesion and spreading.37,48–51 Studies on paxillin functions in paxillin null or knockdown cells using different ECM substrates had conflicting results.28,49,50,52 Therefore, we hypothesized that LPXN and paxillin might be differentially involved in integrin signaling induced by different ECM proteins. In many LPXN-expressing cell lines, the other family members, paxillin and/or Hic-5, are also expressed (Fig. 1). Both LPXN and Hic-5 have been shown to suppress paxillin tyrosine phosphorylation. The counterbalance between paxillin and Hic-5 expression affect integrin-mediated cellular events like cell spreading.36,53 Therefore, it is important to determine the relative expression level of these three family members in one cell line in order to better interpret data obtained from siRNA knockdown studies. We determined the relative expression level of LPXN, paxillin and Hic-5 in PC3, MDA-MB-231 breast cancer cells and Ramos B cell lymphoma using GFP-tagged proteins as references (Sup. Fig. S4). Both PC3 and Ramos cells express vastly (more than 8-fold) different levels of paxillin and LPXN: PC3 cells express more paxillin than LPXN while Ramos cells express more LPXN than Paxillin. In contrast, the difference between LPXN and paxillin expression is much less (about 3 fold different) in MDA-MB-231 cells, making them a suitable model system to compare LPXN and paxillin functions.
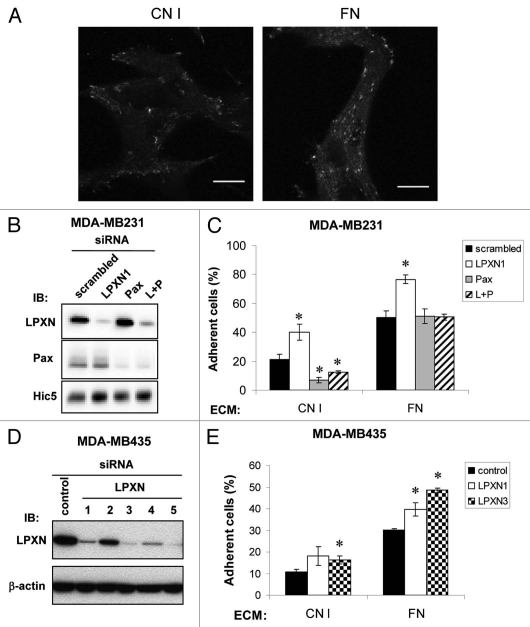
In MDA-MB-231 cells, LPXN localizes to punctate areas consistent with FAs on collagen I (CNI) and fibronectin (FN) (Fig. 6A). To evaluate the roles of LPXN and/or paxillin in adhesion and spreading on different ECM, LPXN and paxillin were knocked down individually or together in MDA-MB-231 cells. 72–96 h after siRNA transfection, LPXN and paxillin expression was suppressed by ∼95 ± 2% and ∼83 ± 5% respectively compared with control (Fig. 6B; Sup. Fig. S5). Simultaneous treatment with LPXN and paxillin siRNAs (L + P) showed a slightly less suppression of each, ∼82 ± 2% for LPXN and ∼77 ± 3% for paxillin (Sup. Fig. S5). A third family member, Hic-5, is also expressed in MDA-MB-231 cells. LPXN and paxillin siRNAs alone caused a small increase in expression of Hic-5 or LPXN respectively (Fig. 6B and Sup. Fig. S5).
Figure 6.
LPXN and paxillin have distinct roles in cell adhesion dependent on ECM. (A) MDA-MB-231 cells were plated onto collagen I (CNI) or fibronectin (FN)-coated (10 µg/ml) coverslips then fixed and stained with anti-LPXN antibody. Data shown are representative images. Bars, 10 µm. (B) siRNA-mediated knockdown of paxillin family members in MDA-MB-231 cells. MDA-MB-231 cells were transiently transfected with scrambled, LPXN, paxillin (Pax) or both LPXN and paxillin (L + P) siRNA duplexes. Expression of each protein was measured by anti-LPXN, anti-paxillin or anti-Hic5 western blotting and quantified relative to scrambled siRNA-treated cells (Sup. Fig. S5). (C) siRNA transfected MDA-MB-231 cells prepared as in (B) were collected by scraping and allowed to adhere for 1 h to 96-well plates coated with 10 µg/ml CNI or FN. BSA-coated wells were included as control. The plates were washed several times. In parallel, the wash step was omitted from at least two wells to determine total input. Cell number was measured by MTT assay. Adhesion was quantified as the absorbance of adherent cells as a percentage of the absorbance of all cells added to an equivalent well. (D) Five independent LPXN siRNAs or a control siRNA were transiently transfected into MDA-MB-435 cells and knockdown efficiency determined by anti-LPXN western blotting. Equal loading was confirmed by anti-β-actin immunoblotting. (E) Adhesion assay was performed with control, LPXN1 and 3 siRNA transfected MDA-MB-435 cells as in (C) except for adhering for 10 min. Results in (C and E) represent the mean ± s.e.m. of at least three experiments. *p < 0.05 compared with scrambled control. In MDA-MB-435 cells, p = 0.055 compared with control for LPXN1 on CN1.
We examined the effect of LPXN and/or paxillin siRNA on MDA-MB-231 adhesion on CNI or FN. On CNI, LPXN knockdown (LPXN1, open bar) enhanced adhesion but paxillin knockdown (Pax, gray bar) suppressed adhesion (Fig. 6C). Therefore, although LPXN and paxillin target to FAs in a similar fashion, they play opposite roles in adhesion to CNI. In addition, concurrent knockdown of both LPXN and Pax (L + P, hatched bar) behaved similarly to paxillin knockdown (Pax, gray bar), suggesting that the function of LPXN in cell adhesion might depend on paxillin. On FN, LPXN knockdown also enhanced adhesion, but knockdown of paxillin alone or together with LPXN had no effect. To rule out the enhancement in adhesion is an off-target effect from the LPXN siRNA used or a phenomenon unique to MDA-MB-231 cells, we employed a second LPXN-targeting siRNA and another cell line, MDA-MB-435 to examine LPXN's anti-adhesive function. MDA-MB-435 cells were considered as a breast cancer model at first but recently proven to be of melanoma origin.54,55 MDA-MB-435 cells express all 3 paxillin members similarly to MDA-MB-231 (Fig. 1). LPXN knockdown by two independent LPXN siRNAs (Fig. 6D, LPXN1 and 3) enhanced MDA-MB-435 adhesion on CNI and FN (Fig. 6E), confirming that LPXN inhibits adhesion.
