Abstract
Currently 1.1 million individuals in the United States of America are living with HIV-1 infection. While this is a relatively small proportion of the global pandemic, the remarkable mix of ancestries in the U.S.A, drawn together over the past two centuries of continuous population migrations, provides an important and unique perspective on adaptive interactions between HIV-1 and human genetic diversity. HIV-1 is a rapidly adaptable organism and mutates within or near immune epitopes which are determined by the human leukocyte antigen (HLA) class I genotype of the infected host. We characterized HLA-associated polymorphisms across the full HIV-1 proteome in a large, ethnically diverse, national U.S cohort of HIV-1 infected individuals. We found a striking divergence in the immunoselection patterns associated with HLA variants which have very similar or identical peptide binding specificities but are differentially distributed among racial/ethnic groups. Though their similarity in peptide binding functionally clusters these HLA variants into supertypes, their differences at sites within the peptide binding groove contributes to ‘race-specific’ selection effects on circulating HIV-1 viruses. This suggests that the interactions between the HLA/HIV peptide complex and the T cell receptor (TCR) varies significantly within HLA supertype groups, which in turn, influences HIV-1 evolution.
Introduction
Whole genome association studies have shown that the most significant genetic determinants of HIV disease outcome are within the major histocompatibility complex (MHC), and more specifically the highly variable human leukocyte antigen (HLA) class I loci (1,2). This builds upon many studies showing associations between HLA class I genotypes and HIV disease progression (3). The HLA class I allele repertoire of an individual determines which peptide epitopes derived from a pathogen may be presented to antigen-specific CD8 T cells. The bulk of genetic variation among more than 2000 known HLA class I subtypes worldwide is within exons 2 and 3, coding for the peptide-binding region of the mature HLA protein (4). Polymorphism in this region provides for the broadest defense against a variety of different pathogens and variability within a pathogen species. There was allelic divergence at MHC class I loci during primate evolution, and the combination of early human colonization history and diverse selection pressures of prevalent microbes in different regions of the world have led to the global diversity of HLA alleles today (5,6). Though the complex interactions between founder effects, pathogen-driven selection and population admixtures are continuous, the timescale of human evolution is such that the HLA allele distribution of modern human populations still reflects in large part the early ancestry of the individuals within them (7).
In the context of this evolutionary underpinning of HLA diversity, the capacity of a relatively new human pathogen such as HIV to evade diverse HLA-restricted T cell responses is remarkable (3). HIV mutations that abrogate HLA-peptide binding (8) or TCR recognition (9) of infected cells or disrupt intracellular epitope processing (10-12) become positively selected in individuals and then apparent as HLA allele-specific viral polymorphisms in a population (13-18). Linked co-variations in the HIV proteome which may compensate or pre-condition primary escape mutations (19-21) are therefore also HLA-allele specific (22). Evidence that adaptive changes in HIV accumulate in populations as a function of the frequency of the selecting HLA alleles has been published (23). The extent to which the HLA-signatures from these studies and their population dynamics can generalize more broadly to other populations depends to a significant extent on how HLA subtypes differentially distributed between different racial groups, even within allelic families, diverge or converge in their interactions with HIV peptides. There has been limited information about this to date from population-based studies, either because low resolution HLA genotyping has been used, because particular molecular splits of broad HLA types predominate in less admixed cohorts or because comparisons across cohorts may be confounded by different viral subtypes. The worldwide clade B epidemic includes immunogenetically distinct populations in Asia, the pacific region, the Caribbean and particularly Central and South America where pathogen diversity has been associated with significant HLA allelic diversification within lineages, particularly at the HLA-B locus (24). Human migrations have also led to increasing racial/ethnic diversity in western countries affected by subtype B HIV, exemplified by the U.S. epidemic (25). The extent to which the selection effects of HLA in distinct ethnic groups leads to population level divergence and subsequent clustering of viral genome or epitope diversity has not been fully elucidated. The U.S. epidemic therefore presents the unique conditions required to characterize differential HIV-1 adaptation patterns in diverse ethnic groupings in the context of one predominant viral subtype.
Conversely, the imprints of selection in HIV should inform an understanding of diversifying selective forces on both HLA peptide binding specificities and more subtle HLA gene variations affecting TCR recognition of HLA/peptide complexes. The latter may be underestimated in more homogenous populations in which a spread of related HLA variants is not present. In this study we utilize high resolution HLA genotyping, self-identified race/ethnicity information, sequences of all HIV genes, and both published and novel analytical methods to examine convergence and divergence of HLA-associated adaptation patterns in HIV. We use a large cohort drawn from 55 centers throughout the country as a ‘snapshot’ of contemporary US genetic and HIV-1 diversity and the adaptive interactions between them.
Materials and Methods
Study populations
The 555 U.S study subjects drawn from 55 participating centers across the U.S. were enrolled in AIDS Clinical Trials Group (ACTG) protocol A5142 (26) and provided DNA under protocol A5128 (27). Protocol A5128 facilitated storage and retrieval of DNA samples from A5142/A5128 participants for ACTG genetics studies. Race/ethnicity groups were pre-defined categories specified by standard ACTG protocol and individuals were self-classified into these categories at study enrollment. Selected analyses also used a comparator population of 245 individuals recruited into the Western Australian (WA) HIV Cohort Study, a population-based, observational cohort study. In both cohorts, patients provided written informed consent to these investigations and studies received approval from their respective IRBs prior to commencement.
HLA genotyping
HLA class I genotypes (at HLA-A, -B and –C loci) were determined based on locus-specific PCR amplification of exons 2–3 and were all resolved to unambiguous four-digit-level resolution using standard DNA sequence-based typing. Ambiguities were resolved following sequencing with allele-specific subtyping primers. Sequence electropherograms were analysed using Assign™ (Conexio Genomics, WA, Australia). HLA allele frequencies in the ACTG study population were compared with published data derived from the U.S. National Marrow Donor Program random sampling of >1000 anonymous unrelated subjects self classified into five major ethnic groups in the United States (7).
HIV Sequencing
Standard bulk sequencing using a nested PCR approach was used for the US cohort HIV-1 sequencing. HIV-1 RNA was isolated from pre-treatment stored plasma using lysis buffer. After reverse transcription (Superscript™ III Reverse Transcriptase; Invitrogen, Carlsberg, CA, USA), two overlapping 5′ and 3′ fragments approximately 6 kb in length and spanning the full HIV-1 genome were amplified by nested-PCR. Bi-directional sequencing on all fragments was performed using an ABI 3130XL Analyzer (Hitachi, Singapore) with iterative gap filling using alternative primers. Electropherograms were analysed and edited using Assign™ (Conexio Genomics, WA, Australia). Standard pre- and post-PCR guidelines were followed and a customised laboratory information management system tracked single PCRs with individual sample numbers including well positions, controls and other PCR details. Due to the nested-PCR approach, contamination was monitored using PCR negative controls for both first and second round PCRs. The large overlapping region between 5′ and 3′ fragments allowed an inbuilt test of sequencing precision and contamination and individual sequence fragments were analysed for phylogenetic relatedness routinely. HIV sequences were aligned against the full genome of reference sequence HXB2 as near full length genomes or partial length sequences with intervening gaps. All insertion/deletion mixtures (predominantly in env, but detected at low frequencies in all proteins) resulting in stretches of unresolvable IUPAC mixture codes were removed, causing gaps in sequence. Areas not sequenced due to repeated sequencing or amplification failures also created gaps. A table indicating the number of sequenced bases, number and proportion of mixtures, number and size of gaps per gene and per individual is provided in the supplemental data. In computation of single site HIV-HLA associations as described below, the US cohort sequences were combined with previously generated WA HIV Cohort data to generate a total sequence dataset of 800 patient-derived sequences and HLA genotypes and associations were adjusted for potential phylogenetic clustering across the two cohorts. For the construction of maximum-likelihood phylogenetic trees in this analysis (28), genome alignments considered HIV-1 genes (for gag: n=746, pol: n=781, nef: n=616 and env: n=742) or 1kb sequence blocks (covering rev: n=708, tat: n=720, vif: n=708, vpr: n=709, vpu: n=715). All gene and sequence blocks included in the analysis were required to have at least 300 nucleotides, however the median level of coverage within all analysed non-env genes was 98.2% to 100% per gene and 79.14% in env.