LPXN knockdown suppresses cell spreading on collagen I but not on fibronectin.
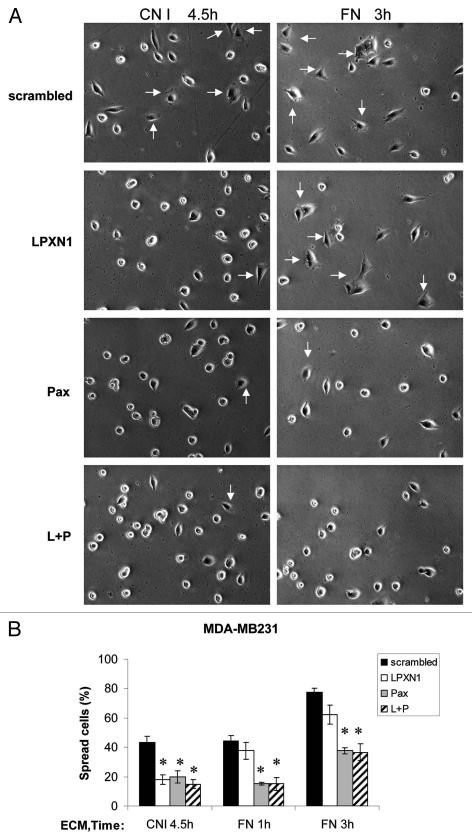
We also examined the roles of LPXN and paxillin in cell spreading. As shown in Figure 7, knockdown of paxillin alone (Pax, gray bar) or together with LPXN (L + P, hatched bar) inhibited MDA-MB-231 spreading on FN but LPXN knockdown (LPXN, open bar) was not different from control. In contrast, on CNI, LPXN knockdown reduced spreading to a similar degree as paxillin and double knockdown. Therefore, LPXN is involved in cell spreading induced by integrins for CNI but not for FN whereas paxillin regulates spreading on both substrates indistinguishably. In addition, although both LPXN and paxillin single knockdown suppressed spreading, simultaneous knockdown of both LPXN and paxillin did not show a stronger suppression, suggesting that LPXN and paxillin likely act on the same pathway to regulate cell spreading.
Figure 7.
LPXN and paxillin have different substrate dependency in regulating cell spreading. siRNA transfected MDA-MB-231 cells were collected as in cell adhesion assay and allowed to spread on 35-mm Petri dishes coated with 10 µg/ml CNI or FN for various times as indicated. Cells were scored for spreading as described under Materials and Methods. “Spread cells (%)” was defined as the number of spread cells as a percentage of the total number of cells plated. In each experiment, 100–250 cells were scored for each siRNA transfectant. Results shown were representative images of the cells (A) and the mean ± s.e.m. of three experiments (B). Arrows indicate some spread cells. *p < 0.05 compared with scrambled control.
Discussion
In this study, we demonstrated that BN-induced GRPr activation stimulated LPXN translocation from cytoplasm to FAs in fibroblasts and human tumor cells. Focal adhesion targeting was mediated primarily through LIM3 domain of LPXN. BN induced LPXN tyrosine phosphorylation on tyrosine residues 22, 62 and 72 at FAs in transfected BALB/c fibroblasts. Despite the similarity between LPXN and paxillin in their regulation of localization and tyrosine phosphorylation, they have distinct functions in cell adhesion and spreading on different ECMs.
Analysis of LPXN mutants and use of inhibitors showed that targeting to FAs is essential for LPXN tyrosine phosphorylation in response to BN. Several lines of evidence showed this principle might also apply to paxillin. Like LPXN, BN-induced paxillin tyrosine phosphorylation requires an intact actin cytoskeleton and Rho signaling.16,25,56 Using a YFP-tagged SH2 domain of pp60c-Src as a phospho-tyrosine moiety reporter, Kirchner et al. demonstrated that the recruitment of several focal adhesion proteins including paxillin to FAs preceded tyrosine phosphorylation after microtubule disruption.57 VBL, which stimulated LPXN FA translocation, also induced LPXN tyrosine phosphorylation, indicating that FA targeting is not only essential but sufficient for LPXN tyrosine phosphorylation. Our data also suggest that activation of GRPr by BN might stimulate LPXN FA targeting and tyrosine phosphorylation through Gα12/13 instead of Gαq. It has been shown that constitutively active mutants of Gα12 or Gα13 stimulate paxillin tyrosine phosphorylation, which is sensitive to cytochalasin D and Rho inhibitor (exoenzyme C3). A constitutively active mutant of Gαq also stimulates paxillin tyrosine phosphorylation but to a less extent compared with Gα12 or Gα13.58 While both Gα12 /13 and Gαq can activate Rho, GPCR-dependent activation through Gαq required agonist concentration about two orders of magnitude higher than activation through Gα12 /13.59 Moreover, although phorbol ester pretreatment inhibits GRPr-catalyzed Gαq signaling including Ins(1,4,5)P3 production and Ca2+ mobilization by ∼80%,30 it did not block LPXN translocation induced by BN (unpublished data), suggesting that BN-stimulated LPXN translocation was likely mediated by Gα12/13 rather than Gαq. Alternatively, it might also be possible that very few BN-occupied GRPr were needed to stimulate LPXN translocation.
LPXN was reported to be phosphorylated only on Y72 upon B-cell antigen receptor stimulation in A20 mouse B lymphoma cells.43 In contrast, our data showed that LPXN is phosphorylated on Y22, 62 and 72 upon GRPr activation in a fibroblast model system. The difference of LPXN tyrosine phosphorylated residues in B cells and adhesive cells might result from the expression of different upstream kinases and/or different stimuli.
FA localization of LPXN, paxillin and Hic-5 is regulated by LIM domains34,60 and they share some common interacting partners, such as Pyk2, focal adhesion kinase (FAK), the paxillin kinase linker p95PKL and PTP-PEST.23,24,60–63 In addition, we have found that many cell types express more than one family member. Our data raise questions as to the functional relationship among these three proteins. In different circumstances, they could have similar or unique functions and might perform their biological roles independently or inter-dependently. Both Hic-5 and LPXN negatively regulate paxillin tyrosine phosphorylation.36,53 Overexpression of Hic-5 inhibits cell spreading on FN in a competitive manner with paxillin, possibly by sequestering FAK from paxillin.53 In this study, we found antagonistic roles of LPXN and paxillin in adhesion by comparing the phenotypes of single and double knockdown of LPXN and paxillin cells. Our data indicate that LPXN normally inhibits cell adhesion whereas paxillin stimulates adhesion. Moreover, the anti-adhesive function of LPXN depends on paxillin since double knockdown of LPXN and paxillin inhibited adhesion to CNI to a similar degree as paxillin single knockdown, suggesting that LPXN may act upstream from paxillin. Interestingly, overexpression of LPXN inhibits adhesion of K562 erythroleukemia cells, supporting the idea of LPXN's anti-adhesive function.36 In fact, cells that predominantly express LPXN-like lymphocytes are non-adherent and highly motile cells. It has been shown that LPXN knockdown inhibits PC3 migration and adenovirus-mediate LPXN overexpression stimulates migration.24 Salgia et al. reported that the expression of paxillin is consistently low in non-adherent small cell lung cancer (SCLC) cell lines, whereas there was paxillin expression in adherent non-small cell lung cancer cells.64 A paxillin antibody-reactive band corresponding to the size of LPXN (∼46 kDa) was concluded to be the dominant form in the non-adherent SCLC cells.64 It is very likely that as in lymphoma, LPXN is the predominant member expressed in non-adherent SCLC cells. It is possible that the expression levels of LPXN and paxillin might determine the adhesion phenotype of a cell: high LPXN expression correlates with non-adherent/amoeboid-like cells and high paxillin expression associates with adherent cells.
In this study, we found the roles of LPXN and paxillin in cell adhesion and spreading are dependent on ECM proteins. LPXN and paxillin stimulate spreading of MDA-MB-231 on CNI in contrast to a report by Tanaka et al. indicating that LPXN suppresses spreading of NIH3T3 cells on FN.36 The apparent discrepancy may be explained by different substrates and cell types. Indeed, LPXN's role in cell spreading was substrate-dependent because on FN, LPXN knockdown cells spread similarly to control cells. In contrast, paxillin regulated spreading on both CNI and FN. The mechanism is unknown whereby LPXN and paxillin have effects that depend upon the specific ECM substrate. Both family members have been shown to directly bind to integrins.65,66 Therefore, specificity of LPXN for CNI signaling in cell spreading might be due to differential affinity of LPXN for individual integrins whereas paxillin might bind to CNI- and FN-binding integrins with comparable affinity. LPXN, paxillin and Hic-5 differ in their mechanism of binding to α9-integrin. Mutations that ablated α9-integrin interaction with paxillin and Hic-5 had a smaller effect on LPXN.65 Alternatively, the association between LPXN and molecules critical for spreading might be regulated differently in response to CNI versus FN. In addition to cell spreading, our results in cell adhesion also provide evidence of ECM substrate dependency of these paxillin family members. Paxillin knockdown only inhibited MDA-MB-231 adhesion to CNI but not to FN. Paradoxically, although paxillin knockdown had no effect on adhesion to FN, in the cells with both LPXN and paxillin knocked down, the enhanced adhesion caused by LPXN knockdown did not occur. We do not know the reason for this finding at this time. It is possible that this unexpected result is due to the expression of Hic-5 in MDA-MB-231 cells. Alternatively, the slightly higher expression of LPXN in cells treated with both siRNAs than cells treated with LPXN siRNA may be above a crucial threshold and allow for adequate signaling.
Finally, the molecular basis for the distinct roles of LPXN and paxillin in cell adhesion remains to be determined. Studies using virus-mediated expression of LPXN and paxillin chimeras in siRNA knockdown cells will likely be able to identify the structural determinant for their functional differences. The more variable amino-terminal regions of paxillin family members probably contain motifs for interacting with different proteins to elicit distinct biological effects. For example, Sheibani et al. recently demonstrated that paxillin has a unique binding partner and functional properties compared with Hic-5 and LPXN.67 These data support the idea that sequence differences among these family members are sufficient to mediate their functional differences. Data presented here provide the first evidence of dynamic regulation of LPXN localization and tyrosine phosphorylation by GPCRs as well as its distinct roles from paxillin in cell adhesion and spreading.
Materials and Methods
Materials.
Dulbecco's modified Eagle's medium (DMEM), fetal bovine serum (FBS), penicillin/streptomycin, G-418 and Lipofectamine 2000 reagent were from Invitrogen (Carlsbad, CA, USA). BN was from Bachem (Torrance, CA, USA). Fibronectin, 3-(4,5-Dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT), anti-Vinculin (hVIN-1) and anti-actin (AC-15) antibodies were from Sigma-Aldrich (St. Louis, MO, USA). Rat tail collagen I, anti-GFP polyclonal and anti-Hic-5 monoclonal antibodies were from BD Biosciences (Palo Alto, CA, USA). Exoenzyme C3, Y27632 and Cytochalasin D were from Calbiochem (San Diego, CA, USA). Vinblastine was a gift from Dr. Susan Horwitz (Albert Einstein College of Medicine; Bronx, NY, USA). Anti-LPXN monoclonal antibody (283c) and pCEP4-GFP-LPXN were gifts from Dr. Donald Staunton (ICOS Corporation, Bothell, WA). GFP-tagged human paxillin and Hic-5 plasmids were gifts from Dr. Ken Jacobson (University of North Carolina; Chapel Hill, NC, USA) and Dr. Lance Terada (University of Texas Southwestern and The Dallas VA Medical Center; Dallas, TX, USA) respectively. Anti-phosphotyrosine monoclonal antibody (pY100) was from Cell Signaling Technology (Danvers, MA, USA). Anti-paxillin polyclonal antibody (H-114) was from Santa Cruz Biotechnology, Inc., (Santa Cruz, CA, USA). PC3, MDA-MB-231, MDA-MB-435 and Ramos cells were obtained from American Type Culture Collection (ATCC; Manassas, VA, USA).
DNA constructs or mutagenesis.
Deletion mutants of GFP-tagged LPXN expression plasmids were generated by PCR amplification of selected regions of a human GFP-tagged LPXN template (pCEP4-GFP-LPXN), followed by introduction of the amplified fragments into pCEP4 vector. The point mutants were created either by using QuikChange site-directed mutagenesis kit from Stratagene (La Jolla, CA) or a “megaprimer” PCR method described before.26 Correct in-frame incorporation with GFP and generation of planned point mutations without any undesired mutations were confirmed by autosequencing.
Cell culture and siRNA-mediated protein knockdown.