Computation of single site HLA-HIV polymorphism associations
Associations between HLA alleles and HIV polymorphisms were tested using methods developed to incorporate viral phylogenetic tree structure and HLA linkage disequilibria (LD) into correlations and have been used in published studies (15, 29, 30). First, standard maximum likelihood phylogenetic trees were constructed from sequences covering Gag, Pol, Nef and Env as separate proteins and 1kb sequence blocks covering Rev, Tat, Vif, Vpr and Vpu. For every viral amino acid and HLA allele combination, a likelihood ratio test was used to evaluate whether a model incorporating both viral phylogenetic structure and HLA-mediated selection pressure explained the observed data better than a model assuming neutral evolution according to the tree only. Two generative models for each residue in viral sequence were created representing the two alternative models and the likelihood of the observations was then maximized over the parameters of both models using an Expectation-Maximization algorithm. A p-value was generated using a likelihood ratio test based on those likelihoods. For every contingency table for a given amino acid-HLA allele pair, associations were defined as denoting “Attraction”: enrichment of the specified amino acid in the specified HLA group, “repulsion”: depletion of the amino acid in the non-HLA allele group, “escape”: depletion of non-amino acid in the HLA group and “reversion”: enrichment of amino acid in the non-HLA group. Thus escape/reversion associations identified the revertant/non-adapted/wildtype amino acid and repulsion/attraction associations identified the adapted amino acid for a given HLA-associated amino acid substitution. The numbers of individuals with/without each HLA allele and with/without each viral polymorphism had to exceed an actual value of three in order to limit instability due to the use of large-sample approximations and the possibility of misclassification. All comparisons were tested using HLA genotypes at both 2-digit and 4-digit resolution to maximize specificity for effects by 4 digit subtypes with different peptide binding but retain sensitivity for effects seen only at 2 digit level because of low frequency subtypes. A multivariate analysis was used to identify and eliminate HLA associations driven by positive LD between HLA alleles (31). For every viral amino acid at each codon, the HLA allele with the strongest association was added to the list of identified associations. Then, individuals expressing this allele were removed from the dataset and the analysis was repeated. This standard forward selection procedure was iterated until no HLA allele yielded an association with uncorrected p<0.05. As there were multiple comparisons made in the analyses, we utilized q-values which estimate the false-discovery rate among identified associations compared with a randomly permuted dataset (32). Only associations with q≤0.2 indicating a 20% false-discovery rate are presented in the paper.
Correlations between HLA matching and viral sequence similarity
Correlations were tested between pair-wise viral sequence dissimilarity and the matching of HLA genotype in individuals carrying those sequences as a more global analysis of the influence of HLA on viral sequence divergence. Analyses were restricted to cases that had at least 90% sequence coverage within the protein window utilized, and to windows for which at least 80 cases satisfied this restriction. Dissimilarity of HLA genotypes was measured by the number of non-matching alleles, counting homozygote alleles as two alleles. Viral sequence dissimilarity was measured by pairwise differences in amino acid using the binary Manhattan metric (which adjusts automatically for missing values). Correlations were tested over full length Gag and then localized correlations were tested over sliding intervals of 10 amino acids across Gag representing an ‘epitope-length’ window. All analyses were carried out using SPlus 8.0 for HLA-A, -B, -C loci combined and separately. As the pair-wise dissimilarity scores were not independent, significance was assessed by randomization tests in which HLA genotypes and viral sequences were permuted and the standard R2 compared with the randomization distribution. 500 permutations were used to estimate p-values, thus truncating p-values at 1/500.
Results
The study population comprised 222 white non-Hispanics (40%), 211 black non-Hispanics (38%), 108 Hispanics (regardless of race) (19.5%), 11 Asians/Pacific Islander (2%), and 3 others (1 Alaskan Native American, 1 identified as of more than one race; and 1 ‘other/unknown’). These proportions were comparable with those reported in the U.S HIV prevalence estimates- white (34.6%), black (46.1%), Hispanic (17.5%) and Asian/Pacific Islander (1.4%)- made by the Centers for Disease Control based on the national HIV/AIDS Reporting System up to 2006 (25). Other epidemiological and clinical data are provided in Supplementary Table 1.
HLA Class I genetic diversity
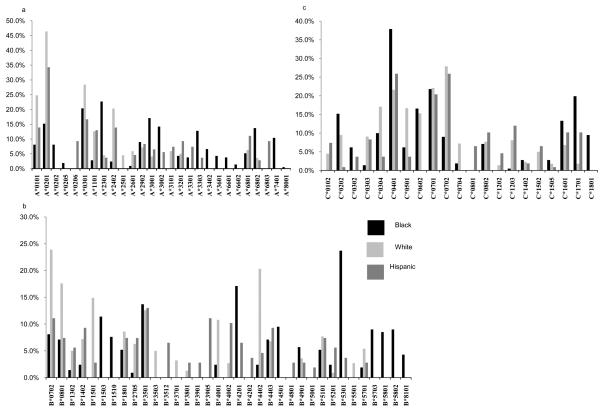
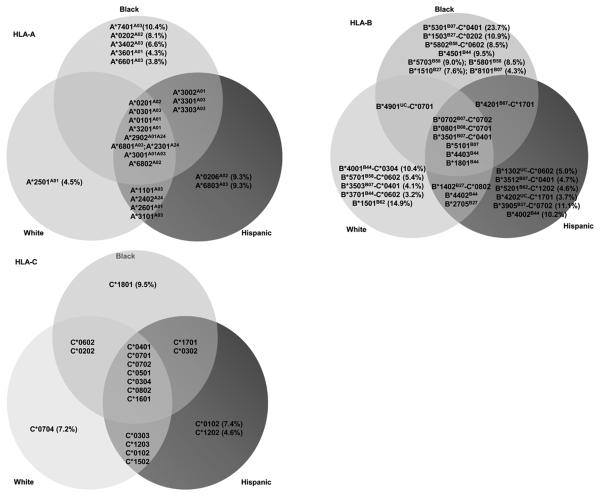
HLA diversity within the study population varied substantially according to race/ethnicity (Figures 1 and 2). Of HLA alleles carried by ≥3% of individuals within at least one ethnic group, 20 HLA-A and 31 HLA-B alleles were identified. Of these, 9 (45%) HLA-A and 6 (18.7%) HLA-B alleles were common to all racial/ethnic groups. The proportion of relatively ‘race-specific’ genotypes was found to be greater within the HLA-B locus than for HLA-A. Hence, 1/13 (7.7%) HLA-A and 5/16 (31.3%) HLA-B alleles could be considered relatively specific to the white population. Similarly, 2/18 (11.1%) HLA-A and 6/16 (37.5%) HLA-B alleles were specific to Hispanics, while the black population provided the largest proportion of race-specific HLA-A alleles (5/17, 29.1%) and HLA-B alleles (8/16, 50.0%). Phylogenetic trees based on the peptide binding regions of all HLA class I alleles were constructed (Supp. Figure 1). Several groups of HLA alleles that appeared more closely associated within a lineage included highly “race-specific” alleles among them, such as HLA-B*5701 and –B*5703 which differ at only two residues in the peptide-binding domain (Supp. Figure 2) but are distributed almost exclusively in whites and blacks respectively (Figure 2). These trees reproduced many previously noted relationships, though strong support for phylogenetic relationships between HLA-A and B alleles are limited by high levels of intra-allelic recombination at these loci through human and primate evolution (33).
Figure 1.
n=555, HLA allele carriage rate in groups defined by self-identified race/ethnicity for (a) HLA-A (b) HLA-B and (c) HLA-C loci. Only HLA alleles with greater than 3% carriage frequency are included. In all plots, black, light grey and dark grey bars represent self-identified black, white and Hispanic groups respectively.
Figure 2.
Prevalence of HLA alleles in relation to specific race/ethnic groups in the U.S cohort. HLA-B-C genotype combinations in which the relevant HLA-B allele is accompanied by its haplotypic HLA-C allele at a frequency of >80% in the cohort are also shown. Supertype groups are in superscript, UC- unclassified supertype.
The gene frequencies of HLA alleles within the study subpopulations were formally compared with frequencies in the only contemporary large-scale population-based sampling of US ethnic groups available (7) (Figure 1, Supp. Table 2). After adjustment for the number of comparisons made over each HLA/ethnic group combination, there were no significant differences in frequency of any HLA-A alleles in whites or blacks, however there was enrichment of HLA-B*5703 in the black study cohort subpopulation (5.07%) compared to African Americans in the healthy reference population (0.4%, p=0.000067) (7). Among Hispanics, HLA-A*6803, -B*4201, -C*0305 and –C*1701 were increased in the HIV-infected cohort compared to background rates (p=0.004, p=0.016, p=0.042 and p=0.00027 respectively) while HLA-C*0304 was depleted (p=0.022).
HIV-1 genetic diversity
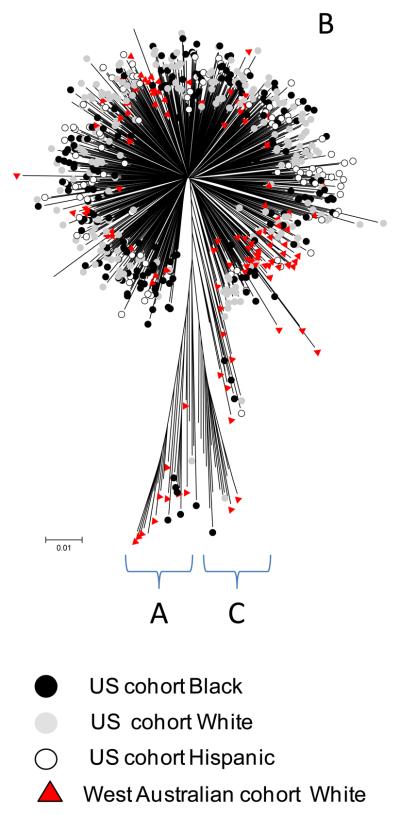
HIV-1 sequences were identified as clade B in 97% of cases based on phylogenetic analysis of all HIV genes. There were six minor clusters with bootstrap values greater than 70% detected in these analyses, all of which had less than five individuals within them with no consistent sharing of study site, city or ethnicity to suggest recent transmission clusters. When U.S gag, pol and nef sequences were compared with those from geographically distant individuals from Western Australia (n=245, 87% subtype B), sequences from Australian individuals self-classified as white inter-digitated with U.S whites, and all other subpopulations from the U.S (Figure 3, Supp Figure 3).
Figure 3.
Example of U.S and WA HIV Cohort HIV-1 sequence phylogenetic trees. HIV-1 pol shown here and gag and nef shown in Supp Figure 3. Only sequences with greater than 75% coverage were included. Neighbor-joining method using Kimura-two-parameter with pair-wise deletion. Cohort and self-identified race/ethnicity (black, white and Hispanic) associated with individual sequences and main subtype assignments are indicated. Other race/ethnicity categories in the WA HIV Cohort and ACTG cohort are not labeled. Analysis performed using MEGA v3.1.
HIV-1 adaptation to HLA
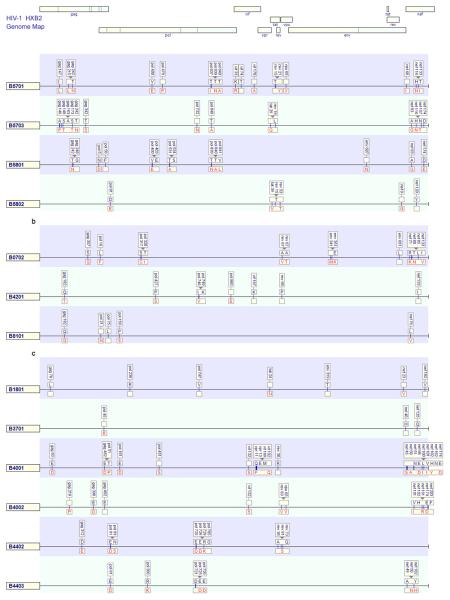
We carried out a comprehensive analysis of HLA-associated viral polymorphisms across the full HIV proteome with adjustment for potential phylogenetic relatedness between viral sequences and linkage disequilibrium between co-inherited HLA alleles. This analysis identified 874 significant associations of unique amino acid polymorphisms and HLA-A (238, 27%), -B (438, 50%) or -C (198, 23%) alleles (q values <0.2) (Fig 4 a-c, Supp. Figure 4 for all full proteome adaptation maps). In all proteins HLA-B associations were most common, except for Vpu where HLA-A dominated. The majority of associations involved substitutions between the population consensus amino acid and a co-dominant dimorphism, however more complex patterns involving minor amino acids at highly polymorphic positions (Env A319T, entropy 0.88), or HLA alleles driving to more than one alternative adaptation (e.g. HLA-A*0301 Gag K28R/Q and HLA-B*0801 Nef K94M/Q/E/N) or away from more than one susceptible amino acid at the same position (e.g. HLA-C*0602 Pol A/N709x) were also apparent. As noted in previous studies, associations frequently overlapped with either the same or opposing effects (13-18). In the latter case, adaptation for one HLA allele corresponded to reversion for other HLA alleles and vice versa. Over the whole proteome, there were 202 statistically significant associations at sites ≤4 amino acids flanking experimentally characterized and published CD8 T cell epitopes with matching HLA restriction (34), and a further 165 associations within or near putative epitopes based on the Epipred epitope prediction algorithm (35). For those associations with corresponding known or high probability putative epitopes, mutation was extra-epitopic in 132 cases (36%). In all cases matching known T cell escape mutations as classically described, Epipred indicated a low prediction score for the adapted epitope sequence relative to the non-adapted. There were other patterns of predicted T cell reactivity in relation to HLA-driven changes, including where immunoreactivity was known or predicted for the adapted epitope. The remaining HLA associations not within or flanking known epitopes may represent viral co-variation associated with a primary HLA-driven adaptation however other factors that contribute to this proportion may include the relative under-representation of known epitopes in polymorphic regions and restricted by less studied HLA alleles and the false discovery rate of 20%.
Figure 4.
Maps of unique HLA-associated adaptations (q-value <0.2) in the HIV-1 genome for selected grouped HLA alleles present in the cohort within the (a) HLA-B58 supertype (b) B7 supertype and (c) B44 supertype. The non-adapted (susceptible/revertant) amino acids are displayed above the line in blue text and adapted amino acid below the line in red text. Blanks indicate where only the non-adapted or the adapted amino acid was evident from the leaf associations in the analysis. Locations of published CD8 T cell epitopes are shown as boxes at association sites.
The first described naturally occurring variant p24 epitopes, GGKKKYKF and DCKTILKAL shown to abolish cytotoxicity of variant expressing cells corresponded to HLA-B*0801 associated polymorphisms Gag L31F and K331R respectively in this study (36). Similarly, many well characterized mutations observed to evolve and escape CD8 T cell responses experimentally in direct immunological studies of individuals were reproduced here as HLA-HIV polymorphism associations (8-12,37-47)(Supp. Figure 4). There was a partial overlap between the associations detected here with the specific associations reported by other studies with other populations (13-18). Complete concordance is not expected however given significant differences in ethnic/HLA distributions, population size and viral subtype composition. Formal concordance and stratification analyses accounting for these differences and involving more populations are underway.
Differential constraints to HLA driven adaptation in the HIV proteome
In 95.7% of identified non-adapted and adapted pairs in HLA-driven polymorphisms, the amino acids were at a minimal single nucleotide distance from each other with the reminder having two nucleotide differences. HLA-associated polymorphisms were dominated by a small group of amino acid pairs with similar physicochemical properties: positively-charged (K/R: 17.8%); negatively-charged (E/D: 14.5%); non-polar aliphatic (V/I: 5.3%, L/I: 4.6%, L/V: 3.1%), and bulky aromatic residues (Y/F: 3.8%), suggesting that protein function and structure contingent upon these properties would tend to be preserved. At a protein level, the density of HLA associations differed between proteins in the following (descending) order: Nef> Vpr> Rev> Vif> Tat> Gag> Pol> Vpu> Env and at sub-protein level: Nef> Vpr> p17> Vif> p2p7p1p6> Rev> p24> Gag/Pol TF> Vpu> Integrase> Protease> Env> Tat> RT (Supp. Figure 5). When average Shannon entropy scores (48) were plotted against HLA association density per protein, both Nef and Env had higher variability (lesser constraint) but levels of HLA selection were dissimilar (Supp. Figure 5). We detected sparse HLA class I associated selection in Env, despite the high degree of general variability. Gag and Pol had less HLA selection than Nef in the face of higher constraint. This may imply that the CD8 T cell responses selecting changes in Gag and Pol epitopes (against constraint) and to a lesser degree Nef (without constraint) are qualitatively stronger. Env, on the other hand, is subject to humoral and other selective forces, and insertions/deletion polymorphisms and other complex variability in Env may reduce the power to detect HLA class I associations. The remaining accessory proteins had higher HLA selection than Gag and Pol, as expected, but the distribution within this group did not indicate a simple direct correlation between entropy and HLA associated selection. Notably, Vpu was the most polymorphic protein but had the least HLA-associated selection after Env.