PC3 cells were maintained at 37°C in F-12K supplemented with 50 units/ml penicillin, 50 µg/ml streptomycin and 10% FBS. MDA-MB-231, MDA-MB-435 cells and Chinese hamster ovary cells expressing polyoma LT antigen (CHOP) cells27 were maintained in DMEM containing penicillin, streptomycin and 10% FBS. Stably transfected BALB/c fibroblasts were maintained in the above-mentioned media plus 200 µg/ml Hygromycin B and 300 µg/ml G-418. siRNA duplexes were purchased from Ambion (Austin, TX, USA) and Thermo Fisher Scientific (Lafayette, CO, USA). The sequences were as follows: LPXN1, 5′-GCU UAA UAG UCU UAU AGA AUU-3′; LPXN2, 5′-AGA GUU AGA UGC CUU AUU G-3′; LPXN3, 5′-CUU CGG AGA UCC UUU CUA U-3′; LPXN4, 5′-GCG CAG CUC GUG UAU ACU A-3′; LPXN5, 5′-GGU ACA AGU UCC AUC CUG A-3′; paxillin, 5′-GUG UGG AGC CUU CUU UGG UUU-3′;28 control, siGENOME Non-Targeting siRNA #2 (Thermo Fisher Scientific) and scrambled (the same nucleotide composition as LPXN siRNA but the sequence is shuffled), 5′-AUA UAU GAU AAC UAG UCU GUU-3′. MDA-MB-231 and MDA-MB-435 cells were transfected with 3 µg siRNA and 300 nM respectively using Amaxa nucleofector system (Amaxa Inc., Gaithersburg, MD, USA) according to the manufacturer's instructions. Pilot experiments revealed that LPXN and paxillin knockdown was maximal between 72 and 96 h after transfection (data not shown). Therefore, functional experiments with siRNA transfected cells were performed 72–96 h following transfection.
Stable expression of LPXN constructs in myc-GRPr-expressing BALB/c 3T3 fibroblasts.
BALB/c 3T3 fibroblasts stably expressing myc-tagged GRPr29 were transfected with different LPXN constructs using Lipofectamine 2000 Reagent (Invitrogen, Carlsbad, CA) following the manufacturer's instructions. Double selection with Hygromycin B (200 µg/ml) and G-418 (300 µg/ml) was begun 48 h after transfection and continued for 2–3 weeks. Stable cell pools containing 3–6 single clones were screened for GFP-tagged LPXN expression by immunofluorescence microscopy. LPXN expression remained stable in a pool for 4–6 weeks (data not shown).
Immunofluorescence staining.
Stably transfected BALB/c fibroblasts or PC3 cells were plated on poly-L-lysine coated coverslips. The next day, PC3 cells were treated with 100 nM BN for various times. Transfected BALB/c cells were serum starved overnight and then treated with 100 nM BN for times indicated in the figure legends. MDA-MB-231 cells were plated on collagen I or fibronectin-coated coverslips. At the end of BN treatment or overnight adhesion onto collagen I or fibronectin-coated coverslips, cells were rinsed once with phosphate-buffered saline (PBS) and fixed with 3% paraformaldehyde. Fixed cells were incubated in 15 mM glycine for 10 min, 50 mM NH4Cl for 2 × 10 min, then permeabilized and blocked with 0.2% saponin, 0.5% BSA and 1% FBS in PBS for 20 min. An anti-vinculin monoclonal antibody (hVIN-1) was used to identify focal adhesions in transfected BALB/c cells and an anti-LPXN (283c) monoclonal antibody was used to detect endogenous LPXN in PC3 and MDA-MB-231 cells. Cells were incubated in primary antibody for 1 h at room temperature. After washing in PBS for 3 × 10 min, cells were labeled with Alexa Fluor 568 or 488 anti-mouse antibody for 1 h at room temperature, washed in PBS and mounted in Vectashield mounting medium (Vector Laboratories, Burlingame, CA).
Immunoprecipitation.
Stably transfected BALB/c fibroblasts were plated on 60-mm polystyrene tissue culture treated dishes at 500,000–600,000 cells/dish. The following day, cells were washed once with PBS and incubated in DMEM (without serum) for 18–24 h at 37°C. Cells were subsequently treated as described in the Figure legends. The media were removed and cells were lysed at 4°C with 0.65 ml of lysis buffer (50 mM HEPES, pH 7.4, 0.75%/0.075% CHAPS/CHS, 50 mM NaCl, 2 mM EDTA, 50 mM NaF, 1 mM Na3VO4, 25 mM sodium pyrophosphate, 100 µM 4-(2-aminoethyl)benzenesulfonyl fluoride, 1 µg/ml leupeptin, 1 µg/ml aprotinin and 1 µg/ml pepstatin). Lysates were clarified by centrifugation at 13,700 rpm in a microcentrifuge for 15 min at 4°C. GFP-LPXN was immunoprecipitated overnight at 4°C with 2.5 µl anti-GFP polyclonal antibody. Immune complexes were washed four times with lysis buffer and with 10 mM Tris pH 8, 140 mM NaCl for the final wash. Proteins were extracted from the complexes by boiling in Laemmli buffer for 5 min. The supernatant was analyzed by 10% SDS-polyacrylamide gel electrophoresis (PAGE) and western blotting.
Western blotting.
Protein concentration was determined with the bicinchoninic acid protein assay from Pierce Chemical (Rockford, IL, USA) using the manufacturer's instructions. Immunoprecipitated samples or total cell lysates were resolved by 10% SDS-PAGE and transferred to nitrocellulose membranes at 30 V overnight at 4°C. For anti-phosphotyrosine western blotting, membranes were blocked in 5% BSA in TBST (0.1% Tween 20 in Tris-buffered saline) and incubated overnight at 4°C in anti-phosphotyrosine antibody (pY100, 1:1,000). Membranes were washed three times with TBST, incubated in anti-mouse antibody conjugated with horseradish peroxidase diluted 1:2,000 in 5% BSA in TBST at room temperature for 1 h, and then washed three times with TBST. For other antibodies, membranes were blocked in blotto (5% non-fat milk, 0.2% NP-40, 50 mM Tris, pH 8, 2 mM CaCl2 and 80 mM NaCl) and incubated with anti-GFP (1:1,000), anti-LPXN (283c, 1 µg/ml), anti-paxillin (1:700) or anti-Hic-5 (1:300) antibody as indicated in the Figure legends. Proteins were visualized by enhanced chemiluminescence (Pierce Chemical; Rockford, IL, USA). Blots were stripped in 2% SDS and 0.1 M β-mercaptoethanol in 62.5 mM Tris-HCl, pH 6.8, at 55°C for 30 min, washed and reprobed with anti-LPXN (283c) or anti-actin antibody to ensure equal loading.
Confocal microscopy.