Cumulative HLA-driven adaptation per individual
We examined, in each study subject all HIV residues at which an HLA association matched their own HLA class I alleles. The residues at these positions were identified as being adapted, non-adapted or missing if within a sequence gap and counted as a proportion of all HLA-associated sites relevant to the individual. While the population level analysis pointed to differences in HLA association density across proteins, it is notable that within individuals, the average proportion of all relevant (individual-specific) HLA-associated sites that were in the adapted state was uniformly high across Vif (66%), Env (64%), Nef (58%), Vpr (57%), Gag (55%), Pol (54%) and Tat (40%) and lower in Rev (28%) and Vpu (27%).
Selection profiles of HLA alleles within supertypes and allelic lineages
HLA associated adaptation in HIV was mapped in a manner analogous to resistance maps for antiretroviral drugs such that the unique “immunoselection profile” across the whole HIV proteome is shown for every given HLA allele in the cohort (Figure 4a-c, Supp. Figure 4). The convergence or divergence in immunoselection patterns associated with different HLA alleles could then be visualized. As the selection imprints on HIV should partly reflect the binding specificity of HLA peptide binding regions, we examined HLA alleles within defined supertypes (49) and more specifically those related in phylogenetic trees based on exon 2 and 3 sequences (Supp. Figure 1). The degree of admixture in the U.S cohort meant that such alleles were comparable in population frequency so that divergence in their immunoselection would not be explained by unequal statistical power. Across all the main relevant groupings, there was some minor overlap in the epitopes targeted by grouped alleles, however overall there was a striking lack of overlap in the selection patterns within those epitopes. For example, HLA-B*5701, -B5703, -B*5801 and -B*5802 are of the B58 supertype and have marked race/ethnic group specificity (Figures 1, 2). HLA-B*5701/3 and –B5801 had clustered adaptations around well known epitopes in p24 (TW10 and IW9), Integrase (SW10), Rev (RY10) and Nef (HW9), however there were only two single codons changes, Gag T242N in TW10 and Pol T840A (or integrase T125A), which was common to all three alleles (Figure 4a). HLA-B*5802, associated with rapid HIV subtype C disease progression had only one shared adaptation with other alleles in the HLA-B58 group which are associated with slow HIV disease progression (HLA-B*5701 associated Rev T15x) and no shared adaptations with the most closely related HLA-B*5801 (50). Within the B7 supertype, HLA-B*0702 is present in both white and black populations, however HLA-B*8101 and -B*4201 are rare in whites. As with the B58 group, the selection profiles of these alleles were largely non-overlapping (Figure 4b). Across selected allele pairs within the B44 supertype (e.g. HLA-B*4001 and -B*4002, HLA-B*4402 and -B4403) which have extremely conserved exon 2 and 3 sequences (Supp data Figure 2) and strong phylogenetic support for close lineage (33), there was little evident overlap in the selection these alleles imposed on HIV (Figure 4c). Differential immunoselection in the Gag KF11 epitope by HLA-B*5701-restricted T cells in subtype B HIV and –B*5703-restricted T cells in subtype C HIV (51) as well as B7 supertype divergence in relation to shared epitopes in HIV subtype C (52) has been shown to be associated with strongly divergent patterns of TCR recruitment and qualitatively different T cell responses. Here the patterns of HIV subtype B adaptations associated with all HLA-A and –B alleles within all defined HLA supertypes and broad allelic families for all HLA-C alleles relevant to this population (Supp. Figure 4) suggest that this is indeed a widespread, general phenomenon. As such, subtle HLA gene variations can make a significant impact on viral diversity at the population level, not just through diversification of viral peptide sampling, but perhaps more extensively through differential TCR usage by the same peptides.
The influence of HLA on viral sequence divergence
We then sought to reconcile the observations that, on the one hand, race-specific HLA alleles have very divergent viral adaptation patterns in terms of single site associations; while on the other hand, there was no clustering of viral sequences by race/ethnic group in the phylogenetic trees (Figure 3). Given the multiple diversifying forces on viral sequences and underlying constraint, we analysed the correlations between pair-wise viral sequence divergence and HLA divergence (or non-matching of HLA alleles) in individuals carrying those sequences. When whole protein lengths were considered, HLA-A/B/C matching did not correlate significantly with greater viral sequence similarity (e.g p=0.06 in Gag), consistent with the phylogenetic trees. However, when shorter sliding intervals of ten amino acids within viral proteins were considered, the localized correlations within a number of intervals became significant, with p-values based on 500 randomizations of the data (Figure 5a). Thus the influence of HLA diversity on HIV diversity is demonstrably localized within ‘epitope-length’ windows and the correlation plots effectively visualized a landscape of HLA-driven divergent evolution of HIV. In contrast to phylogenetic relationships calculated using full protein sequences and encompassing all influences on diversity, these profiles represent the CD8 T cell view of HIV, not as a whole replicating virus or functional proteins, but as an assortment of 8-11mer peptides. Given our analyses capture both primary polymorphisms and co-varying adaptation patterns, this also suggests that most co-variation is highly localized and proximal to the primary adaptation site in HIV. This is supported by explicit population-based analyses of amino acid co-variation in HIV (22) and is the case in most experimentally characterized examples of HIV immune escape and compensatory mutation (8, 19-21).
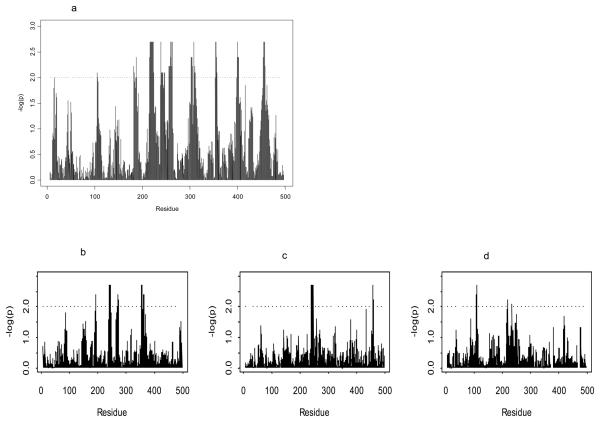
Figure 5.
Plots of significance (−logP) of correlations between non-matching in HLA genotype and viral sequence dissimilarity over sliding windows of ten amino acids in HIV-1 Gag. P-values determined by randomizations are plotted for correlation analyses of (a) HLA-A, -B and –C loci combined in all US cohort individuals. Correlation significance values are plotted for HLA-B locus non-matching in particular in (b) white n=222, (c) black n=209 and (d) Hispanic n= 107 subpopulations.
Having established the areas of strongest HLA-driven epitope diversity, we split the correlations by race/ethnic group and by HLA loci (Figure 5b-d, Supp. Fig 6). We chose to focus on the example of Gag for these detailed analyses given the importance of this protein to immunological control and as a common vaccine antigen (53). In these plots, the only strongly significant HLA-B peak (–log P>2) shared between white and black subpopulations corresponded to clustered HLA-B*57/5801 associated Gag T242N/ I247 and G248 as described in previous analyses. Otherwise the HLA-HIV correlation landscape was strikingly different between all racial/ethnic groups. The absence of the T242N-associated peak in Hispanics may be in part related to the complete absence of the B57/58 group in Amerindians. There were unique peaks in the Hispanics corresponding to HLA alleles more common in Hispanics and adaptations detected in phylogeny-corrected analyses (e.g HLA-B*4002 Gag 219P in Hispanics alone) as in other groups (e.g. HLA-B*0702 Gag in whites alone). Peaks not corresponding to specific HLA associations could also be caused by HLA-lineage correlations in this analysis or by HLA-selection effects not sufficiently strong enough to be detected with the current level of statistical power. In either case, these plots show the extent to which “race-specific” HLA alleles could shape viral epitope diversity, and in turn the CD8 T cell responses they elicit, in distinct ways in different human populations.