For live cell imaging, stably transfected BALB/c fibroblasts were plated in glass bottom 35 mm dishes (MatTek; Ashland, MA, USA) at 150,000 cells/dish and cultured overnight. The next day, cells were rinsed once with PBS and binding solution (98 mM NaCl, 59 mM KCl, 5 mM pyruvate, 6 mM fumarate, 5 mM glutamate, 11 mM glucose, 25 mM HEPES, 2.2 mM KH2PO4, 0.1% BSA, 0.02% bacitracin, 1.5 mM CaCl2 and 1 mM MgCl2 adjusted to pH 7.4) was added. Cells were then stimulated with 100 nM BN and imaged in real time on the stage of a Bio-Rad Radiance 2000 laser scanning confocal microscope preheated to 37°C. Immunofluorescence microscopy of fixed samples was carried out using a Bio-Rad Radiance 2000 laser scanning confocal microscope without temperature control. The fluorescence signal was recorded using parameters published previously.30
Cell adhesion assay.
24 h after siRNA transfection, cells were trypsinized then plated in 60-mm polystyrene tissue culture treated dishes at 1.8 × 106 cells/dish. Two days later, nearly confluent siRNA transfected cells were wash twice with PBS without Ca2+/Mg2+ and incubated in 0.54 mM EDTA in PBS for 5 min at 4°C. Cell were collected by gently scraping and resuspended in binding solution. A 96-well plate (PerkinElmer; Waltham, MA, USA) precoated with 10 µg/ml collagen I, fibronectin or BSA was blocked by incubating with 150 µl 0.2% heat-inactivated BSA in PBS for 1 h at 37°C. 52,000 cells in a volume of 100 µl were added to each well and the 96-well plates were incubated at 37°C for 1 h (MDA-MB-231) or for 10 min (MDA-MB-435). The plates were then washed three times with 100 µl 0.2% heat-inactivated BSA in PBS to remove unbound cells, followed by addition of 100 µl binding solution. In parallel, the wash step was omitted from at least two wells for use to determine total input. Cell number was measured by MTT assay with minor modifications.31 Briefly, 10 µl MTT reagent was added to each well and the plate incubated at 37°C for 4 h. 100 µl MTT lysis buffer (10% SDS, 50% dimethylformamide, pH 4.0) was then added to each well and the plate incubated at 37°C overnight. Cell number was determined by reading OD540.
Cell spreading assay.
72 h after siRNA transfection, nearly confluent MDA-MB-231 cells were collected by scraping as described under “Cell adhesion assay.” 150,000 cells were plated onto 35 mm Petri dishes precoated with 10 µg/ml collagen I or fibronectin and allowed to spread for the times indicated at 37°C. Five pictures were taken randomly from different fields of cells plated on each substrate. Cells were scored with the investigator blinded to the treatment conditions as (1) spread, defined as a non-rounded shape with extended membrane protrusions, (2) partially spread, defined as a non-rounded shape but few membrane protrusions, and (3) non-spread, defined as round and phase bright without any membrane protrusions. In each experiment, 100–250 cells were scored under each condition. Results analyzed with or without “partially spread” cells grouped with “spread” cells yielded the same conclusion and only “spread” cells results are presented.
Image analysis and statistics.
LPXN translocation was reported as “BN-induced translocation (% of total cells),” “cells with LPXN at focal adhesions (FAs) (%)” or “LPXN-containing FAs/cell” according to different experimental methods in order to best illustrate LPXN translocation for both live cells and fixed cells samples. “BN-induced translocation (% of total cells)” was defined as the percentage of cells demonstrating a BN-induced increase in LPXN-containing FA number or size relative to the total number of cells with detectable GFP signal. “Cells with LPXN at FAs (%)” was defined as the percentage of cells with LPXN localized at FAs relative to the total cell number. Five images were taken randomly from different fields and approximately 50–100 cells were scored blindly under each treatment. “LPXN-containing FAs/cell” was analyzed using ImageJ (Rasband, W.S., ImageJ, U.S. National Institutes of Health; Bethesda, MD, USA, http://rsb.info.nih.gov/ij/, 1997–2007). Images were high-pass filtered to approximate nearest neighbor deconvolution. This method works well when the features (such as focal adhesions) are within a known size range and are high contrast from the background. A Gaussian blurred image was subtracted from each frame in the live cell movies and the entire volume was linearly intensity rescaled based on the intensities within the entire volume. Particles were counted when greater than 1 µm in size. For western blotting analysis, band intensity was quantified using Kodak Digital Science Image Station 440CF with Kodak 1D 3.5 Image Analysis Software. Data were calculated using Microsoft Excel and expressed as means ± s.e.m. Difference between two treatments were analyzed by Student's t-test, with p < 0.05 considered to be significant.
Acknowledgements
The authors thank Michael Cammer (Analytical Imaging Facility, Albert Einstein College of Medicine) for invaluable assistance with image analysis. Authors also thank Drs. Paul Randazzo (Laboratory of Cellular and Molecular Biology, NCI, NIH) and Jeffrey Segall (Albert Einstein College of Medicine) for their support and help on completing this work.
Grants: This work was supported by National Institutes of Health grant K22CA098102, the Albert Einstein Cancer Center and the Intramural Research Program of the National Cancer Institute, National Institutes of Health.