Discussion
The global diversity of HLA derives from the unique demographic histories of human ancestral groups spanning 100-200,000 years as well as the selection imposed by the diverse microbial environments into which these groups migrated and survived in isolation (5,6). In this context, the convergence of African, European, Asian and Amerindian ancestries into a single United States population as sampled here brings together a diversity of HLA alleles across multiple allelic lineages and with widely differing evolutionary histories. This type of population sampling, with ascertainment not specifically based on race/ethnicity provides a more relevant sampling of HLA diversity for HIV vaccine and therapeutics research and the major subpopulations studied here are comparable in size to those in the largest, national healthy population study of contemporary HLA genetic diversity in the United States (7). Though the different HLA distributions in this study cohort of HIV-infected individuals predominantly reflects ancestry groups as seen in the background population, the statistically significant enrichment of HLA-B*5703 which is associated with slower HIV disease progression is notable and could be evidence of HIV infection as a selective influence on the frequency of particular HLA alleles in a HIV-prevalent population within a contemporary timeframe. Given that those individuals with the extremes of slow disease progression are not likely to be enrolled in this cohort, such effects may only be underestimated in this study. It is not clear what underpins the differences in other alleles among Hispanics however. It is also possible that all these differences are caused by some differential ascertainment of certain groups into the study cohort, or the background population study, though the basis for these effects for these particular alleles is unknown.
By examination of the selection effects of these diverse HLA alleles on HIV sequences, aspects of ancestral HLA evolution are apparent. There is a dominance of the HLA-B in selection, as seen in immunological studies of HIV (50) and adding to accumulating evidence of the strongest pathogen-driven balancing selection operating at the HLA-B locus (5,6). As most serological/broad allelic families of HLAs and supertypes contain subtypes that traverse human ancestral groups and there is the adequate statistical power to compare these subtypes here, we show that there is striking divergence across most alleles. Here it is HIV itself that reveals the functional differences between HLA alleles in a way that is independent of, and without specific reference to their phylogenetic relatedness or supertype. These results suggest that alleles within a supertype are functionally very different in terms of the HLA-peptide-TCR interaction, even those apparently very closely related phylogenetically and within the same 2 digit genotype. The parallel selection effects of HLA-B*5701 and –B*5703 which have been a strong focus in HIV research appear to be the exception rather than the rule. This is consistent with the markedly different effects on HIV disease progression associated with HLA-B58 (50) and -B35 subtypes (54) and suggests that the HLA allelic variation across the many other HLA groupings, particularly at HLA-B, have a functional evolutionary basis. It is therefore important for clinical applications that HLA alleles of a supertype or family are not assumed to elicit qualitatively equal T cell responses, despite promiscuously bound epitopes.
Just as these analyses provide a view of “immunology taught by viruses”, the counter-evolution of HIV provides an approach to “virology taught by the immune system” (55). The large number of HLA associations across all HIV proteins reveals the enormous scope of subtype B HIV-1 adaptability against a wide span of global HLA diversity. Cumulative per-individual adaptation, as a proportion of sites relevant to the autologous HLA repertoire, is high across most HIV proteins, though Vpu stands out as an exception to the rule in this and other respects having high entropy but low density of HLA associations and dominant HLA-A locus selection (Supp. Figure 5). Finally the analyses of HLA-viral sequence dissimilarity correlations show the extent to which overall HLA repertoire drives viral sequence divergence in a global sense, given the degree of constraint and the other immune and non-immune influences on viral epitope diversity at any one site. Ultimately it is diversity within the viral ‘peptidome’ that directly affects applications in CD8 T cell immunotherapy or vaccines. Within these ‘immunologically relevant’ sequence windows, diversity is influenced by HLA to the extent that the landscape of HLA-driven viral diversity differs between race/ethnicity groups (Figure 5). Such divergence could impact HIV vaccine efficacy across these groups, or the extrapolation of efficacy in one group based on viral sequence data from another group, if not informed by knowledge of variances in HLA alleles at high resolution and their divergent imprints on viral sequence. It is important to recognize that this divergence is clear within epitope windows and driven by definable changes at single residues (Figure 4, Supp. Figure 4) but is not visible in standard phylogenetic trees (Figure 3). Given the functional relevance of these changes, this could be important in understanding the optimal CD8 T cell responses in certain populations where strong population substructures exist such as in Asia or Central and South America. The host genetic admixture within the U.S therefore provides the unique opportunity to characterize and measure these effects, and would be further enhanced by studies of specifically targeted populations that are likely to shape circulating viral epitopes in distinct ways (56).
While studies of larger and combined HLA-viral sequence datasets from different countries and regions will benefit from the general increase in statistical power, this study illustrates the value of broad population-based sampling, inclusion of immunogenetically distinct groupings, individual level ancestry information and high resolution HLA genotyping in characterizing the specific interactions underpinning HLA-HIV co-evolution. By mapping these interactions as proteome-wide putative immunoselection profiles of individual HLA alleles (Figure 4, Supp. Figure 4) as well as ‘hotspots’ of HLA-driven epitope divergence (Figure 5), it is hoped that this information can be more readily utilized, not only for the significant amount of HIV immunology, virology and vaccine research involving study subjects within the U.S where this genetic data is directly drawn, but also populations within the broader HIV subtype B epidemic in which many of these HLA alleles are represented.
Supplementary Material
Acknowledgments
We thank study teams, study sites, and participants of the US AACTG A5142 and A5128 protocols and the WA HIV Cohort Study and colleagues in the Centre for Clinical Immunology and Biomedical Statistics. We also thank Susan Herrmann, Shay Leary and Anthony Fordham for their assistance in manuscript preparation.
Footnotes
Publisher's Disclaimer: Disclaimer: This is an author-produced version of a manuscript accepted for publication in The Journal of Immunology (The JI). The American Association of Immunologists, Inc. (AAI), publisher of The JI, holds the copyright to this manuscript. This manuscript has not yet been copyedited or subjected to editorial proofreading by The JI; hence it may differ from the final version published in The JI (online and in print). AAI (The JI) is not liable for errors or omissions in this author-produced version of the manuscript or in any version derived from it by the United States National Institutes of Health or any other third party. The final, citable version of record can be found at www.jimmunol.org.”
Web resources
Source code for the program used to compute HLA-HIV polymorphism associations is available at http://www.microsoft.com/science.
The CD8 T cell epitope prediction program cited in this paper is available at http://atom.research.microsoft.com/bio/epipred.aspx
Accession numbers
The GenBank (http://www.ncbi.nlm.nih.gov/Genbank/) accession numbers for AACTG 5142/5128 cohort HIV-1 sequences reported in this paper are GQ371216-GQ371763 (gag), GQ371764-GQ372317 (pol), GQ372318-GQ372824 and GQ398382-GQ398387 (nef) and GU727870-GU731062 (remaining genes). Patient specific genetic (HLA) data cannot be posted publically however data may be shared with collaborators for specific research projects subject to approval by the appropriate study teams and IRBs governing ACTG A5128 and the WAHCS materials and data. Please contact the corresponding author.
Funding support: This project was supported by grant number RO1 AI060460 from the National Institute of Allergy and Infectious Diseases (NIAID). The ACTG is supported by grant number AI-68636 and the Vanderbilt DNA Resources Core by grant number RR024975. The ACTG Clinical Trials Sites that collected DNA were supported by NIH grants AI64086, AI68636, AI68634, AI069471, AI27661, AI069439, AI25859, AI069477, AI069513, AI069452, AI27673, AI069419, AI069474, AI69411, AI69423, AI69494, AI069484, AI069472, AI069501, AI69467, AI069450, AI32782, AI69465, AI069424, AI38858, AI069447, AI069495, AI069502, AI069556, AI069432, AI46370, AI069532, AI046376, AI34853, and AI069434. The content of this study is the responsibility of the authors and does not necessarily represent the official views of NIAID or the National Institutes of Health (US). The project was additionally supported by the Australian National Health and Medical Research Council and the Bill and Melinda Gates Foundation.
References
- 1.Fellay J, Shianna KV, Ge D, Colombo S, Ledergerber B, Weale M, Zhang K, Gumbs C, Castagna A, Cossarizza A, Cozzi-Lepri A, De Luca A, Easterbrook P, Francioli P, Mallal S, Martinez-Picado J, Miro JM, Obel N, Smith JP, Wyniger J, Descombes P, Antonarakis SE, Letvin NL, McMichael AJ, Haynes BF, Telenti A, Goldstein DB. A whole-genome association study of major determinants for host control of HIV-1. Science. 2007;317(5840):944–947. doi: 10.1126/science.1143767. 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Limou S, Le Clerc S, Coulonges C, Carpentier W, Dina C, Delaneau O, Labib T, Taing L, Sladek R, Deveau C, Ratsimandresy R, Montes M, Spadoni JL, Lelièvre JD, Lévy Y, Therwath A, Schächter F, Matsuda F, Gut I, Froguel P, Delfraissy JF, Hercberg S, Zagury JF, ANRS Genomic Group Genomewide association study of an AIDS non-progression cohort emphasizes the role played by HLA genes (ANRS Genomewide Association Study 02) JID. 2009;199:419–426. doi: 10.1086/596067. [DOI] [PubMed] [Google Scholar]
- 3.Goulder PJ, Watkins DI. Impact of MHC class I diversity on immune control of immunodeficiency virus replication. Nature Reviews Immunology. 2008;8(8):619–630. doi: 10.1038/nri2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hughes AL, Nei M. Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals over-dominant selection. Nature. 1988;335(6186):167–170. doi: 10.1038/335167a0. 8. [DOI] [PubMed] [Google Scholar]
- 5.Meyer D, Single RM, Mack SJ, Erlich HA, Thomson G. Signatures of demographic history and natural selection in the human major histocompatibility complex Loci. Genetics. 2006;173:2121–2142. doi: 10.1534/genetics.105.052837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prugnolle F, Manica A, Charpentier M, Guégan JF, Guernier V, Balloux F. Pathogen-driven selection and worldwide HLA class I diversity. Curr Biol. 2005;15(11):1022–1027. doi: 10.1016/j.cub.2005.04.050. [DOI] [PubMed] [Google Scholar]
- 7.Cao K, Hollenbach J, Xuejiang S, Wenxia S, Chopek M, Fernandez-Vina MA. Analysis of the frequencies of HLA-A, B, and C alleles and haplotypes in the five major ethnic groups of the United States reveals high levels of diversity in these loci and contrasting distribution patterns in these populations. Human Immunology. 2001;62:1009–1030. doi: 10.1016/s0198-8859(01)00298-1. [DOI] [PubMed] [Google Scholar]
- 8.Kelleher AD, Long C, Holmes EC, Allen RL, Wilson J, Conlon C, Workman C, Shaunak S, Olson K, Goulder P, Brander C, Ogg G, Sullivan JS, Dyer W, Jones I, McMichael AJ, Rowland-Jones S, Phillips RE. Clustered mutations in HIV-1 gag are consistently required for escape from HLA-B27-restricted cytotoxic T lymphocyte responses. J Exp Med. 2001;193:375–386. doi: 10.1084/jem.193.3.375. P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leslie AJ, Pfafferott KJ, Chetty P, Draenert R, Addo MM, Feeney M, Tang Y, Holmes EC, Allen T, Prado JG, Altfeld M, Brander C, Dixon C, Ramduth D, Jeena P, Thomas SA, John A, St, Roach TA, Kupfer B, Luzzi G, Edwards A, Taylor G, Lyall H, Tudor-Williams G, Novelli V, Martinez-Picado J, Kiepiela P, Walker BD, Goulder PJ. Nat Med. 2004;10(3):282–289. doi: 10.1038/nm992. [DOI] [PubMed] [Google Scholar]
- 10.Draenert R, Le Gall S, Pfafferott KJ, Leslie AJ, Chetty P, Brander C, Holmes EC, Chang SC, Feeney ME, Addo MM, Ruiz L, Ramduth D, Jeena P, Altfeld M, Thomas S, Tang Y, Verrill CL, Dixon C, Prado JG, Kiepiela P, Martinez-Picado J, Walker BD, Goulder PJ. Immune selection for altered antigen processing leads to cytotoxic T lymphocyte escape in chronic HIV-1 infection. J Exp Med. 2004;199:905–915. doi: 10.1084/jem.20031982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allen TM, Altfeld M, Yu XG, O'Sullivan KM, Lichterfeld M, Le Gall S, John M, Mothe BR, Lee PK, Kalife ET, Cohen DE, Freedberg KA, Strick DA, Johnston MN, Sette A, Rosenberg ES, Mallal SA, Goulder PJ, Brander C. Selection, transmission, and reversion of an antigen-processing cytotoxic T-lymphocyte escape mutation in human immunodeficiency virus type 1 infection. J Virol. 2004;78:7069–7078. doi: 10.1128/JVI.78.13.7069-7078.2004. B.D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yokomaku Y, Miura H, Tomiyama H, Kawana-Tachikawa A, Takiguchi M, Kojima A, Nagai Y, Iwamoto A, Matsuda Z, Ariyoshi K. Impaired processing and presentation of cytotoxic-T-lymphocyte (CTL) epitopes are major escape mechanisms from CTL immune pressure in human immunodeficiency virus type 1 infection. J Virol. 2004;78:1324–1332. doi: 10.1128/JVI.78.3.1324-1332.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moore CB, John M, James IR, Christiansen FT, Witt CS, Mallal SA. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science. 2002;296:1439–1443. doi: 10.1126/science.1069660. [DOI] [PubMed] [Google Scholar]
- 14.Brumme ZL, Brumme CJ, Heckerman D, Korber BT, Daniels M, Carlson JM, Kadie C, Bhattacharya T, Chui C, Szinger J, Mo T, Hogg RS, Montaner JS, Frahm N, Brander C, Walker BD, Harrigan PR. Evidence of Differential HLA Class I-Mediated Viral Evolution in Functional and Accessory/Regulatory Genes of HIV-1. PLoS Pathog. 2007;3:e94. doi: 10.1371/journal.ppat.0030094. doi:10.1371/journal.ppat.0030094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brumme ZL, Tao I, Szeto S, Brumme CJ, Carlson JM, Chan D, Kadie C, Frahm N, Brander C, Walker BD, Heckerman D, Harrigan PR. Human leukocyte antigen-specific polymorphisms in HIV-1 Gag and their association with viral load in chronic untreated infection. AIDS. 2008;22(11):1277–1286. doi: 10.1097/QAD.0b013e3283021a8c. 11. [DOI] [PubMed] [Google Scholar]
- 16.Rousseau CM, Daniels MG, Carlson JM, Kadie C, Crawford H, Prendergast A, Matthews P, Payne R, Rolland M, Raugi DN, Maust BS, Learn GH, Nickle DC, Coovadia H, Ndung'u T, Frahm N, Brander C, Walker BD, Goulder PJ, Bhattacharya T, Heckerman DE, Korber BT, Mullins JI. J Virol. 2008;82(13):6434–6446. doi: 10.1128/JVI.02455-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang YE, Li B, Carlson JM, Streeck H, Gladden AD, Goodman R, Schneidewind A, Power KA, Toth I, Frahm N, Alter G, Brander C, Carrington M, Walker BD, Altfeld M, Heckerman D, Allen TM. Protective HLA class I alleles that restrict acute-phase CD8+ T-cell responses are associated with viral escape mutations located in highly conserved regions of human immunodeficiency virus type 1. J Virol. 