Abbreviation
- FA
focal adhesion
- LPXN
leupaxin
- GRPr
gastrin-releasing peptide receptor
- BN
bombesin
- CNI
collagen I
- FN
fibronectin
- ECM
extracellular matrix
- FX
focal complex
- FB
fibrillar adhesion
- GPCR
G protein-coupled receptor
- MTT
3-(4,5-dimethyl-2-thiazolyl)-,5-diphenyl-2H-tetrazolium bromide
- VBL
vinblastine
- Tyr
tyrosine
- NES
nuclear export signal
- cytoD
cytochalasin D
- ROCK
Rho-associated kinase
- CHOP
Chinese hamster ovary cells expressing polyoma LT antigen
- CHAPS
3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate
- CHS
cholesteryl hemisuccinate tris salt
- Pax
paxillin
- SCLC
small cell lung cancer
Footnotes
Previously published online: www.landesbioscience.com/journals/celladhesion/article/12399
Supplementary Material
References
- 1.Juliano RL. Signal transduction by cell adhesion receptors and the cytoskeleton: functions of integrins, cadherins, selectins and immunoglobulin-superfamily members. Annu Rev Pharmacol Toxicol. 2002;42:283–323. doi: 10.1146/annurev.pharmtox.42.090401.151133. [DOI] [PubMed] [Google Scholar]
- 2.Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 3.Plow EF, Haas TA, Zhang L, Loftus J, Smith JW. Ligand binding to integrins. J Biol Chem. 2000;275:21785–21788. doi: 10.1074/jbc.R000003200. [DOI] [PubMed] [Google Scholar]
- 4.Burridge K, Fath K, Kelly T, Nuckolls G, Turner C. Focal adhesions: transmembrane junctions between the extracellular matrix and the cytoskeleton. Annu Rev Cell Biol. 1988;4:487–525. doi: 10.1146/annurev.cb.04.110188.002415. [DOI] [PubMed] [Google Scholar]
- 5.Jockusch BM, Bubeck P, Giehl K, Kroemker M, Moschner J, Rothkegel M, et al. The molecular architecture of focal adhesions. Annu Rev Cell Dev Biol. 1995;11:379–416. doi: 10.1146/annurev.cb.11.110195.002115. [DOI] [PubMed] [Google Scholar]
- 6.Schwartz MA, Schaller MD, Ginsberg MH. Integrins: emerging paradigms of signal transduction. Annu Rev Cell Dev Biol. 1995;11:549–599. doi: 10.1146/annurev.cb.11.110195.003001. [DOI] [PubMed] [Google Scholar]
- 7.Zamir E, Geiger B. Molecular complexity and dynamics of cell-matrix adhesions. J Cell Sci. 2001;114:3583–3590. doi: 10.1242/jcs.114.20.3583. [DOI] [PubMed] [Google Scholar]
- 8.Zamir E, Katz M, Posen Y, Erez N, Yamada KM, Katz BZ, et al. Dynamics and segregation of cell-matrix adhesions in cultured fibroblasts. Nat Cell Biol. 2000;2:191–196. doi: 10.1038/35008607. [DOI] [PubMed] [Google Scholar]
- 9.Nobes CD, Hall A. Rho, rac and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia and filopodia. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 10.Rottner K, Hall A, Small JV. Interplay between Rac and Rho in the control of substrate contact dynamics. Curr Biol. 1999;9:640–648. doi: 10.1016/s0960-9822(99)80286-3. [DOI] [PubMed] [Google Scholar]
- 11.Wierzbicka-Patynowski I, Schwarzbauer JE. The ins and outs of fibronectin matrix assembly. J Cell Sci. 2003;116:3269–3276. doi: 10.1242/jcs.00670. [DOI] [PubMed] [Google Scholar]
- 12.Zaidel-Bar R, Milo R, Kam Z, Geiger B. A paxillin tyrosine phosphorylation switch regulates the assembly and form of cell-matrix adhesions. J Cell Sci. 2007;120:137–148. doi: 10.1242/jcs.03314. [DOI] [PubMed] [Google Scholar]
- 13.Ridley AJ, Hall A. The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell. 1992;70:389–399. doi: 10.1016/0092-8674(92)90163-7. [DOI] [PubMed] [Google Scholar]
- 14.Seufferlein T, Rozengurt E. Lysophosphatidic acid stimulates tyrosine phosphorylation of focal adhesion kinase, paxillin and p130. Signaling pathways and cross-talk with platelet-derived growth factor. J Biol Chem. 1994;269:9345–9351. [PubMed] [Google Scholar]
- 15.Ridley AJ, Paterson HF, Johnston CL, Diekmann D, Hall A. The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell. 1992;70:401–410. doi: 10.1016/0092-8674(92)90164-8. [DOI] [PubMed] [Google Scholar]
- 16.Sinnett-Smith J, Lunn JA, Leopoldt D, Rozengurt E. Y-27632, an inhibitor of Rho-associated kinases, prevents tyrosine phosphorylation of focal adhesion kinase and paxillin induced by bombesin: dissociation from tyrosine phosphorylation of p130(CAS) Exp Cell Res. 2001;266:292–302. doi: 10.1006/excr.2001.5219. [DOI] [PubMed] [Google Scholar]
- 17.Kroog GS, Jensen RT, Battey JF. Mammalian bombesin receptors. Med Res Rev. 1995;15:389–417. doi: 10.1002/med.2610150502. [DOI] [PubMed] [Google Scholar]
- 18.Sinnett-Smith J, Santiskulvong C, Duque J, Rozengurt E. [D-Arg(1),D-Trp(5,7,9),Leu(11)]Substance P inhibits bombesin-induced mitogenic signal transduction mediated by both G(q) and G(12) in Swiss 3T3cells. J Biol Chem. 2000;275:30644–30652. doi: 10.1074/jbc.M003702200. [DOI] [PubMed] [Google Scholar]
- 19.De la Fuente M, Del Rio M, Ferrandez MD, Hernanz A. Modulation of phagocytic function in murine peritoneal macrophages by bombesin, gastrin-releasing peptide and neuromedin C. Immunology. 1991;73:205–211. [PMC free article] [PubMed] [Google Scholar]
- 20.Del Rio M, De la Fuente M. Chemoattractant capacity of bombesin, gastrin-releasing peptide and neuromedin C is mediated through PKC activation in murine peritoneal leukocytes. Regul Pept. 1994;49:185–193. doi: 10.1016/0167-0115(94)90140-6. [DOI] [PubMed] [Google Scholar]
- 21.Ruff M, Schiffmann E, Terranova V, Pert CB. Neuropeptides are chemoattractants for human tumor cells and monocytes: a possible mechanism for metastasis. Clin Immunol Immunopathol. 1985;37:387–396. doi: 10.1016/0090-1229(85)90108-4. [DOI] [PubMed] [Google Scholar]
- 22.Lipsky BP, Beals CR, Staunton DE. Leupaxin is a novel LIM domain protein that forms a complex with PYK2. J Biol Chem. 1998;273:11709–11713. doi: 10.1074/jbc.273.19.11709. [DOI] [PubMed] [Google Scholar]
- 23.Gupta A, Lee BS, Khadeer MA, Tang Z, Chellaiah M, Abu-Amer Y, et al. Leupaxin is a critical adaptor protein in the adhesion zone of the osteoclast. J Bone Miner Res. 2003;18:669–685. doi: 10.1359/jbmr.2003.18.4.669. [DOI] [PubMed] [Google Scholar]