2009;83(4):1845–1855. doi: 10.1128/JVI.01061-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brumme ZL, John M, Carlson JM, Brumme CJ, Chan D, Brockman MA, Swenson LC, Tao I, Szeto S, Rosato P, Sela J, Kadie CM, Frahm N, Brander C, Haas DW, Riddler SA, Haubrich R, Walker BD, Harrigan PR, Heckerman D, Mallal S. HLA-associated immune escape pathways in HIV-1 subtype B Gag, Pol and Nef Proteins. PLoS ONE. 2009;4(8):e6687. doi: 10.1371/journal.pone.0006687. doi: 10.1371/journal.pone.0006687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schneidewind A, Brockman MA, Yang R, Adam RI, Li B, Le Gall S, Rinaldo CR, Craggs SL, Allgaier RL, Power KA, Kuntzen T, Tung CS, LaBute MX, Mueller SM, Harrer T, McMichael AJ, Goulder PJ, Aiken C, Brander C, Kelleher AD, Allen TM. Escape from the Dominant HLA-B27-restricted cytotoxic T-lymphocyte response in Gag is associated with a dramatic reduction in human immunodeficiency virus type 1 replication. J. Virol. 2007;81:12382–12393. doi: 10.1128/JVI.01543-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brockman MA, Schneidewind A, Lahaie M, Schmidt A, Miura T, Desouza I, Ryvkin F, Derdeyn CA, Allen S, Hunter E, Mulenga J, Goepfert PA, Walker BD, Allen TM. Escape and compensation from early HLA-B57-mediated cytotoxic T lymphocyte pressure on human immunodeficiency virus type 1 Gag alter capsid interactions with cyclophilin A. J. Virol. 2007;81:12608–12618. doi: 10.1128/JVI.01369-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crawford H, Prado JG, Leslie A, Hué S, Honeyborne I, Reddy S, van der Stok M, Mncube Z, Brander C, Rousseau C, Mullins JI, Kaslow R, Goepfert P, Allen S, Hunter E, Mulenga J, Kiepiela P, Walker BD, Goulder PJ. Compensatory mutation partially restores fitness and delays reversion of escape mutation within the immunodominant HLA-B* 5703-restricted Gag epitope in chronic human immunodeficiency virus type 1 infection. J Virol. 2007;81:8346–8351. doi: 10.1128/JVI.00465-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carlson JM, Brumme ZL, Rousseau CM, Brumme CJ, Matthews P, Kadie C, Mullins JI, Walker BD, Harrigan PR, Goulder PJ, Heckerman D. Phylogenetic dependency networks: inferring patterns of CTL escape and codon covariation in HIV-1 Gag. PLoS Comput Biol. 2008;4:e100022. doi: 10.1371/journal.pcbi.1000225. doi:10.1371/journal.pcbi.1000225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawashima Y, Pfafferott K, Frater J, Matthews P, Payne R, Addo M, Gatanaga H, Fujiwara M, Hachiya A, Koizumi H, Kuse N, Oka S, Duda A, Prendergast A, Crawford H, Leslie A, Brumme ZL, Brumme C, Allen T, Brander C, Kaslow R, Tang J, Hunter E, Allen S, Mulenga J, Branch S, Roach T, John M, Mallal S, Ogwu A, Shapiro R, Prado JG, Fidler S, Weber J, Pybus OG, Klenerman P, Ndung'u T, Phillips R, Heckerman D, Harrigan PR, Walker BD, Takiguchi M, Goulder PJ. Adaptation of HIV-1 to human leukocyte antigen class I. Nature. 2009;458:641–645. doi: 10.1038/nature07746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fernández-Viña MA, Lázaro AM, Marcos CY, Nulf C, Raimondi E, Haas EJ, Stastny P, Fernández-Viña MA. Dissimilar evolution of B-locus versus A-locus and class II loci of the HLA region in South American Indian tribes. Tissue Antigens. 1997;50(3):233–250. doi: 10.1111/j.1399-0039.1997.tb02867.x. [DOI] [PubMed] [Google Scholar]
- 25.CDC HIV prevalence estimates, United States, 2006. MMWR. 2008;57(39):1073–1076. [PubMed] [Google Scholar]
- 26.Riddler SA, Haubrich R, DiRienzo AG, Peeples L, Powderly WG, Klingman KL, Garren KW, George T, Rooney JF, Brizz B, Lalloo UG, Murphy RL, Swindells S, Havlir D, Mellors JW, AIDS Clinical Trials Group Study A5142 Team Class-sparing regimens for initial treatment of HIV-1 infection. N Engl J Med. 2008;358(20):2095–2106. doi: 10.1056/NEJMoa074609. 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haas DW, Wilkinson GR, Kuritzkes DR, Richman DD, Nicotera J, Mahon LF, Sutcliffe C, Siminski S, Andersen J, Coughlin K, Clayton EW, Haines J, Marshak A, Saag M, Lawrence J, Gustavson J, Anne Bennett J, Christensen R, Matula MA, Wood AJ, Adult AIDS Clinical Trials Group A multi-investigator/institutional DNA bank for AIDS-related human genetic studies: AACTG Protocol A5128. HIV Clin Trials. 2003;4:287–300. doi: 10.1310/MUQC-QXBC-8118-BPM5. [DOI] [PubMed] [Google Scholar]
- 28.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 29.Bhattacharya T, Daniels M, Heckerman D, Foley B, Frahm N, Kadie C, Carlson J, Yusim K, McMahon B, Gaschen B, Mallal S, Mullins JI, Nickle DC, Herbeck J, Rousseau C, Learn GH, Miura T, Brander C, Walker BD, Korber BT. Founder effects in the assessment of HIV polymorphisms and HLA allele associations. Science. 2007;315:1583–1586. doi: 10.1126/science.1131528. [DOI] [PubMed] [Google Scholar]
- 30.Carlson JM, Kadie C, Mallal S, Heckerman D. Leveraging Hierarchical Population Structure in Discrete Association Studies. PLoS ONE. 2007;2:e591. doi: 10.1371/journal.pone.0000591. doi:10.1371/journal.pone.0000591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cattley SK, Williamson JF, Tay GK, Martinez OP, Gaudieri S, Dawkins RL. Further characterization of MHC haplotypes demonstrates conservation telomeric of HLA-A: update of the 4AOH and 10IHW cell panels. Eur J Immunogenetics. 2000;27:397–426. doi: 10.1046/j.1365-2370.2000.00226.x. [DOI] [PubMed] [Google Scholar]
- 32.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci. 2003;100(16):9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McKenzie LM, Pecon-Slattery J, Carrington M, O'Brien SJ. Taxonomic hierarchy of HLA class I allele sequences. Genes and Immunity. 1999;1:120–129. doi: 10.1038/sj.gene.6363648. [DOI] [PubMed] [Google Scholar]
- 34.Korber BT, Brander C, Haynes BF, Koup R, Moore JP, Walker BD, Watkins DI, editors. HIV Molecular Immunology. Los Alamos National Laboratory, Theoretical Biology and Biophysics; Los Alamos, New Mexico: 2006/2007. pp. 07–4752. LA-UR. [Google Scholar]
- 35.Heckerman D, Kadie C, Listgarden J. Leveraging information across HLA alleles/Supertypes improves Epitope Prediction. RECOMB. 2006;3903:296–308. LNBI. [Google Scholar]
- 36.Phillips RE, Rowland-Jones S, Nixon DF, Gotch FM, Edwards JP, Ogunlesi AO, Elvin JG, Rothbard JA, Bangham CRM, Rizza CR, McMichael AJ. Human immunodeficiency virus genetic variation that can escape cytotoxic T cell recognition. Nature. 1991;354:453–459. doi: 10.1038/354453a0. [DOI] [PubMed] [Google Scholar]
- 37.Price DA, Goulder PJ, Klenerman P, Sewell AK, Easterbrook PJ, Troop M, Bangham CR, Phillips RE. Positive selection of HIV-1 cytotoxic T lymphocyte escape variants during primary infection. Proc Natl Acad Sci. 1997;94:1890–1895. doi: 10.1073/pnas.94.5.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Milicic A, Edwards CT, Hué S, Fox J, Brown H, Pillay T, Drijfhout JW, Weber JN, Holmes EC, Fidler SJ, Zhang HT, Phillips RE. Sexual transmission of single human immunodeficiency virus type 1 virions encoding highly polymorphic multisite cytotoxic T-lymphocyte escape variants. J Virol. 2005;79:13953–13962. doi: 10.1128/JVI.79.22.13953-13962.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goulder PJ, Phillips RE, Colbert RA, McAdam S, Ogg G, Nowak MA, Giangrande P, Luzzi G, Morgan B, Edwards A, McMichael AJ, Rowland-Jones S. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat Med. 1997;3:212–217. doi: 10.1038/nm0297-212. [DOI] [PubMed] [Google Scholar]