- 24.Sahu SN, Nunez S, Bai G, Gupta A. Interaction of Pyk2 and PTP-PEST with leupaxin in prostate cancer cells. Am J Physiol Cell Physiol. 2007;292:2288–2296. doi: 10.1152/ajpcell.00503.2006. [DOI] [PubMed] [Google Scholar]
- 25.Zachary I, Sinnett-Smith J, Turner CE, Rozengurt E. Bombesin, vasopressin and endothelin rapidly stimulate tyrosine phosphorylation of the focal adhesion-associated protein paxillin in Swiss 3T3 cells. J Biol Chem. 1993;268:22060–22065. [PubMed] [Google Scholar]
- 26.Ke SH, Madison EL. Rapid and efficient site-directed mutagenesis by single-tube ‘megaprimer' PCR method. Nucleic Acids Res. 1997;25:3371–3372. doi: 10.1093/nar/25.16.3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heffernan M, Dennis JW. Polyoma and hamster papovavirus large T antigen-mediated replication of expression shuttle vectors in Chinese hamster ovary cells. Nucleic Acids Res. 1991;19:85–92. doi: 10.1093/nar/19.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanders MA, Basson MD. p130cas but not paxillin is essential for Caco-2 intestinal epithelial cell spreading and migration on collagen IV. J Biol Chem. 2005;280:23516–23522. doi: 10.1074/jbc.M413165200. [DOI] [PubMed] [Google Scholar]
- 29.Kroog GS, Sainz E, Worland PJ, Akeson MA, Benya RV, Jensen RT, et al. The gastrin-releasing peptide receptor is rapidly phosphorylated by a kinase other than protein kinase C after exposure to agonist. J Biol Chem. 1995;270:8217–8224. doi: 10.1074/jbc.270.14.8217. [DOI] [PubMed] [Google Scholar]
- 30.Ally RA, Ives KL, Traube E, Eltounsi I, Chen PW, Cahill PJ, et al. Agonist- and protein kinase C-induced phosphorylation have similar functional consequences for gastrin-releasing peptide receptor signaling via Gq. Mol Pharmacol. 2003;64:890–904. doi: 10.1124/mol.64.4.890. [DOI] [PubMed] [Google Scholar]
- 31.Konstantinov SM, Topashka-Ancheva M, Benner A, Berger MR. Alkylphosphocholines: Effects on human leukemic cell lines and normal bone marrow cells. Int J Cancer. 1998;77:778–786. doi: 10.1002/(sici)1097-0215(19980831)77:5<778::aid-ijc18>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 32.Kaulfuss S, Grzmil M, Hemmerlein B, Thelen P, Schweyer S, Neesen J, et al. Leupaxin, a novel coactivator of the androgen receptor, is expressed in prostate cancer and plays a role in adhesion and invasion of prostate carcinoma cells. Mol Endocrinol. 2008;22:1606–1621. doi: 10.1210/me.2006-0546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bologna M, Festuccia C, Muzi P, Biordi L, Ciomei M. Bombesin stimulates growth of human prostatic cancer cells in vitro. Cancer. 1989;63:1714–1720. doi: 10.1002/1097-0142(19900501)63:9<1714::aid-cncr2820630912>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 34.Brown MC, Perrotta JA, Turner CE. Identification of LIM3 as the principal determinant of paxillin focal adhesion localization and characterization of a novel motif on paxillin directing vinculin and focal adhesion kinase binding. J Cell Biol. 1996;135:1109–1123. doi: 10.1083/jcb.135.4.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feuerstein R, Wang X, Song D, Cooke NE, Liebhaber SA. The LIM/double zinc-finger motif functions as a protein dimerization domain. Proc Natl Acad Sci USA. 1994;91:10655–10659. doi: 10.1073/pnas.91.22.10655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tanaka T, Moriwaki K, Murata S, Miyasaka M. LIM domain-containing adaptor, leupaxin, localizes in focal adhesion and suppresses the integrin-induced tyrosine phosphorylation of paxillin. Cancer Sci. 101:363–368. doi: 10.1111/j.1349-7006.2009.01398.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brown MC, Perrotta JA, Turner CE. Serine and threonine phosphorylation of the paxillin LIM domains regulates paxillin focal adhesion localization and cell adhesion to fibronectin. Mol Biol Cell. 1998;9:1803–1816. doi: 10.1091/mbc.9.7.1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bellis SL, Perrotta JA, Curtis MS, Turner CE. Adhesion of fibroblasts to fibronectin stimulates both serine and tyrosine phosphorylation of paxillin. Biochem J. 1997;325:375–381. doi: 10.1042/bj3250375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schaller MD, Schaefer EM. Multiple stimuli induce tyrosine phosphorylation of the Crk-binding sites of paxillin. Biochem J. 2001;360:57–66. doi: 10.1042/0264-6021:3600057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brill LM, Salomon AR, Ficarro SB, Mukherji M, Stettler-Gill M, Peters EC. Robust phosphoproteomic profiling of tyrosine phosphorylation sites from human T cells using immobilized metal affinity chromatography and tandem mass spectrometry. Anal Chem. 2004;76:2763–2772. doi: 10.1021/ac035352d. [DOI] [PubMed] [Google Scholar]
- 41.Rush J, Moritz A, Lee KA, Guo A, Goss VL, Spek EJ, et al. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat Biotechnol. 2005;23:94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- 42.Tao WA, Wollscheid B, O'Brien R, Eng JK, Li XJ, Bodenmiller B, et al. Quantitative phosphoproteome analysis using a dendrimer conjugation chemistry and tandem mass spectrometry. Nat Methods. 2005;2:591–598. doi: 10.1038/nmeth776. [DOI] [PubMed] [Google Scholar]
- 43.Chew VS, Lam KP. Leupaxin negatively regulates B cell receptor signaling. J Biol Chem. 2007 doi: 10.1074/jbc.M704625200. [DOI] [PubMed] [Google Scholar]
- 44.Bershadsky A, Chausovsky A, Becker E, Lyubimova A, Geiger B. Involvement of microtubules in the control of adhesion-dependent signal transduction. Curr Biol. 1996;6:1279–1289. doi: 10.1016/s0960-9822(02)70714-8. [DOI] [PubMed] [Google Scholar]