- 40.Feeney ME, Tang Y, Pfafferott K, Roosevelt KA, Draenert R, Trocha A, Yu XG, Verrill C, Allen T, Moore C, Mallal S, Burchett S, McIntosh K, Pelton SI, John MA, St, Hazra R, Klenerman P, Altfeld M, Walker BD, Goulder PJ. HIV-1 viral escape in infancy followed by emergence of a variant-specific CTL response. J Immunol. 2005;174:7524–7530. doi: 10.4049/jimmunol.174.12.7524. [DOI] [PubMed] [Google Scholar]
- 41.Sipsas NV, Kalams SA, Trocha A, He S, Blattner WA, Walker BD, Johnson RP. Identification of type-specific cytotoxic T lymphocyte responses to homologous viral proteins in laboratory workers accidentally infected with HIV-1. J Clin Invest. 1997;99:752–762. doi: 10.1172/JCI119221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Geels MJ, Cornelissen M, Schuitemaker H, Anderson K, Kwa D, Maas J, Dekker JT, Baan E, Zorgdrager F, van den Burg R, van Beelen M, Lukashov VV, Fu TM, Paxton WA, van der Hoek L, Dubey SA, Shiver JW, Goudsmit J. Identification of sequential viral escape mutants associated with altered T-cell responses in a human immunodeficiency virus type 1-infected individual. J Virol. 2003;77:12430–12440. doi: 10.1128/JVI.77.23.12430-12440.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jones NA, Wei X, Flower DR, Wong M, Michor F, Saag MS, Hahn BH, Nowak MA, Shaw GM, Borrow P. Determinants of human immunodeficiency virus type 1 escape from the primary CD8+ cytotoxic T lymphocyte response. J Exp Med. 2004;200:1243–1256. doi: 10.1084/jem.20040511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Furutsuki T, Hosoya N, Kawana-Tachikawa A, Tomizawa M, Odawara T, Goto M, Kitamura Y, Nakamura T, Kelleher AD, Cooper DA, Iwamoto A. Frequent transmission of cytotoxic-T-lymphocyte escape mutants of human immunodeficiency virus type 1 in the highly HLA-A24-positive Japanese population. J Virol. 2004;78:8437–8445. doi: 10.1128/JVI.78.16.8437-8445.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Allen TM, Yu XG, Kalife ET, Reyor LL, Lichterfeld M, John M, Cheng M, Allgaier RL, Mui S, Frahm N, Alter G, Brown NV, Johnston MN, Rosenberg ES, Mallal SA, Brander C, Walker BD, Altfeld M. De novo generation of escape variant-specific CD8+ T-cell responses following cytotoxic T-lymphocyte escape in chronic human immunodeficiency virus type 1infection. J Virol. 2005;79:12952–12960. doi: 10.1128/JVI.79.20.12952-12960.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leslie A, Kavanagh D, Honeyborne I, Pfafferott K, Edwards C, Pillay T, Hilton L, Thobakgale C, Ramduth D, Draenert R, Le Gall S, Luzzi G, Edwards A, Brander C, Sewell AK, Moore S, Mullins J, Moore C, Mallal S, Bhardwaj N, Yusim K, Phillips R, Klenerman P, Korber B, Kiepiela P, Walker BD, Goulder PJ. Transmission and accumulation of CTL escape variants drive negative associations between HIV polymorphisms and HLA. J Exp Med. 2005;201:891–902. doi: 10.1084/jem.20041455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pillay T, Zhang HT, Drijfhout JW, Robinson N, Brown H, Khan M, Moodley J, Adhikari M, Pfafferott K, Feeney ME, John A, St, Holmes EC, Coovadia HM, Klenerman P, Goulder PJ, Phillips RE. Unique acquisition of cytotoxic T-lymphocyte escape mutants in infant human immunodeficiency virus type 1 infection. J Virol. 2005;79:12100–12105. doi: 10.1128/JVI.79.18.12100-12105.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shannon CE. A mathematical theory of communication. Bell Syst. Tech. J. 1948;27:623–656. [Google Scholar]
- 49.Sidney J, Peters B, Frahm N, Brander C, Sette A. HLA class I Supertypes: a revised and updated classification. BMC Immunology. 2008;9:1. doi: 10.1186/1471-2172-9-1. doi: 10.1186/1471-2172-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kiepiela P, Leslie AJ, Honeyborne I, Ramduth D, Thobakgale C, Chetty S, Rathnavalu P, Moore C, Pfafferott KJ, Hilton L, Zimbwa P, Moore S, Allen T, Brander C, Addo MM, Altfeld M, James I, Mallal S, Bunce M, Barber LD, Szinger J, Day C, Klenerman P, Mullins J, Korber BT, Coovadia HM, Walker BD, Goulder PJ. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature. 2004;432:769–775. doi: 10.1038/nature03113. [DOI] [PubMed] [Google Scholar]
- 51.Yu XG, Lichterfeld M, Chetty S, Williams KL, Mui SK, Miura T, Frahm N, Feeney ME, Tang Y, Pereyra F, Labute MX, Pfafferott K, Leslie A, Crawford H, Allgaier R, Hildebrand W, Kaslow R, Brander C, Allen TM, Rosenberg ES, Kiepiela P, Vajpayee M, Goepfert PA, Altfeld M, Goulder PJ, Walker BD. Mutually Exclusive T-Cell Receptor Induction and Differential Susceptibility to Human Immunodeficiency Virus Type 1 Mutational Escape Associated with a Two-Amino-Acid Difference between HLA Class I Subtypes. J Virol. 2007;18(4):1619–1631. doi: 10.1128/JVI.01580-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leslie A, Price DA, Mkhize P, Bishop K, Rathod A, Day C, Crawford H, Honeyborne I, Asher TE, Luzzi G, Edwards A, Rousseau CM, Mullins JI, Tudor-Williams G, Novelli V, Brander C, Douek DC, Kiepiela P, Walker BD, Goulder PJ. Differential selection pressure exerted on HIV by CTL targeting identical epitopes but restricted by distinct HLA alleles from the same HLA supertype. J. Immunol. 2006;177:4699–4708. doi: 10.4049/jimmunol.177.7.4699. [DOI] [PubMed] [Google Scholar]
- 53.Kiepiela P, Ngumbela K, Thobakgale C, Ramduth D, Honeyborne I, Moodley E, Reddy S, de Pierres C, Mncube Z, Mkhwanazi N, Bishop K, van der Stok M, Nair K, Khan N, Crawford H, Payne R, Leslie A, Prado J, Prendergast A, Frater J, McCarthy N, Brander C, Learn GH, Nickle D, Rousseau C, Coovadia H, Mullins JI, Heckerman D, Walker BD, Goulder PJ. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat Med. 2007;13(1):46–53. doi: 10.1038/nm1520. [DOI] [PubMed] [Google Scholar]
- 54.Gao X, Nelson GW, Karacki P, Martin MP, Phair J, Kaslow R, Goedert JJ, Buchbinder S, Hoots K, Vlahov D, O'Brien SJ, Carrington M. Effect of a single amino acid change in MHC class I molecules on the rate of progression to AIDS. N. Engl. J. Med. 2001;344:1668–1675. doi: 10.1056/NEJM200105313442203. [DOI] [PubMed] [Google Scholar]
- 55.Nolan D, Gaudieri S, Mallal S. Host genetics and viral infections: immunology taught by viruses, virology taught by the immune system. Curr Opin Immunol. 2006;18(4):413–421. doi: 10.1016/j.coi.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 56.Avila-Rios S, Ormsby CE, Carlson JM, Valenzuela-Ponce H, Blanco-Heredia J, Garrido-Rodriguez D, Garcia-Morales C, Heckerman D, Brumme ZL, Mallal S, John M, Espinosa E, Reyes-Teran G. Unique features of HLA-mediated HIV evolution in a Mexican cohort: a comparative study. Retrovirology. 2009;6:72. doi: 10.1186/1742-4690-6-72. doi:10.1186/1742-4690-6-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.