- 45.Enomoto T. Microtubule disruption induces the formation of actin stress fibers and focal adhesions in cultured cells: possible involvement of the rho signal cascade. Cell Struct Funct. 1996;21:317–326. doi: 10.1247/csf.21.317. [DOI] [PubMed] [Google Scholar]
- 46.Liu BP, Chrzanowska-Wodnicka M, Burridge K. Microtubule depolymerization induces stress fibers, focal adhesions and DNA synthesis via the GTP-binding protein Rho. Cell Adhes Commun. 1998;5:249–255. doi: 10.3109/15419069809040295. [DOI] [PubMed] [Google Scholar]
- 47.Zhang Q, Magnusson MK, Mosher DF. Lysophosphatidic acid and microtubule-destabilizing agents stimulate fibronectin matrix assembly through Rho-dependent actin stress fiber formation and cell contraction. Mol Biol Cell. 1997;8:1415–1425. doi: 10.1091/mbc.8.8.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brown MC, Turner CE. Roles for the tubulin- and PTP-PEST-binding paxillin LIM domains in cell adhesion and motility. Int J Biochem Cell Biol. 2002;34:855–863. doi: 10.1016/s1357-2725(01)00154-6. [DOI] [PubMed] [Google Scholar]
- 49.Hagel M, George EL, Kim A, Tamimi R, Opitz SL, Turner CE, et al. The adaptor protein paxillin is essential for normal development in the mouse and is a critical transducer of fibronectin signaling. Mol Cell Biol. 2002;22:901–915. doi: 10.1128/MCB.22.3.901-915.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wade R, Bohl J, Vande Pol S. Paxillin null embryonic stem cells are impaired in cell spreading and tyrosine phosphorylation of focal adhesion kinase. Oncogene. 2002;21:96–107. doi: 10.1038/sj.onc.1205013. [DOI] [PubMed] [Google Scholar]
- 51.Woods AJ, Kantidakis T, Sabe H, Critchley DR, Norman JC. Interaction of paxillin with poly(A)-binding protein 1 and its role in focal adhesion turnover and cell migration. Mol Cell Biol. 2005;25:3763–3773. doi: 10.1128/MCB.25.9.3763-3773.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yano H, Mazaki Y, Kurokawa K, Hanks SK, Matsuda M, Sabe H. Roles played by a subset of integrin signaling molecules in cadherin-based cell-cell adhesion. J Cell Biol. 2004;166:283–295. doi: 10.1083/jcb.200312013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nishiya N, Tachibana K, Shibanuma M, Mashimo JI, Nose K. Hic-5-reduced cell spreading on fibronectin: competitive effects between paxillin and Hic-5 through interaction with focal adhesion kinase. Mol Cell Biol. 2001;21:5332–5345. doi: 10.1128/MCB.21.16.5332-5345.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rae JM, Creighton CJ, Meck JM, Haddad BR, Johnson MD. MDA-MB-435 cells are derived from M14 melanoma cells—a loss for breast cancer, but a boon for melanoma research. Breast Cancer Res Treat. 2007;104:13–19. doi: 10.1007/s10549-006-9392-8. [DOI] [PubMed] [Google Scholar]
- 55.Ross DT, Scherf U, Eisen MB, Perou CM, Rees C, Spellman P, et al. Systematic variation in gene expression patterns in human cancer cell lines. Nat Genet. 2000;24:227–235. doi: 10.1038/73432. [DOI] [PubMed] [Google Scholar]
- 56.Rankin S, Morii N, Narumiya S, Rozengurt E. Botulinum C3 exoenzyme blocks the tyrosine phosphorylation of p125FAK and paxillin induced by bombesin and endothelin. FEBS Lett. 1994;354:315–319. doi: 10.1016/0014-5793(94)01148-6. [DOI] [PubMed] [Google Scholar]
- 57.Kirchner J, Kam Z, Tzur G, Bershadsky AD, Geiger B. Live-cell monitoring of tyrosine phosphorylation in focal adhesions following microtubule disruption. J Cell Sci. 2003;116:975–986. doi: 10.1242/jcs.00284. [DOI] [PubMed] [Google Scholar]
- 58.Needham LK, Rozengurt E. Galpha12 and Galpha13 stimulate Rho-dependent tyrosine phosphorylation of focal adhesion kinase, paxillin and p130 Crk-associated substrate. J Biol Chem. 1998;273:14626–14632. doi: 10.1074/jbc.273.23.14626. [DOI] [PubMed] [Google Scholar]
- 59.Vogt S, Grosse R, Schultz G, Offermanns S. Receptor-dependent RhoA activation in G12/G13-deficient cells: genetic evidence for an involvement of Gq/G11. J Biol Chem. 2003;278:28743–28749. doi: 10.1074/jbc.M304570200. [DOI] [PubMed] [Google Scholar]
- 60.Nishiya N, Iwabuchi Y, Shibanuma M, Cote JF, Tremblay ML, Nose K. Hic-5, a paxillin homologue, binds to the protein-tyrosine phosphatase PEST (PTP-PEST) through its LIM 3 domain. J Biol Chem. 1999;274:9847–9853. doi: 10.1074/jbc.274.14.9847. [DOI] [PubMed] [Google Scholar]
- 61.Fujita H, Kamiguchi K, Cho D, Shibanuma M, Morimoto C, Tachibana K. Interaction of Hic-5, A senescence-related protein, with focal adhesion kinase. J Biol Chem. 1998;273:26516–26521. doi: 10.1074/jbc.273.41.26516. [DOI] [PubMed] [Google Scholar]
- 62.Matsuya M, Sasaki H, Aoto H, Mitaka T, Nagura K, Ohba T, et al. Cell adhesion kinase beta forms a complex with a new member, Hic-5, of proteins localized at focal adhesions. J Biol Chem. 1998;273:1003–1014. doi: 10.1074/jbc.273.2.1003. [DOI] [PubMed] [Google Scholar]
- 63.Brown MC, Turner CE. Paxillin: adapting to change. Physiol Rev. 2004;84:1315–1339. doi: 10.1152/physrev.00002.2004. [DOI] [PubMed] [Google Scholar]
- 64.Salgia R, Li JL, Ewaniuk DS, Wang YB, Sattler M, Chen WC, et al. Expression of the focal adhesion protein paxillin in lung cancer and its relation to cell motility. Oncogene. 1999;18:67–77. doi: 10.1038/sj.onc.1202273. [DOI] [PubMed] [Google Scholar]
- 65.Liu S, Slepak M, Ginsberg MH. Binding of paxillin to the alpha 9 integrin cytoplasmic domain inhibits cell spreading. J Biol Chem. 2001;276:37086–37092. doi: 10.1074/jbc.M105114200. [DOI] [PubMed] [Google Scholar]
- 66.Liu S, Thomas SM, Woodside DG, Rose DM, Kiosses WB, Pfaff M, et al. Binding of paxillin to alpha4 integrins modifies integrin-dependent biological responses. Nature. 1999;402:676–681. doi: 10.1038/45264. [DOI] [PubMed] [Google Scholar]
- 67.Sheibani N, Tang Y, Sorenson CM. Paxillin's LD4 motif interacts with bcl-2. J Cell Physiol. 2008;214:655–661. doi: 10.1002/jcp.21256. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.